

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.11 - phase: 0
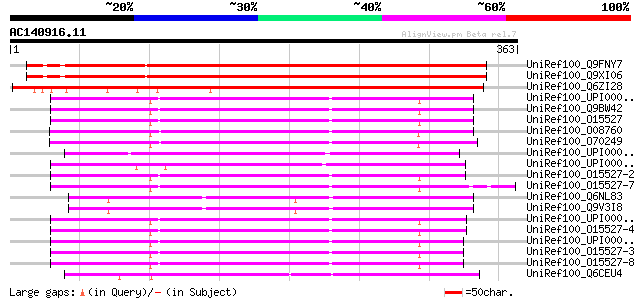
(363 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FNY7 8-oxoguanine DNA glycosylase [Arabidopsis thali... 426 e-118
UniRef100_Q9XI06 F8K7.14 protein [Arabidopsis thaliana] 426 e-118
UniRef100_Q6ZI28 Putative 8-oxoguanine DNA glycosylase isoform 1... 359 6e-98
UniRef100_UPI000036B382 UPI000036B382 UniRef100 entry 198 2e-49
UniRef100_Q9BW42 8-oxoguanine DNA glycosylase, isoform 1a [Homo ... 198 2e-49
UniRef100_O15527 N-glycosylase/DNA lyase [Includes: 8-oxoguanine... 198 2e-49
UniRef100_O08760 N-glycosylase/DNA lyase [Includes: 8-oxoguanine... 197 3e-49
UniRef100_O70249 N-glycosylase/DNA lyase [Includes: 8-oxoguanine... 196 6e-49
UniRef100_UPI00003AA902 UPI00003AA902 UniRef100 entry 196 1e-48
UniRef100_UPI000001A584 UPI000001A584 UniRef100 entry 195 2e-48
UniRef100_O15527-2 Splice isoform 1B of O15527 [Homo sapiens] 191 3e-47
UniRef100_O15527-7 Splice isoform 2D of O15527 [Homo sapiens] 191 3e-47
UniRef100_Q6NL83 RE57519p [Drosophila melanogaster] 191 4e-47
UniRef100_Q9V3I8 N-glycosylase/DNA lyase (dOgg1) [Includes: 8-ox... 191 4e-47
UniRef100_UPI000036B381 UPI000036B381 UniRef100 entry 190 6e-47
UniRef100_O15527-4 Splice isoform 2A of O15527 [Homo sapiens] 190 6e-47
UniRef100_UPI000036B37D OGG1 [Mus spretus] 189 8e-47
UniRef100_O15527-3 Splice isoform 1C of O15527 [Homo sapiens] 189 8e-47
UniRef100_O15527-8 Splice isoform 2E of O15527 [Homo sapiens] 189 8e-47
UniRef100_Q6CEU4 Yarrowia lipolytica chromosome B of strain CLIB... 185 1e-45
>UniRef100_Q9FNY7 8-oxoguanine DNA glycosylase [Arabidopsis thaliana]
Length = 365
Score = 426 bits (1096), Expect = e-118
Identities = 212/330 (64%), Positives = 255/330 (77%), Gaps = 7/330 (2%)
Query: 13 PPPLPPSTPPTPHTQTRHSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQY 72
PP PP TP Q H T K W PL +T EL+LPLTFPTGQTFRWK T QY
Sbjct: 18 PPLSPPVTPILK--QKLHRTGTPK---WFPLKLTHTELTLPLTFPTGQTFRWKKTGAIQY 72
Query: 73 TGVVGSHLISLKHLHNGD-VCYTLHSQSPSNDDCKTALLDFLNADVSLADTWKVFSDSDE 131
+G +G HL+SL+ D V Y +H S S + ALLDFLNA++SLA+ W FS D
Sbjct: 73 SGTIGPHLVSLRQRPGDDAVSYCVHC-STSPKSAELALLDFLNAEISLAELWSDFSKKDP 131
Query: 132 RFAELAQHLSGARVLRQDPFECLIQFMCSSNNHISRITKMVDYVSSLGTYLGCVEGFDFH 191
RF ELA+HL GARVLRQDP ECLIQF+CSSNN+I+RITKMVD+VSSLG +LG ++GF+FH
Sbjct: 132 RFGELARHLRGARVLRQDPLECLIQFLCSSNNNIARITKMVDFVSSLGLHLGDIDGFEFH 191
Query: 192 AFPTLNQLSLVSEQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISE 251
FP+L++LS VSE++ R AGFGYRAKYITGTVN LQ+KP GG EWL SLRK+EL++ ++
Sbjct: 192 QFPSLDRLSRVSEEEFRKAGFGYRAKYITGTVNALQAKPGGGNEWLLSLRKVELQEAVAA 251
Query: 252 LIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQKYILPELAGSKLTQKLHSRVAEA 311
L LPGVGPKVAACIAL+SLDQH AIPVD H+W+IA Y+LP+LAG+KLT KLH RVAEA
Sbjct: 252 LCTLPGVGPKVAACIALFSLDQHSAIPVDTHVWQIATNYLLPDLAGAKLTPKLHGRVAEA 311
Query: 312 FVTKFGKYAGWAQAVLFIAELPSQKAILPS 341
FV+K+G+YAGWAQ +LFIAELP+QK +L S
Sbjct: 312 FVSKYGEYAGWAQTLLFIAELPAQKTLLQS 341
>UniRef100_Q9XI06 F8K7.14 protein [Arabidopsis thaliana]
Length = 391
Score = 426 bits (1096), Expect = e-118
Identities = 212/330 (64%), Positives = 255/330 (77%), Gaps = 7/330 (2%)
Query: 13 PPPLPPSTPPTPHTQTRHSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQY 72
PP PP TP Q H T K W PL +T EL+LPLTFPTGQTFRWK T QY
Sbjct: 18 PPLSPPVTPILK--QKLHRTGTPK---WFPLKLTHTELTLPLTFPTGQTFRWKKTGAIQY 72
Query: 73 TGVVGSHLISLKHLHNGD-VCYTLHSQSPSNDDCKTALLDFLNADVSLADTWKVFSDSDE 131
+G +G HL+SL+ D V Y +H S S + ALLDFLNA++SLA+ W FS D
Sbjct: 73 SGTIGPHLVSLRQRPGDDAVSYCVHC-STSPKSAELALLDFLNAEISLAELWSDFSKKDP 131
Query: 132 RFAELAQHLSGARVLRQDPFECLIQFMCSSNNHISRITKMVDYVSSLGTYLGCVEGFDFH 191
RF ELA+HL GARVLRQDP ECLIQF+CSSNN+I+RITKMVD+VSSLG +LG ++GF+FH
Sbjct: 132 RFGELARHLRGARVLRQDPLECLIQFLCSSNNNIARITKMVDFVSSLGLHLGDIDGFEFH 191
Query: 192 AFPTLNQLSLVSEQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISE 251
FP+L++LS VSE++ R AGFGYRAKYITGTVN LQ+KP GG EWL SLRK+EL++ ++
Sbjct: 192 QFPSLDRLSRVSEEEFRKAGFGYRAKYITGTVNALQAKPGGGNEWLLSLRKVELQEAVAA 251
Query: 252 LIKLPGVGPKVAACIALYSLDQHHAIPVDVHIWRIAQKYILPELAGSKLTQKLHSRVAEA 311
L LPGVGPKVAACIAL+SLDQH AIPVD H+W+IA Y+LP+LAG+KLT KLH RVAEA
Sbjct: 252 LCTLPGVGPKVAACIALFSLDQHSAIPVDTHVWQIATNYLLPDLAGAKLTPKLHGRVAEA 311
Query: 312 FVTKFGKYAGWAQAVLFIAELPSQKAILPS 341
FV+K+G+YAGWAQ +LFIAELP+QK +L S
Sbjct: 312 FVSKYGEYAGWAQTLLFIAELPAQKTLLQS 341
>UniRef100_Q6ZI28 Putative 8-oxoguanine DNA glycosylase isoform 1a; 8-hydroxyguanine
DNA glycosylase [Oryza sativa]
Length = 399
Score = 359 bits (922), Expect = 6e-98
Identities = 194/371 (52%), Positives = 248/371 (66%), Gaps = 34/371 (9%)
Query: 3 KRKRSPIKSLPPPL------PPSTPP--TPHTQT--------RHSTNLSKSRA------- 39
+R R P+ PP L PP TPP TP ++ R L + A
Sbjct: 7 RRVRPPLPPSPPLLSSTTATPPPTPPVGTPKSEAADRRPPPARRRLPLVSAAAVEEEDGE 66
Query: 40 WIPLNITRQELSLPLTFPTGQTFRWKNTA--PSQYTGVVGSHLISLKHLHNGD--VCYTL 95
W PL ++ +LSLPLT PTGQTF W+ T+ P ++TG VG HL+SL HL + D + + L
Sbjct: 67 WHPLPLSAADLSLPLTLPTGQTFLWRRTSLSPLRFTGAVGPHLVSLSHLPSSDGRLAFLL 126
Query: 96 HSQSPSNDD-----CKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSG--ARVLRQ 148
H+ S+ + AL D+LNA V LAD W+ F+ +D RFAE++ L G ARVLRQ
Sbjct: 127 HNNGGSSSSSVPAAARAALSDYLNAAVPLADLWRRFAAADARFAEVSARLGGGGARVLRQ 186
Query: 149 DPFECLIQFMCSSNNHISRITKMVDYVSSLGTYLGCVEGFDFHAFPTLNQLSLVSEQQLR 208
DP EC+ QF+CSSNN+I+RI KMV ++ G LG V G+ FH FPT+ +L+ VSEQ+LR
Sbjct: 187 DPVECVFQFLCSSNNNIARIEKMVWALAGYGERLGEVGGYQFHQFPTIERLARVSEQELR 246
Query: 209 DAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIAL 268
DAGFGYRAKYI GT +LQ+KP GGE+WL SLR EL +VI L LPGVGPKVAAC+AL
Sbjct: 247 DAGFGYRAKYIVGTAKILQAKPGGGEKWLASLRTRELPEVIEALCTLPGVGPKVAACVAL 306
Query: 269 YSLDQHHAIPVDVHIWRIAQKYILPELAGSKLTQKLHSRVAEAFVTKFGKYAGWAQAVLF 328
+SLDQ+HAIPVD H+W++A +Y++PELAG LT KL VA+AFV KFG YAGWAQ VLF
Sbjct: 307 FSLDQNHAIPVDTHVWKVATQYLMPELAGKSLTPKLSVAVADAFVAKFGNYAGWAQNVLF 366
Query: 329 IAELPSQKAIL 339
I +L +QK ++
Sbjct: 367 IGQLSAQKLMV 377
>UniRef100_UPI000036B382 UPI000036B382 UniRef100 entry
Length = 345
Score = 198 bits (503), Expect = 2e-49
Identities = 117/313 (37%), Positives = 172/313 (54%), Gaps = 12/313 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVLFIAEL 332
AGWAQAVLF A+L
Sbjct: 311 AGWAQAVLFSADL 323
>UniRef100_Q9BW42 8-oxoguanine DNA glycosylase, isoform 1a [Homo sapiens]
Length = 345
Score = 198 bits (503), Expect = 2e-49
Identities = 117/313 (37%), Positives = 172/313 (54%), Gaps = 12/313 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVLFIAEL 332
AGWAQAVLF A+L
Sbjct: 311 AGWAQAVLFSADL 323
>UniRef100_O15527 N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC
3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP lyase)] [Homo sapiens]
Length = 345
Score = 198 bits (503), Expect = 2e-49
Identities = 117/313 (37%), Positives = 172/313 (54%), Gaps = 12/313 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVLFIAEL 332
AGWAQAVLF A+L
Sbjct: 311 AGWAQAVLFSADL 323
>UniRef100_O08760 N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC
3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP lyase)] [Mus musculus]
Length = 345
Score = 197 bits (502), Expect = 3e-49
Identities = 118/314 (37%), Positives = 171/314 (53%), Gaps = 12/314 (3%)
Query: 29 RHSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHN 88
RH T S W + R EL L L +GQ+FRWK +P+ ++GV+ + +L +
Sbjct: 12 RHRTLSSSPALWASIPCPRSELRLDLVLASGQSFRWKEQSPAHWSGVLADQVWTLTQTED 71
Query: 89 GDVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGA 143
C P+ ++ +T L + DVSLA + ++ D F +AQ G
Sbjct: 72 QLYCTVYRGDDSQVSRPTLEELET-LHKYFQLDVSLAQLYSHWASVDSHFQRVAQKFQGV 130
Query: 144 RVLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLS-L 201
R+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP L+ L+
Sbjct: 131 RLLRQDPTECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPNLHALAGP 190
Query: 202 VSEQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPK 261
+E LR G GYRA+Y+ + + + +GG WL LR E+ L LPGVG K
Sbjct: 191 EAETHLRKLGLGYRARYVRASAKAILEE-QGGPAWLQQLRVAPYEEAHKALCTLPGVGAK 249
Query: 262 VAACIALYSLDQHHAIPVDVHIWRIAQKYI--LPELAGSKLTQKL-HSRVAEAFVTKFGK 318
VA CI L +LD+ A+PVDVH+W+IA + P+ + +K L + + F +G
Sbjct: 250 VADCICLMALDKPQAVPVDVHVWQIAHRDYGWHPKTSQAKGPSPLANKELGNFFRNLWGP 309
Query: 319 YAGWAQAVLFIAEL 332
YAGWAQAVLF A+L
Sbjct: 310 YAGWAQAVLFSADL 323
>UniRef100_O70249 N-glycosylase/DNA lyase [Includes: 8-oxoguanine DNA glycosylase (EC
3.2.2.-); DNA-(apurinic or apyrimidinic site) lyase (EC
4.2.99.18) (AP lyase)] [Rattus norvegicus]
Length = 345
Score = 196 bits (499), Expect = 6e-49
Identities = 116/317 (36%), Positives = 170/317 (53%), Gaps = 12/317 (3%)
Query: 29 RHSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHN 88
RH T S W + R EL L L +GQ+FRW+ +P+ ++GV+ + +L +
Sbjct: 12 RHRTLTSSPALWASIPCPRSELRLDLVLASGQSFRWREQSPAHWSGVLADQVWTLTQTED 71
Query: 89 GDVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGA 143
C P+ ++ +T L + DVSL + ++ D F +AQ G
Sbjct: 72 QLYCTVYRGDKGQVGRPTLEELET-LHKYFQLDVSLTQLYSHWASVDSHFQSVAQKFQGV 130
Query: 144 RVLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLV 202
R+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP L+ L+
Sbjct: 131 RLLRQDPTECLFSFICSSNNNIARITGMVERLCQAFGPRLVQLDDVTYHGFPNLHALAGP 190
Query: 203 S-EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPK 261
E LR G GYRA+Y+ + + + +GG WL LR E+ L LPGVG K
Sbjct: 191 EVETHLRKLGLGYRARYVCASAKAILEE-QGGPAWLQQLRVASYEEAHKALCTLPGVGTK 249
Query: 262 VAACIALYSLDQHHAIPVDVHIWRIAQKYI--LPELAGSKLTQKL-HSRVAEAFVTKFGK 318
VA CI L +LD+ A+PVD+H+W+IA + P+ + +K L + + F +G
Sbjct: 250 VADCICLMALDKPQAVPVDIHVWQIAHRDYGWQPKTSQTKGPSPLANKELGNFFRNLWGP 309
Query: 319 YAGWAQAVLFIAELPSQ 335
YAGWAQAVLF A+L Q
Sbjct: 310 YAGWAQAVLFSADLRQQ 326
>UniRef100_UPI00003AA902 UPI00003AA902 UniRef100 entry
Length = 297
Score = 196 bits (497), Expect = 1e-48
Identities = 106/286 (37%), Positives = 162/286 (56%), Gaps = 7/286 (2%)
Query: 40 WIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNGDVCYTLHSQS 99
W L EL L L +GQ FRW+ ++P +TGV+G + +L+ + YT++ +
Sbjct: 16 WRWLRCPPAELRLDLVLASGQAFRWRESSPGAWTGVLGDRVWTLRQ-ERDRLWYTVYGRE 74
Query: 100 PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGARVLRQDPFECLIQFMC 159
+ L D+ DV LA ++ + +D F + A G RVLRQDP ECL+ F+C
Sbjct: 75 TPGPETDRILRDYFQLDVGLAALYRTWGAADPLFCQTATAFPGVRVLRQDPVECLLSFIC 134
Query: 160 SSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLS-LVSEQQLRDAGFGYRAK 217
+SNNH++RIT M++ + + G +L ++ FHAFP+L L+ +E +LR GFGYRA+
Sbjct: 135 TSNNHVARITTMIERLCQAFGQHLCSLDEQPFHAFPSLAALAGSEAEAKLRALGFGYRAR 194
Query: 218 YITGTVNVLQSKPEG-GEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQHHA 276
+++GT + EG G + L LR + + L LPGVG KVA C+ L +LD+ A
Sbjct: 195 FVSGTARAI---AEGMGAKGLCQLRAVPYAEARRVLCALPGVGAKVADCVCLMALDKAEA 251
Query: 277 IPVDVHIWRIAQKYILPELAGSKLTQKLHSRVAEAFVTKFGKYAGW 322
+PVD H+W IA++ L LT ++H + + F +G YAGW
Sbjct: 252 VPVDTHVWHIARQRYGAALGARSLTARVHQEIGDFFRELWGPYAGW 297
>UniRef100_UPI000001A584 UPI000001A584 UniRef100 entry
Length = 304
Score = 195 bits (495), Expect = 2e-48
Identities = 120/306 (39%), Positives = 171/306 (55%), Gaps = 11/306 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H+ + S+ W L + EL L LT GQ+FRW+ TA +TGV+ + +L +
Sbjct: 1 HAILSTGSKMWRSLPCAKSELRLDLTLECGQSFRWRKTAEGHWTGVMKGRVWTLTQTDDT 60
Query: 90 ---DVCYTLHSQSPSNDDCKTALL--DFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
V T Q ND + ++ D+ DV+L + + + +D F +A +G R
Sbjct: 61 LWYHVYRTQERQRKGNDRKRKTIMLRDYFQLDVNLGELYNKWGAADPHFRSVANVFTGVR 120
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+C+SNNHISRI MV+ + +LGT L ++ +++FPTL LS S
Sbjct: 121 MLRQDPVECLFSFICTSNNHISRIQGMVERLCQTLGTPLCELDQDSYYSFPTLAALSGKS 180
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E QLRD GFGYRA+++ + Q G +WL LR + L LPGVG KV
Sbjct: 181 VEAQLRDLGFGYRARFLQQSAK--QILDTHGMQWLDGLRTVPYVQARDALRTLPGVGTKV 238
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQK-YILPELAGSK-LTQKLHSRVAEAFVTKFGKYA 320
A C+ L SLD+ A+PVD H+W+IA++ Y G K LT K+H + + F +G YA
Sbjct: 239 ADCVCLMSLDKAEAVPVDTHVWQIAKRDYNCAFGNGQKSLTDKVHRDIGDFFRKLWGPYA 298
Query: 321 GWAQAV 326
GWAQ+V
Sbjct: 299 GWAQSV 304
>UniRef100_O15527-2 Splice isoform 1B of O15527 [Homo sapiens]
Length = 324
Score = 191 bits (485), Expect = 3e-47
Identities = 113/307 (36%), Positives = 167/307 (53%), Gaps = 12/307 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAV 326
AGWAQAV
Sbjct: 311 AGWAQAV 317
>UniRef100_O15527-7 Splice isoform 2D of O15527 [Homo sapiens]
Length = 349
Score = 191 bits (485), Expect = 3e-47
Identities = 118/343 (34%), Positives = 179/343 (51%), Gaps = 16/343 (4%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVLFIAELPSQKAILPSHLRATKQQSPANIENSEEEIG 362
AGWAQA+ + + + L H Q P ++ S +G
Sbjct: 311 AGWAQALCQV--ITTFMTFLGPH--RLDQMPPEELQTSSSRLG 349
>UniRef100_Q6NL83 RE57519p [Drosophila melanogaster]
Length = 359
Score = 191 bits (484), Expect = 4e-47
Identities = 112/300 (37%), Positives = 168/300 (55%), Gaps = 14/300 (4%)
Query: 43 LNITRQELSLPLTFPTGQTFRWKNTAP---SQYTGVVGSHLISLKHLHNGDVCYTLHSQS 99
+ ++ +E L T GQ+FRW++ ++Y GVV + L+ + + S
Sbjct: 27 IGLSLEECDLERTLLGGQSFRWRSICDGNRTKYGGVVFNTYWVLQQEESFITYEAYGTSS 86
Query: 100 P-SNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGARVLRQDPFECLIQFM 158
P + D + + D+L D L K + D+ F + R+L Q+PFE + F+
Sbjct: 87 PLATKDYSSLISDYLRVDFDLKVNQKDWLSKDDNFVKFLS--KPVRLLSQEPFENIFSFL 144
Query: 159 CSSNNHISRITKMVD-YVSSLGTYLGCVEGFDFHAFPTLNQLSLVS----EQQLRDAGFG 213
CS NN+I RI+ M++ + ++ GT +G G D + FPT+N+ + QLR A FG
Sbjct: 145 CSQNNNIKRISSMIEWFCATFGTKIGHFNGADAYTFPTINRFHDIPCEDLNAQLRAAKFG 204
Query: 214 YRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQ 273
YRAK+I T+ +Q K GG+ W SL+ + E EL LPG+G KVA CI L S+
Sbjct: 205 YRAKFIAQTLQEIQKK--GGQNWFISLKSMPFEKAREELTLLPGIGYKVADCICLMSMGH 262
Query: 274 HHAIPVDVHIWRIAQKYILPELAGSK-LTQKLHSRVAEAFVTKFGKYAGWAQAVLFIAEL 332
++PVD+HI+RIAQ Y LP L G K +T+K++ V++ F GKYAGWAQA+LF A+L
Sbjct: 263 LESVPVDIHIYRIAQNYYLPHLTGQKNVTKKIYEEVSKHFQKLHGKYAGWAQAILFSADL 322
>UniRef100_Q9V3I8 N-glycosylase/DNA lyase (dOgg1) [Includes: 8-oxoguanine DNA
glycosylase (EC 3.2.2.-); DNA-(apurinic or apyrimidinic
site) lyase (EC 4.2.99.18) (AP lyase)] [Drosophila
melanogaster]
Length = 343
Score = 191 bits (484), Expect = 4e-47
Identities = 112/300 (37%), Positives = 168/300 (55%), Gaps = 14/300 (4%)
Query: 43 LNITRQELSLPLTFPTGQTFRWKNTAP---SQYTGVVGSHLISLKHLHNGDVCYTLHSQS 99
+ ++ +E L T GQ+FRW++ ++Y GVV + L+ + + S
Sbjct: 27 IGLSLEECDLERTLLGGQSFRWRSICDGNRTKYGGVVFNTYWVLQQEESFITYEAYGTSS 86
Query: 100 P-SNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGARVLRQDPFECLIQFM 158
P + D + + D+L D L K + D+ F + R+L Q+PFE + F+
Sbjct: 87 PLATKDYSSLISDYLRVDFDLKVNQKDWLSKDDNFVKFLS--KPVRLLSQEPFENIFSFL 144
Query: 159 CSSNNHISRITKMVD-YVSSLGTYLGCVEGFDFHAFPTLNQLSLVS----EQQLRDAGFG 213
CS NN+I RI+ M++ + ++ GT +G G D + FPT+N+ + QLR A FG
Sbjct: 145 CSQNNNIKRISSMIEWFCATFGTKIGHFNGADAYTFPTINRFHDIPCEDLNAQLRAAKFG 204
Query: 214 YRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQ 273
YRAK+I T+ +Q K GG+ W SL+ + E EL LPG+G KVA CI L S+
Sbjct: 205 YRAKFIAQTLQEIQKK--GGQNWFISLKSMPFEKAREELTLLPGIGYKVADCICLMSMGH 262
Query: 274 HHAIPVDVHIWRIAQKYILPELAGSK-LTQKLHSRVAEAFVTKFGKYAGWAQAVLFIAEL 332
++PVD+HI+RIAQ Y LP L G K +T+K++ V++ F GKYAGWAQA+LF A+L
Sbjct: 263 LESVPVDIHIYRIAQNYYLPHLTGQKNVTKKIYEEVSKHFQKLHGKYAGWAQAILFSADL 322
>UniRef100_UPI000036B381 UPI000036B381 UniRef100 entry
Length = 424
Score = 190 bits (482), Expect = 6e-47
Identities = 113/308 (36%), Positives = 167/308 (53%), Gaps = 12/308 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVL 327
AGWAQA L
Sbjct: 311 AGWAQAGL 318
>UniRef100_O15527-4 Splice isoform 2A of O15527 [Homo sapiens]
Length = 424
Score = 190 bits (482), Expect = 6e-47
Identities = 113/308 (36%), Positives = 167/308 (53%), Gaps = 12/308 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQAVL 327
AGWAQA L
Sbjct: 311 AGWAQAGL 318
>UniRef100_UPI000036B37D OGG1 [Mus spretus]
Length = 410
Score = 189 bits (481), Expect = 8e-47
Identities = 112/306 (36%), Positives = 166/306 (53%), Gaps = 12/306 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQA 325
AGWAQA
Sbjct: 311 AGWAQA 316
>UniRef100_O15527-3 Splice isoform 1C of O15527 [Homo sapiens]
Length = 410
Score = 189 bits (481), Expect = 8e-47
Identities = 112/306 (36%), Positives = 166/306 (53%), Gaps = 12/306 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQA 325
AGWAQA
Sbjct: 311 AGWAQA 316
>UniRef100_O15527-8 Splice isoform 2E of O15527 [Homo sapiens]
Length = 322
Score = 189 bits (481), Expect = 8e-47
Identities = 112/306 (36%), Positives = 166/306 (53%), Gaps = 12/306 (3%)
Query: 30 HSTNLSKSRAWIPLNITRQELSLPLTFPTGQTFRWKNTAPSQYTGVVGSHLISLKHLHNG 89
H T S W + R EL L L P+GQ+FRW+ +P+ ++GV+ + +L
Sbjct: 13 HRTLASTPALWASIPCPRSELRLDLVLPSGQSFRWREQSPAHWSGVLADQVWTLTQTEEQ 72
Query: 90 DVCYTLHSQS-----PSNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGAR 144
C P+ D+ + A+ + DV+LA + + D F E+AQ G R
Sbjct: 73 LHCTVYRGDKSQASRPTPDELE-AVRKYFQLDVTLAQLYHHWGSVDSHFQEVAQKFQGVR 131
Query: 145 VLRQDPFECLIQFMCSSNNHISRITKMVDYV-SSLGTYLGCVEGFDFHAFPTLNQLSLVS 203
+LRQDP ECL F+CSSNN+I+RIT MV+ + + G L ++ +H FP+L L+
Sbjct: 132 LLRQDPIECLFSFICSSNNNIARITGMVERLCQAFGPRLIQLDDVTYHGFPSLQALAGPE 191
Query: 204 -EQQLRDAGFGYRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKV 262
E LR G GYRA+Y++ + + + +GG WL LR+ E+ L LPGVG KV
Sbjct: 192 VEAHLRKLGLGYRARYVSASARAILEE-QGGLAWLQQLRESSYEEAHKALCILPGVGTKV 250
Query: 263 AACIALYSLDQHHAIPVDVHIWRIAQKYIL--PELAGSK-LTQKLHSRVAEAFVTKFGKY 319
A CI L +LD+ A+PVDVH+W IAQ+ P + +K + + + + F + +G Y
Sbjct: 251 ADCICLMALDKPQAVPVDVHMWHIAQRDYSWHPTTSQAKGPSPQTNKELGNFFRSLWGPY 310
Query: 320 AGWAQA 325
AGWAQA
Sbjct: 311 AGWAQA 316
>UniRef100_Q6CEU4 Yarrowia lipolytica chromosome B of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 403
Score = 185 bits (470), Expect = 1e-45
Identities = 106/303 (34%), Positives = 171/303 (55%), Gaps = 8/303 (2%)
Query: 40 WIPLNITRQELSLPLTFPTGQTFRW-KNTAPSQYTGVVG--SHLISLKHLHNGDVCYTLH 96
W L + + + +L + GQ+FRW K P+ ++G I L + + +
Sbjct: 13 WNRLGLAKTDANLEILLKCGQSFRWTKIEHPNNNYWIIGMEGRGIVLNQKDDDTMWAEVS 72
Query: 97 SQSP--SNDDCKTALLDFLNADVSLADTWKVFSDSDERFAELAQHLSGARVLRQDPFECL 154
+ + D L D+ N ++ +S D+ F + G RVLRQDP+E L
Sbjct: 73 DKGKPVKSRDTAAILNDYFNISTDTIKLYEDWSSRDDHFKNKSIKYLGIRVLRQDPWENL 132
Query: 155 IQFMCSSNNHISRITKMVDYVS-SLGTYLGCVEGFDFHAFPTLNQLSLVSEQQLRDAGFG 213
F+CSSNN++ RI+++V ++ + G ++ ++ H+FP+ ++L+ +E LR+ G G
Sbjct: 133 CSFICSSNNNVKRISQLVQKMTITFGDHVATLDDLKIHSFPSPDKLA-DTEPILRELGLG 191
Query: 214 YRAKYITGTVNVLQSKPEGGEEWLYSLRKLELEDVISELIKLPGVGPKVAACIALYSLDQ 273
YRAKYI+ T +L +KP GGE++L+ LR ++ S +++ GVGPKVA C+ L+SLD+
Sbjct: 192 YRAKYISKTAEMLLTKP-GGEQFLHELRDASFDEAKSSIMEFLGVGPKVADCVCLFSLDK 250
Query: 274 HHAIPVDVHIWRIAQKYILPELAGSKLTQKLHSRVAEAFVTKFGKYAGWAQAVLFIAELP 333
H +PVD H+W+IAQK + +T K +++V E F K+G YAGWA VLF A+L
Sbjct: 251 HDTVPVDTHVWQIAQKDYGVARSAKTMTPKAYAQVQEFFREKWGPYAGWAHCVLFAAQLS 310
Query: 334 SQK 336
K
Sbjct: 311 DFK 313
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 627,656,841
Number of Sequences: 2790947
Number of extensions: 26264443
Number of successful extensions: 123918
Number of sequences better than 10.0: 512
Number of HSP's better than 10.0 without gapping: 134
Number of HSP's successfully gapped in prelim test: 381
Number of HSP's that attempted gapping in prelim test: 123120
Number of HSP's gapped (non-prelim): 661
length of query: 363
length of database: 848,049,833
effective HSP length: 129
effective length of query: 234
effective length of database: 488,017,670
effective search space: 114196134780
effective search space used: 114196134780
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC140916.11


