

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
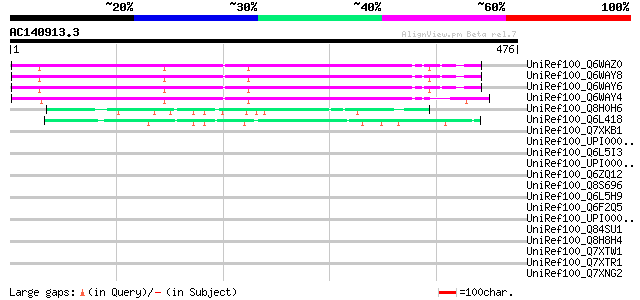
Query= AC140913.3 - phase: 0
(476 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum] 274 5e-72
UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum] 273 6e-72
UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum] 272 2e-71
UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum] 269 1e-70
UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum] 61 8e-08
UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demi... 60 1e-07
UniRef100_Q7XKB1 OSJNBa0064G10.20 protein [Oryza sativa] 43 0.023
UniRef100_UPI0000360333 UPI0000360333 UniRef100 entry 39 0.43
UniRef100_Q6L5I3 Putative mutator-like transposase [Oryza sativa] 39 0.43
UniRef100_UPI0000437B8E UPI0000437B8E UniRef100 entry 38 0.73
UniRef100_Q6ZQ12 MKIAA0980 protein [Mus musculus] 38 0.73
UniRef100_Q8S696 Putative mutator-like transposase [Oryza sativa] 37 0.95
UniRef100_Q6L5H9 Hypothetical protein OJ1741_B01.15 [Oryza sativa] 37 0.95
UniRef100_Q6F2Q5 Hypothetical protein OSJNBa0088M06.4 [Oryza sat... 37 0.95
UniRef100_UPI000031882B UPI000031882B UniRef100 entry 37 1.2
UniRef100_Q84SU1 Putative transposase [Oryza sativa] 37 1.2
UniRef100_Q8H8H4 Putative mutator-like transposase [Oryza sativa] 37 1.2
UniRef100_Q7XTW1 OSJNBa0010D21.4 protein [Oryza sativa] 37 1.2
UniRef100_Q7XTR1 OSJNBb0085C12.10 protein [Oryza sativa] 37 1.2
UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa] 37 1.2
>UniRef100_Q6WAZ0 Hypothetical protein [Pisum sativum]
Length = 562
Score = 274 bits (700), Expect = 5e-72
Identities = 172/457 (37%), Positives = 249/457 (53%), Gaps = 27/457 (5%)
Query: 2 KKTKCYKFKEVDLVSLRELALKVKS--QTGFRLRYGGLLTLLRTDVEEKLVHTLVQFYDP 59
+KT Y F L SL EL+ + S Q GF +YG +LTLL+ V+ + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 60 SFRCFTFPDFQLVPTLEAYSNLVGLPIAEKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHL 119
CFTF D+QL PTLE YS L+ +P+ + PF + V+A+ L+L +VS++
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDNW 144
Query: 120 ITKSHIRGFTSKYLLDQANLSTTRQ--DTLEAILALLIYGLLLFPNLDNFVDMNAIEIFH 177
+ + G K+LL A + + A LA++IYG++LFP++ NFVD A+ IF
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 178 SKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLPSS--FHDNSENWSYSQR 235
NPVPTLLADTY+AIH R KG G I CC+ LL RWF+S LP S F D ++QR
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 236 IMALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYNPELAMRQFGFPMKSKPI 295
+M+LT ++ W + V+ +IM CG F NVPL+G RG INYNP L++RQ GF MK +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 296 NLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEIG 355
+ +F + + + AW + K LG + VA YT WV +R E +
Sbjct: 324 EAEIAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 356 MPYPAMRYVSSSTPSMPLPLLPATQDMYQEHLAMESR---------EKQVWKARYNQAEN 406
+PY M + P + +PA + Y++ L ME+R + +++KA+ + N
Sbjct: 384 LPYDRMEPLQEQPPLILAESVPA--EHYKQAL-MENRRLREKEQDTQMELYKAKAGRL-N 439
Query: 407 LIMTLDGRDEQKTHENLMLKKELAKARRELEEKDELL 443
L L G E+ L +R EE + +L
Sbjct: 440 LAHQLRGVREEDA-------SRLRSKKRSYEEMESML 469
>UniRef100_Q6WAY8 Hypothetical protein [Pisum sativum]
Length = 562
Score = 273 bits (699), Expect = 6e-72
Identities = 172/457 (37%), Positives = 247/457 (53%), Gaps = 27/457 (5%)
Query: 2 KKTKCYKFKEVDLVSLRELALKVKS--QTGFRLRYGGLLTLLRTDVEEKLVHTLVQFYDP 59
+KT Y F L SL EL+ + S Q GF +YG +LTLL+ V+ + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQYGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 60 SFRCFTFPDFQLVPTLEAYSNLVGLPIAEKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHL 119
CFTF D+QL PTLE YS L+ +P+ + PF + V+A+ L+L +VS+
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFRVVARALHLGIKEVSDSW 144
Query: 120 ITKSHIRGFTSKYLLDQANLSTTRQ--DTLEAILALLIYGLLLFPNLDNFVDMNAIEIFH 177
+ + G K+LL A + + A LA++IYG++LFP++ NFVD A+ IF
Sbjct: 145 KSSGDVVGLPLKFLLRVAREEAEKGSWEAFHAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 178 SKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLPSS--FHDNSENWSYSQR 235
NPVPTLLADTY+AIH R KG G I CC+ LL RWF+S LP S F D ++QR
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 236 IMALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYNPELAMRQFGFPMKSKPI 295
+M+LT ++ W + V+ +IM CG F NVPL+G RG INYNP L++RQ GF MK +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLIGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 296 NLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEIG 355
+ F + + + AW + K LG + VA YT WV +R E +
Sbjct: 324 EAEVAESVCFEKRSDPARLEQIGRAWKSIGMKDGSVLGKKFAVAMPDYTDWVKERVETLL 383
Query: 356 MPYPAMRYVSSSTPSMPLPLLPATQDMYQEHLAMESR---------EKQVWKARYNQAEN 406
+PY M + P + +PA + Y++ L ME+R + +++KA+ + N
Sbjct: 384 LPYDRMEPLQEQPPLILAESVPA--EHYKQAL-MENRRLREKEQDTQMELYKAKADSL-N 439
Query: 407 LIMTLDGRDEQKTHENLMLKKELAKARRELEEKDELL 443
L L G E+ L +R EE + +L
Sbjct: 440 LAHQLRGVREEDA-------SRLRSKKRSYEEVESML 469
>UniRef100_Q6WAY6 Hypothetical protein [Pisum sativum]
Length = 562
Score = 272 bits (695), Expect = 2e-71
Identities = 172/457 (37%), Positives = 249/457 (53%), Gaps = 27/457 (5%)
Query: 2 KKTKCYKFKEVDLVSLRELALKVKS--QTGFRLRYGGLLTLLRTDVEEKLVHTLVQFYDP 59
+KT Y F L SL EL+ + S Q GF ++G +LTLL+ V+ + TL+QFYDP
Sbjct: 25 RKTCSYSFHREPLTSLIELSNLMTSGNQKGFVDQHGDILTLLKMVVDPVPLQTLLQFYDP 84
Query: 60 SFRCFTFPDFQLVPTLEAYSNLVGLPIAEKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHL 119
CFTF D+QL PTLE YS L+ +P+ + PF + V+A+ L+L +VS+
Sbjct: 85 ELHCFTFQDYQLAPTLEEYSILLSVPVRYQVPFLDVPKEVDFGVVARALHLGIKEVSDSW 144
Query: 120 ITKSHIRGFTSKYLLDQANLSTTRQ--DTLEAILALLIYGLLLFPNLDNFVDMNAIEIFH 177
+ G K+LL A + + A LA++IYG++LFP++ NFVD A+ IF
Sbjct: 145 KYSGDVVGLPLKFLLRVAREEAEKGSWEAFRAQLAVMIYGIVLFPSMPNFVDYAAVSIFI 204
Query: 178 SKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLPSS--FHDNSENWSYSQR 235
NPVPTLLADTY+AIH R KG G I CC+ LL RWF+S LP S F D ++QR
Sbjct: 205 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFLSLLPVSGPFVDAQSTHKWTQR 263
Query: 236 IMALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYNPELAMRQFGFPMKSKPI 295
+M+LT ++ W + V+ +IM CG F NVPL+G RG INYNP L++RQ GF MK +P+
Sbjct: 264 VMSLTSYDIRWQSYRMDVRNVIMSCGGFRNVPLVGTRGCINYNPVLSLRQLGFVMKGRPL 323
Query: 296 NLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEIG 355
+ +F + + + AW + K LG + VA YT WV +R E +
Sbjct: 324 EAEVAESVYFEKQSDPARLEQIGRAWKSIGVKDGSVLGKKFVVAMPDYTDWVKERVETLL 383
Query: 356 MPYPAMRYVSSSTPSMPLPLLPATQDMYQEHLAMESR---------EKQVWKARYNQAEN 406
+PY M + PS+ +PA + Y++ L ME+R + +++KA+ ++ N
Sbjct: 384 LPYDRMEPLQEQPPSILAESVPA--EHYKQAL-MENRRLRGKEQDTQLELYKAKADKL-N 439
Query: 407 LIMTLDGRDEQKTHENLMLKKELAKARRELEEKDELL 443
L L G E+ L +R EE + +L
Sbjct: 440 LAHQLRGVREEDA-------SRLRSKKRSYEEMESML 469
>UniRef100_Q6WAY4 Hypothetical protein [Pisum sativum]
Length = 546
Score = 269 bits (687), Expect = 1e-70
Identities = 170/459 (37%), Positives = 240/459 (52%), Gaps = 32/459 (6%)
Query: 2 KKTKCYKFKEVDLVSLRELALKVKSQT--GFRLRYGGLLTLLRTDVEEKLVHTLVQFYDP 59
+KT Y F L SL EL + S GF ++G +LTLL+T V+ + TL+QFYDP
Sbjct: 9 RKTCSYSFYREPLTSLIELGSLMPSDQLKGFVGQFGDILTLLKTVVDPVPLQTLLQFYDP 68
Query: 60 SFRCFTFPDFQLVPTLEAYSNLVGLPIAEKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHL 119
RCFTF D+QL PTLE YS L+ +PI + PF + VIA+ L + +V ++
Sbjct: 69 ELRCFTFQDYQLAPTLEEYSILMSIPIQHQVPFLDVPKEVDFRVIARALRMSVKEVCDNW 128
Query: 120 ITKSHIRGFTSKYLLDQANLSTTRQ--DTLEAILALLIYGLLLFPNLDNFVDMNAIEIFH 177
+ G K+LL A + + A LA +IYG++LFP++ NF+D AI IF
Sbjct: 129 KPSGEVVGMPLKFLLRVAREEAEKGNWEVFHAQLAAMIYGIVLFPSMPNFIDHAAISIFI 188
Query: 178 SKNPVPTLLADTYHAIHDRTLKGRGYILCCISLLYRWFISHLPSS--FHDNSENWSYSQR 235
NPVPTLLADTY+AIH R KG G I CC+ LL RWF+S LP+S F D ++QR
Sbjct: 189 GGNPVPTLLADTYYAIHSRHGKG-GAIRCCLPLLLRWFMSLLPASGPFMDTQSTLKWTQR 247
Query: 236 IMALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYNPELAMRQFGFPMKSKPI 295
+M+LT ++ W + VK+++M CG+F NVPL+G +G INYNP L++RQ GF M +P+
Sbjct: 248 VMSLTSYDIRWQSYRMDVKDVVMSCGEFRNVPLVGTKGCINYNPVLSLRQLGFIMSRRPL 307
Query: 296 NLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSVRHLGVRSGVAHEAYTQWVIDRAEEIG 355
+F + + AW + K LG + +A YT WV R E +
Sbjct: 308 EAEIVESVYFEKRTDPVRLEQIGRAWKSIGVKDGSVLGKKFAIAMPDYTDWVKKRVETLL 367
Query: 356 MPYPAMRYVSSSTPSMPLPLLPATQDMYQEHLAMESREKQVWKARYNQAENLIMTLDGRD 415
+PY M + P + +PA + Y++ L ME+R +
Sbjct: 368 LPYDRMEPLQEQPPLILADSVPA--EHYKQAL-MENRRLR------------------EK 406
Query: 416 EQKTHENLMLKK----ELAKARRELEEKDELLMRDSKRA 450
EQ T L K LA RE++ +D R KR+
Sbjct: 407 EQDTRMELYKAKADKLNLAHQLREMQGEDASRARSKKRS 445
>UniRef100_Q8H0H6 Hypothetical protein B27 [Nicotiana tabacum]
Length = 464
Score = 60.8 bits (146), Expect = 8e-08
Identities = 92/419 (21%), Positives = 156/419 (36%), Gaps = 85/419 (20%)
Query: 35 GGLLTLLRTDVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSNLVGLPIAEKAPFTG 94
G L +++ + L+ LV F+DP F F DF+L PTLE ++G
Sbjct: 39 GALTDIMKIKPRDDLIEALVTFWDPVHNVFCFSDFELTPTLEEIDG-----------YSG 87
Query: 95 PGTSLT------PLVIAKDLYLKTSDVSNHLITKSHIRGFTSKYLL-------------D 135
G L P ++ + ++S + + ++G S Y L +
Sbjct: 88 FGRDLRNQELIFPRALSVHRFFDLLNISKQIRKTNVVKGCCSFYFLYSRFGQPNGFEMHE 147
Query: 136 QANLSTTRQDTLEA------ILALLIYGLLLFPNLDNFVDMN---AIEIFHSKNP---VP 183
+ + +DT I+A L G+++FPN + +D +++ +K P
Sbjct: 148 KGLNNKQNKDTWHIHRCFAFIMAFL--GIMVFPNRERTIDTRIARVVQVLTTKEHHTLAP 205
Query: 184 TLLADTYHAIHDRTL--KGRGYILCCISLLYRWFISHLPS-----SFHDNSENW--SYSQ 234
+L+D Y A+ TL G + C LL W I HL S+ + +N+ SY +
Sbjct: 206 IILSDIYRAL---TLCKSGAKFFEGCNILLQMWLIEHLRHHPKFMSYGPSKDNFIDSYEE 262
Query: 235 RIM----------------ALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYN 278
R+ +L ++ W ++E+I ++ LLG+R Y
Sbjct: 263 RVKEYNSPEGVETWISHLRSLNACQIEWTLGWLPLREVIHMSALKSHLLLLGLRSVQPYM 322
Query: 279 PELAMRQFG-FPMKSKPINLATSPEFFFYTNAPTGQRKAFMDAWSKVR--RKSVRHLGVR 335
P +RQ G + + K +L+ + AP + W+ R + +
Sbjct: 323 PHRVLRQLGRYQVVPKDEDLSVQ-VIELHPKAPLPE-ALIQQIWNGCRYLKGDTQVPDPA 380
Query: 336 SGVAHEAYTQWVIDRAEEIGMPYPAMRYVSSSTPSMPLPLLPATQDMYQEHLAMESREK 394
G Y W R+ +P P P + A D QE LA REK
Sbjct: 381 RGEVDPGYAIWFGKRSRVDDVPEPKR--------PTKRPHVQAFDDKIQERLAWGEREK 431
>UniRef100_Q6L418 Hypothetical protein PGEC989P08.9 [Solanum demissum]
Length = 607
Score = 60.1 bits (144), Expect = 1e-07
Identities = 107/486 (22%), Positives = 172/486 (35%), Gaps = 87/486 (17%)
Query: 33 RYGGLLTLLRTDVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSNLVGLPIAEKAPF 92
R G L +LL + L+ LV F+DP F F DF+L PTLE +G +
Sbjct: 47 RLGFLRSLLYVEPRRDLIEALVHFWDPIKNVFHFSDFELTPTLEEIGAFIG-----RGKN 101
Query: 93 TGPGTSLTPLVIAKDLYLKTSDVSNHLITKSHIRGFT------------------SKYLL 134
G + P I +L+ ++ + I G+ K L
Sbjct: 102 LHEGEPMIPKHINGRRFLELLHINENEIGGCLDNGWVPLEFLYKRYGRKDGFELYGKKLH 161
Query: 135 DQANLSTTRQDTLEAILALLIYGLLLFPNLDNFVDMN--AIEIFHSKNP----VPTLLAD 188
+ T + +AI + G+++F +++N A+ K P VP +LAD
Sbjct: 162 NNGCRLTWETHSYDAITVAFL-GIMVFLKRGGKININLAAVITAFEKKPNITLVPMILAD 220
Query: 189 TYHAIHDRTLKGRGYILCCISLLYRWFISHL-------------------------PSSF 223
Y A+ G Y C LL W I H+ SF
Sbjct: 221 IYRAL-TICKNGGDYFEGCNMLLQLWMIEHIRHHPYVVDFKVECNDYIGGHEERIKDHSF 279
Query: 224 HDNSENWSYSQRIMALTPNEVVWLTPAAQVKEIIMGCGDFLNVPLLGIRGGINYNPELAM 283
E W + + LT +++VW E+I + L+G+RG Y P M
Sbjct: 280 PKGIEAW--KKYLNNLTADKIVWNYHWFPSAEVIYMSTFRSFIVLMGLRGVQPYMPLRVM 337
Query: 284 RQFGFPMKSKPINLATSPEFFFYTNAPTGQRKAFMDAWSKVRRKSV--RHLGVRSGVAHE 341
RQ G P + F+ P + + F W S R G +
Sbjct: 338 RQLGRRQFLPPNEDIREFMYEFHPEIPLRKSEIF-KIWGGCMLSSPHDRVEDRTKGEVDQ 396
Query: 342 AYTQWV-----------------IDRAEEIGMPYPAMRY-----VSSSTPSMPLPLLPAT 379
AY +WV +DR EI + R S+ + L A
Sbjct: 397 AYLEWVHDQPSPKVLPEGSVKGPVDREAEIEVRIKQARLEVERSYRSTLDCLSNDLKNAK 456
Query: 380 QDMYQEHLAMESREKQVWKARYNQAENL---IMTLDGRDEQKTHENLMLKKELAKARREL 436
+++ Q E R ++ E+L I ++ ++E+ T E + + E+ + +R +
Sbjct: 457 EELAQRDAIFEVRVRKHRSTILTLQEDLGIVISAMEQQEEEYTKEKVQAECEVERGQR-I 515
Query: 437 EEKDEL 442
E+D L
Sbjct: 516 AERDAL 521
>UniRef100_Q7XKB1 OSJNBa0064G10.20 protein [Oryza sativa]
Length = 1602
Score = 42.7 bits (99), Expect = 0.023
Identities = 53/189 (28%), Positives = 80/189 (42%), Gaps = 31/189 (16%)
Query: 42 RTDVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSNLVGLPI--AEKAPFTGPGTSL 99
R D + L+ LV + P F P ++ PTL+ S L+GLP+ A P GP T+L
Sbjct: 920 RWDADRSLLAALVDRWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGPTTTL 979
Query: 100 TPLVIAKDLYLKTSDVSNHLITKSHIRGFTSKYLLDQANLSTTRQDTLEAILALLIYGLL 159
P S +K+ + FT+ L A+ + R+ +LEA L L ++G +
Sbjct: 980 PP-------------YSTVGPSKAWLLQFTADLLHPDADDYSVRR-SLEAYL-LWLFGWV 1024
Query: 160 LFPNL-DNFVDMNAIEIFHS-----KNPVP------TLLADTYHAIHDRTLK--GRGYIL 205
+F + + VD + S VP +LA TY A+ + K I
Sbjct: 1025 MFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAGAIIA 1084
Query: 206 CCISLLYRW 214
C LL W
Sbjct: 1085 GCPMLLQLW 1093
>UniRef100_UPI0000360333 UPI0000360333 UniRef100 entry
Length = 896
Score = 38.5 bits (88), Expect = 0.43
Identities = 27/87 (31%), Positives = 38/87 (43%), Gaps = 1/87 (1%)
Query: 363 YVSSSTPSMPLPLLPATQDMYQEHLAMESREKQVWKAR-YNQAENLIMTLDGRDEQKTHE 421
+ + S P+ P+ T Y+ + +E K R Y E + D E
Sbjct: 1 FAAPSFPAKMSPIKALTMKDYENQITALKKENFNLKLRIYFMEERMQQKCDDSTEDIFKT 60
Query: 422 NLMLKKELAKARRELEEKDELLMRDSK 448
N+ LK EL +REL EK ELL+ SK
Sbjct: 61 NIELKVELESMKRELAEKQELLVSASK 87
>UniRef100_Q6L5I3 Putative mutator-like transposase [Oryza sativa]
Length = 1684
Score = 38.5 bits (88), Expect = 0.43
Identities = 61/237 (25%), Positives = 89/237 (36%), Gaps = 67/237 (28%)
Query: 30 FRLRYGGLLTLLRT--------------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTL 75
+RLR GLL L R D + L+ LV + P F P ++ PTL
Sbjct: 938 YRLRAAGLLPLCRLVEAAADDRDPAKRWDADRSLLAALVDRWRPETHTFHLPCGEMAPTL 997
Query: 76 EAYSNLVGLPIAEKA------------------------PFTGPGTSLTPLVIAKDLYLK 111
+ S L+GLP+A A P GP T+L P
Sbjct: 998 QDVSYLLGLPLAGAAVGPVDGVFGWKEDITARFEQVMRLPHLGPTTTLPP---------- 1047
Query: 112 TSDVSNHLITKSHIRGFTSKYLLDQANLSTTRQDTLEAILALLIYGLLLFPNL-DNFVDM 170
S +K+ + FT+ L A+ + R+ +LEA L L ++G ++F + + VD
Sbjct: 1048 ---YSTVGPSKAWLLQFTADLLHPDADDYSVRR-SLEAYL-LWLFGWVMFTSTHGHAVDF 1102
Query: 171 NAIEIFHS-----KNPVP------TLLADTYHAIHDRTLK--GRGYILCCISLLYRW 214
+ S VP +LA TY A+ + K I C LL W
Sbjct: 1103 RLVHYARSIADAQPQDVPQWSWGSAVLAATYRALCEACTKTDAGAIIAGCPMLLQLW 1159
>UniRef100_UPI0000437B8E UPI0000437B8E UniRef100 entry
Length = 786
Score = 37.7 bits (86), Expect = 0.73
Identities = 22/82 (26%), Positives = 41/82 (49%), Gaps = 1/82 (1%)
Query: 367 STPSMPLPLLPATQDMYQEHLAMESREKQVWKARYNQAENLIMTLDGRDEQKTHENLMLK 426
+TP+ P P + AT + +E +++ + R ++ E L E+K E +K
Sbjct: 195 TTPASPAPPIAATCPLSEEKTKTSEEQEEKQRQR-DEKERLKQEAKAAKEKKKEEARKMK 253
Query: 427 KELAKARRELEEKDELLMRDSK 448
+E + ++E +EKDE R+ K
Sbjct: 254 EEKEREKKEKKEKDEKERREKK 275
>UniRef100_Q6ZQ12 MKIAA0980 protein [Mus musculus]
Length = 1292
Score = 37.7 bits (86), Expect = 0.73
Identities = 30/101 (29%), Positives = 48/101 (46%), Gaps = 20/101 (19%)
Query: 369 PSMPLPLLPATQDMYQEHLAMESREKQVWKARYNQAENLIMTLDGRDEQKTH-------- 420
PS+P LP Q+ ES K V K + Q E+++ L+ D ++++
Sbjct: 935 PSLPCSELPNPQEATVMPAMSESEMKDV-KTKLLQLEDVVRALEKADSRESYRAELQRLS 993
Query: 421 -ENLMLKKELAKARRELEEKD----------ELLMRDSKRA 450
ENL+LK +L K + ELE + E+L RD ++A
Sbjct: 994 EENLVLKSDLGKIQLELETSESKNEVQRQEIEVLKRDKEQA 1034
>UniRef100_Q8S696 Putative mutator-like transposase [Oryza sativa]
Length = 1530
Score = 37.4 bits (85), Expect = 0.95
Identities = 28/82 (34%), Positives = 38/82 (46%), Gaps = 16/82 (19%)
Query: 31 RLRYGGLLTLLRT--------------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLE 76
RLR GLL + R V+ L+ TLV + P F P ++ PTL+
Sbjct: 937 RLREAGLLPMCRLVEAAAGDADPARRWSVDRSLLATLVDRWRPETHTFHLPCGEVAPTLQ 996
Query: 77 AYSNLVGLPIAEKAPFTGPGTS 98
S L+GLP+A A GP T+
Sbjct: 997 DVSYLLGLPLAGDA--VGPVTT 1016
>UniRef100_Q6L5H9 Hypothetical protein OJ1741_B01.15 [Oryza sativa]
Length = 896
Score = 37.4 bits (85), Expect = 0.95
Identities = 29/88 (32%), Positives = 40/88 (44%), Gaps = 16/88 (18%)
Query: 25 KSQTGFRLRYGGLLTLLRT--------------DVEEKLVHTLVQFYDPSFRCFTFPDFQ 70
+SQ RLR GLL + R V+ L+ LV + P F P +
Sbjct: 257 RSQYEVRLREAGLLPMCRLVEAAAGDADPARRWTVDRSLLAALVDRWRPETHTFHLPCGE 316
Query: 71 LVPTLEAYSNLVGLPIAEKAPFTGPGTS 98
+ PTL+ S L+GLP+A A GP T+
Sbjct: 317 VAPTLQDVSYLLGLPLAGDA--VGPVTT 342
>UniRef100_Q6F2Q5 Hypothetical protein OSJNBa0088M06.4 [Oryza sativa]
Length = 1635
Score = 37.4 bits (85), Expect = 0.95
Identities = 27/76 (35%), Positives = 35/76 (45%), Gaps = 13/76 (17%)
Query: 31 RLRYGGLLTLLRT-----------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYS 79
RLR GLLT R ++ L+ LV + P F ++VPTL+ S
Sbjct: 900 RLRAAGLLTFARLVEPSRARSERIHIDAALLSALVDRWRPETHTFHLTVGEMVPTLQDVS 959
Query: 80 NLVGLPIAEKAPFTGP 95
L+GLPIA P GP
Sbjct: 960 YLLGLPIA--GPVVGP 973
>UniRef100_UPI000031882B UPI000031882B UniRef100 entry
Length = 883
Score = 37.0 bits (84), Expect = 1.2
Identities = 19/64 (29%), Positives = 34/64 (52%)
Query: 388 AMESREKQVWKARYNQAENLIMTLDGRDEQKTHENLMLKKELAKARRELEEKDELLMRDS 447
A + E+Q W+ + + + R +Q+ E L+++LA+ RRELE + E +D
Sbjct: 606 AQQRLEQQRWEREREKHRQRVEATEARLQQREEECRRLEEQLAEERRELESQRETYQQDL 665
Query: 448 KRAR 451
+R R
Sbjct: 666 ERLR 669
>UniRef100_Q84SU1 Putative transposase [Oryza sativa]
Length = 1511
Score = 37.0 bits (84), Expect = 1.2
Identities = 27/76 (35%), Positives = 35/76 (45%), Gaps = 13/76 (17%)
Query: 31 RLRYGGLLTLLRT-----------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYS 79
RLR GLLT R ++ L+ LV + P F ++VPTL+ S
Sbjct: 868 RLRAAGLLTFARLVEPSRARSERIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVS 927
Query: 80 NLVGLPIAEKAPFTGP 95
L+GLPIA P GP
Sbjct: 928 YLLGLPIA--GPAVGP 941
>UniRef100_Q8H8H4 Putative mutator-like transposase [Oryza sativa]
Length = 1436
Score = 37.0 bits (84), Expect = 1.2
Identities = 27/76 (35%), Positives = 35/76 (45%), Gaps = 13/76 (17%)
Query: 31 RLRYGGLLTLLRT-----------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYS 79
RLR GLLT R ++ L+ LV + P F ++VPTL+ S
Sbjct: 922 RLRAAGLLTFARLVEPSRARSERIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVS 981
Query: 80 NLVGLPIAEKAPFTGP 95
L+GLPIA P GP
Sbjct: 982 YLLGLPIA--GPAVGP 995
>UniRef100_Q7XTW1 OSJNBa0010D21.4 protein [Oryza sativa]
Length = 1609
Score = 37.0 bits (84), Expect = 1.2
Identities = 27/76 (35%), Positives = 35/76 (45%), Gaps = 13/76 (17%)
Query: 31 RLRYGGLLTLLRT-----------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYS 79
RLR GLLT R ++ L+ LV + P F ++VPTL+ S
Sbjct: 913 RLRAAGLLTFARLVEPSRARSERIHIDAALMSALVDRWRPETHTFHLTVGEMVPTLQDVS 972
Query: 80 NLVGLPIAEKAPFTGP 95
L+GLPIA P GP
Sbjct: 973 YLLGLPIA--GPAVGP 986
>UniRef100_Q7XTR1 OSJNBb0085C12.10 protein [Oryza sativa]
Length = 960
Score = 37.0 bits (84), Expect = 1.2
Identities = 27/76 (35%), Positives = 35/76 (45%), Gaps = 13/76 (17%)
Query: 31 RLRYGGLLTLLRT-----------DVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYS 79
RLR GLLT R ++ L+ LV + P F ++VPTL+ S
Sbjct: 584 RLRAAGLLTFARLVEPSRARSERIHIDAALLSALVDRWRPETHTFHLTVGEMVPTLQDVS 643
Query: 80 NLVGLPIAEKAPFTGP 95
L+GLPIA P GP
Sbjct: 644 YLLGLPIA--GPAVGP 657
>UniRef100_Q7XNG2 OSJNBa0096F01.11 protein [Oryza sativa]
Length = 1365
Score = 37.0 bits (84), Expect = 1.2
Identities = 53/211 (25%), Positives = 80/211 (37%), Gaps = 53/211 (25%)
Query: 42 RTDVEEKLVHTLVQFYDPSFRCFTFPDFQLVPTLEAYSNLVGLPIA-------------- 87
R D + L+ LV + P F P ++ PTL+ S L+GLP+A
Sbjct: 801 RWDADRSLLAALVDHWRPETHTFHLPCGEMAPTLQDVSYLLGLPLAGAPVGPVDGVFGWK 860
Query: 88 ----------EKAPFTGPGTSLTPLVIAKDLYLKTSDVSNHLITKSHIRGFTSKYLLDQA 137
+ P GP +L P S +K+ + FT+ L A
Sbjct: 861 EDITARFEQVMRLPHLGPANTLPP-------------YSTVGPSKAWLLQFTADLLHPDA 907
Query: 138 NLSTTRQDTLEAILALLIYGLLLFPNL-DNFVDMNAIEIFHS-----KNPVP------TL 185
+ R+ +LEA L L ++G ++F + + VD + S VP +
Sbjct: 908 DDYLVRR-SLEAYL-LWLFGWVMFTSTHGHAVDFRLVHYARSIADAQPQDVPQWSWGSAV 965
Query: 186 LADTYHAIHDRTLK--GRGYILCCISLLYRW 214
LA TYHA+ + K I C LL W
Sbjct: 966 LAATYHALCEACTKTDAGAIIAGCPMLLQLW 996
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 826,574,471
Number of Sequences: 2790947
Number of extensions: 36157652
Number of successful extensions: 93886
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 93763
Number of HSP's gapped (non-prelim): 146
length of query: 476
length of database: 848,049,833
effective HSP length: 131
effective length of query: 345
effective length of database: 482,435,776
effective search space: 166440342720
effective search space used: 166440342720
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC140913.3


