

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.13 + phase: 0 /pseudo
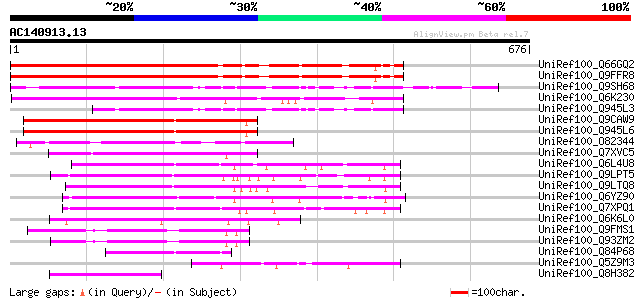
(676 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q66GQ2 At5g41620 [Arabidopsis thaliana] 443 e-123
UniRef100_Q9FFR8 Gb|AAF24581.1 [Arabidopsis thaliana] 443 e-123
UniRef100_Q9SH68 F22C12.6 [Arabidopsis thaliana] 377 e-103
UniRef100_Q6K230 Intracellular protein transport protein USO1-li... 329 1e-88
UniRef100_Q945L3 At1g64180/F22C12_1 [Arabidopsis thaliana] 234 8e-60
UniRef100_Q9CAW9 Hypothetical protein F24K9.26 [Arabidopsis thal... 197 8e-49
UniRef100_Q945L6 AT3g11590/F24K9_26 [Arabidopsis thaliana] 196 2e-48
UniRef100_O82344 Hypothetical protein At2g46250 [Arabidopsis tha... 183 1e-44
UniRef100_Q7XVC5 OSJNBa0072D21.15 protein [Oryza sativa] 159 3e-37
UniRef100_Q6L4U8 Hypothetical protein P0010D04.11 [Oryza sativa] 132 3e-29
UniRef100_Q9LPT5 F11F12.2 protein [Arabidopsis thaliana] 130 2e-28
UniRef100_Q9LTQ8 Gb|AAF00675.1 [Arabidopsis thaliana] 127 1e-27
UniRef100_Q6YZ90 Hypothetical protein B1099H05.52 [Oryza sativa] 124 9e-27
UniRef100_Q7XPQ1 OSJNBa0053K19.22 protein [Oryza sativa] 124 9e-27
UniRef100_Q6K6L0 Hypothetical protein P0047E05.7 [Oryza sativa] 122 5e-26
UniRef100_Q9FMS1 Gb|AAF00675.1 [Arabidopsis thaliana] 117 1e-24
UniRef100_Q93ZM2 AT5g22310/MWD9_9 [Arabidopsis thaliana] 113 2e-23
UniRef100_Q84P68 Hypothetical protein [Oryza sativa] 90 2e-16
UniRef100_Q5Z9M3 Intracellular protein transport protein USO1-li... 84 1e-14
UniRef100_Q8H382 Hypothetical protein OSJNBa0061L20.108 [Oryza s... 84 1e-14
>UniRef100_Q66GQ2 At5g41620 [Arabidopsis thaliana]
Length = 543
Score = 443 bits (1140), Expect = e-123
Identities = 258/521 (49%), Positives = 342/521 (65%), Gaps = 44/521 (8%)
Query: 1 MQHHRAIDRNNHALQPLSPASYGSSMEMTPYNPAATPNSSLDFKGRIG-EPHYSLKTSTE 59
++HH++IDRNNHALQP+SPASYGSS+E+T YN A TP+SSL+F+GR EPHY+LKTSTE
Sbjct: 59 IKHHQSIDRNNHALQPVSPASYGSSLEVTTYNKAVTPSSSLEFRGRPSREPHYNLKTSTE 118
Query: 60 LLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAED 119
LLKVLNRIWSLEEQH SNISLIKALKTE+ +R+++KELLR +QADRHE+D ++KQ+AE+
Sbjct: 119 LLKVLNRIWSLEEQHVSNISLIKALKTEVAHSRVRIKELLRYQQADRHELDSVVKQLAEE 178
Query: 120 KLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQ 179
KL+ K+KE +R+ +AVQSVR LEDERKLRKRSES+HRK+ARELSE+KSS ++ +K+LE+
Sbjct: 179 KLLSKNKEVERMSSAVQSVRKALEDERKLRKRSESLHRKMARELSEVKSSLSNCVKELER 238
Query: 180 ERTRRKLLEDLCDEFARGINEYEQEVHTLKQKS-EKDWVQRADHDRLVLHISESWLDERM 238
K++E LCDEFA+GI YE+E+H LK+K+ +KDW R D+LVLHI+ESWLDERM
Sbjct: 239 GSKSNKMMELLCDEFAKGIKSYEEEIHGLKKKNLDKDWAGRGGGDQLVLHIAESWLDERM 298
Query: 239 QMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVS 298
QM+LE S++DKL +EIETF++ K+ N++ R+RRNSLESVP N +
Sbjct: 299 QMRLEGGDTLNGKNRSVLDKLEVEIETFLQEKR------NEIPRNRRNSLESVPFNTLSA 352
Query: 299 APQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKSVPGE 358
P+ + D ++DS GSDS+CFEL KP+ + DET K N K E
Sbjct: 353 PPRDV--DCEEDSGGSDSNCFELKKPA------------ESYGDETKKPNQHNKDGSIDE 398
Query: 359 GFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPEHFEISESGGFERKNNLSELH 418
K+ N + Q A L++N + I +E E ++ K +E
Sbjct: 399 KPKSPSSFQVNFEDQMAWA---LSSNGKKKTTRAIEDEEEEEDVKPENSNNNKKPENECA 455
Query: 419 STSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASPVRQWMAS---- 474
+T+KN ++ +IR +E EASC+ R QASPVRQW++
Sbjct: 456 TTNKNDVMGEMIRTH--------RRLLSETREIDEASCNFPSSRRQASPVRQWISRTVAP 507
Query: 475 --LDISEASKVHSGSKDNTLKAKLLEARSKGQRSRLRTLKG 513
L SE + H G KDNTLK KL + +SRLR KG
Sbjct: 508 DLLGPSEIAIAH-GVKDNTLKTKL----ANSSKSRLRLFKG 543
>UniRef100_Q9FFR8 Gb|AAF24581.1 [Arabidopsis thaliana]
Length = 623
Score = 443 bits (1140), Expect = e-123
Identities = 258/521 (49%), Positives = 342/521 (65%), Gaps = 44/521 (8%)
Query: 1 MQHHRAIDRNNHALQPLSPASYGSSMEMTPYNPAATPNSSLDFKGRIG-EPHYSLKTSTE 59
++HH++IDRNNHALQP+SPASYGSS+E+T YN A TP+SSL+F+GR EPHY+LKTSTE
Sbjct: 139 IKHHQSIDRNNHALQPVSPASYGSSLEVTTYNKAVTPSSSLEFRGRPSREPHYNLKTSTE 198
Query: 60 LLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAED 119
LLKVLNRIWSLEEQH SNISLIKALKTE+ +R+++KELLR +QADRHE+D ++KQ+AE+
Sbjct: 199 LLKVLNRIWSLEEQHVSNISLIKALKTEVAHSRVRIKELLRYQQADRHELDSVVKQLAEE 258
Query: 120 KLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQ 179
KL+ K+KE +R+ +AVQSVR LEDERKLRKRSES+HRK+ARELSE+KSS ++ +K+LE+
Sbjct: 259 KLLSKNKEVERMSSAVQSVRKALEDERKLRKRSESLHRKMARELSEVKSSLSNCVKELER 318
Query: 180 ERTRRKLLEDLCDEFARGINEYEQEVHTLKQKS-EKDWVQRADHDRLVLHISESWLDERM 238
K++E LCDEFA+GI YE+E+H LK+K+ +KDW R D+LVLHI+ESWLDERM
Sbjct: 319 GSKSNKMMELLCDEFAKGIKSYEEEIHGLKKKNLDKDWAGRGGGDQLVLHIAESWLDERM 378
Query: 239 QMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVS 298
QM+LE S++DKL +EIETF++ K+ N++ R+RRNSLESVP N +
Sbjct: 379 QMRLEGGDTLNGKNRSVLDKLEVEIETFLQEKR------NEIPRNRRNSLESVPFNTLSA 432
Query: 299 APQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKSVPGE 358
P+ + D ++DS GSDS+CFEL KP+ + DET K N K E
Sbjct: 433 PPRDV--DCEEDSGGSDSNCFELKKPA------------ESYGDETKKPNQHNKDGSIDE 478
Query: 359 GFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPEHFEISESGGFERKNNLSELH 418
K+ N + Q A L++N + I +E E ++ K +E
Sbjct: 479 KPKSPSSFQVNFEDQMAWA---LSSNGKKKTTRAIEDEEEEEDVKPENSNNNKKPENECA 535
Query: 419 STSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASPVRQWMAS---- 474
+T+KN ++ +IR +E EASC+ R QASPVRQW++
Sbjct: 536 TTNKNDVMGEMIRTH--------RRLLSETREIDEASCNFPSSRRQASPVRQWISRTVAP 587
Query: 475 --LDISEASKVHSGSKDNTLKAKLLEARSKGQRSRLRTLKG 513
L SE + H G KDNTLK KL + +SRLR KG
Sbjct: 588 DLLGPSEIAIAH-GVKDNTLKTKL----ANSSKSRLRLFKG 623
>UniRef100_Q9SH68 F22C12.6 [Arabidopsis thaliana]
Length = 776
Score = 377 bits (967), Expect = e-103
Identities = 269/640 (42%), Positives = 372/640 (58%), Gaps = 98/640 (15%)
Query: 1 MQHHRAIDRNNHALQPLSPASYGSSMEMTPYNPAATPNSSLDFKGR--IGEPHYSLKTST 58
M+HH +RN+HALQP+SP SY +SSL+F+GR GEP+ ++KTST
Sbjct: 204 MKHHHLTERNDHALQPVSPTSY---------------DSSLEFRGRRRAGEPNNNIKTST 248
Query: 59 ELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAE 118
ELLKVLNRIW LEEQHS+NISLIK+LKTEL +R ++K+LLR +QAD+ ++DD +KQ+AE
Sbjct: 249 ELLKVLNRIWILEEQHSANISLIKSLKTELAHSRARIKDLLRCKQADKRDMDDFVKQLAE 308
Query: 119 DKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLE 178
+KL + +KE DRL +AVQS LEDERKLRKRSES++RKLA+ELSE+KS+ ++ +K++E
Sbjct: 309 EKLSKGTKEHDRLSSAVQS----LEDERKLRKRSESLYRKLAQELSEVKSTLSNCVKEME 364
Query: 179 QERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERM 238
+ +K+LE LCDEFA+GI YE+E+H LKQK +K+W + D ++L I+ESWLDER+
Sbjct: 365 RGTESKKILERLCDEFAKGIKSYEREIHGLKQKLDKNWKGWDEQDHMILCIAESWLDERI 424
Query: 239 QMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVS 298
Q + NG S ++KL EIETF+K QN+ S N++ R+RR SLESVP N A+S
Sbjct: 425 Q-----SGNG-----SALEKLEFEIETFLKTNQNADS--NEIARNRRTSLESVPFN-AMS 471
Query: 299 AP-QAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKSVPG 357
AP + ++++DS GS S+CFEL K AK + DET K + K
Sbjct: 472 APIWEVDREEEEDSGGSGSNCFELKKHGSDVAKPPRG-------DETEKPELIKVGVSER 524
Query: 358 EGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISE-EPEHFEISESGGFERKNNLSE 416
++ PS+ K + A +++N + + TR +E EPE +E G E N + E
Sbjct: 525 PQRRSQSPSSLQVKFEDQMA-WAMSSNEK--KKTRANEMEPE----TEKCGKETNNVVGE 577
Query: 417 LHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASPVRQWMASLD 476
+ T + L+SE+ EASCS R + SP+RQW +
Sbjct: 578 MIRTHRR-----------LSSET---------REIDEASCSYPS-RRRESPIRQW-NTRT 615
Query: 477 ISEASKVHSGSKDNTLKAKLLEARSKGQRSRLRTLKGPKFK**PNPI*KQRSE*DTNPNQ 536
++ G KDNTLK KL EAR+ R R R + +
Sbjct: 616 VTPDLGAPRGVKDNTLKTKLSEARTTSSRPRSEMATA-----------SFRWILQLHRDV 664
Query: 537 CKKIRNTKRSVADGSVVQVVITTTQRCSKSCTFLFRRLRLHSQRLHSPLG*TSICDQSTQ 596
K R ++ + D SV V+T +S ++ S+ + S G +S
Sbjct: 665 PKAARFYEKGL-DFSV--NVVTLRWAELQSGPLKLALMQAPSEHVMSEKGYSS------- 714
Query: 597 KGYSSLLSFTVTDINSTVTKLMALGAELDGPIKYEVHGKV 636
LLSFTV DIN+T++KLM LGAELDG IKYEVHGKV
Sbjct: 715 -----LLSFTVADINTTISKLMELGAELDGSIKYEVHGKV 749
>UniRef100_Q6K230 Intracellular protein transport protein USO1-like [Oryza sativa]
Length = 711
Score = 329 bits (844), Expect = 1e-88
Identities = 218/546 (39%), Positives = 301/546 (54%), Gaps = 63/546 (11%)
Query: 3 HHRAIDRNNHALQPLSPASYGSSMEMTPYNPAATPNSSLDFKGRIGEPHYSLKTSTELLK 62
H + N QP SP SY SS+ + N A +P SLD KGR Y+LKTSTELLK
Sbjct: 187 HQKLNQARNCTAQPFSPGSYRSSVGDSSINQAISPARSLDIKGRFRGADYNLKTSTELLK 246
Query: 63 VLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLV 122
VLNRIWSLEEQH++++S I LK EL + ++EL +R+ RH+V L++Q++EDKLV
Sbjct: 247 VLNRIWSLEEQHTADMSAINGLKLELQHAQEHIQELKCERRGYRHDVASLVRQLSEDKLV 306
Query: 123 RKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERT 182
RK+K+++++ A + S++DELEDER+LR+ SE +HRK +ELSE+KS+F A+KDLE+E+
Sbjct: 307 RKNKDKEKIAADIHSLQDELEDERRLRRHSEDLHRKFGKELSEIKSAFVKAVKDLEKEKK 366
Query: 183 RRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQL 242
+ LLEDLCD+FA GI +YE+EV LKQ+ Q D+ VLH+SE+WLDERMQMQ
Sbjct: 367 TKNLLEDLCDQFAMGIRDYEEEVRALKQRHVNYEYQ---FDKSVLHVSEAWLDERMQMQN 423
Query: 243 EAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQ--------VIRDRRNSLESVPLN 294
+ + K++I ++L EIE F+ AK++ N R RR SLESV N
Sbjct: 424 TDVKEDSLKKSTITERLRSEIEAFLLAKRSVSFKNNDNYMHDSRPNARLRRQSLESVHFN 483
Query: 295 DAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKK- 353
A SAPQ + +DDDDDSV SD HCFELN G+ + + T + K++
Sbjct: 484 GATSAPQ-LAEDDDDDSVASDLHCFELNM---HGSSIQMHDHTGPRRSYTGNMDAPKRRT 539
Query: 354 ----SVPGEGFK----------NHIPSNSNK-----KSQSVDAEEGLTTNIRLVEGTRIS 394
SV GE N S+S++ ++Q +D++ + R V +
Sbjct: 540 EYSHSVVGESSHMSDVQIYSQCNKARSSSSRPWHATRTQEIDSQ----ASARTVPADEQN 595
Query: 395 EEP-EHFEISESGGFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGE 453
E P H G KNNL +D+ R L
Sbjct: 596 EIPCPHISQGYHNGTTSKNNLGAHADCLGQESLDHYSRASLFCD---------------- 639
Query: 454 ASCSNAGWRNQASPVRQW---MASL--DISEASK-VHSGSKDNTLKAKLLEARSKGQRSR 507
++ N SP RQ ASL DI E S + G K+NTLKAKLL+AR +G+ +R
Sbjct: 640 -GTTSGDLCNPHSPSRQLDYPSASLGHDIGECSTGLLVGMKENTLKAKLLQARLEGRHAR 698
Query: 508 LRTLKG 513
L+ G
Sbjct: 699 LKASGG 704
>UniRef100_Q945L3 At1g64180/F22C12_1 [Arabidopsis thaliana]
Length = 354
Score = 234 bits (596), Expect = 8e-60
Identities = 167/406 (41%), Positives = 237/406 (58%), Gaps = 55/406 (13%)
Query: 109 VDDLMKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKS 168
+DD +KQ+AE+KL + +KE DRL +AVQS LEDERKLRKRSES++RKLA+ELSE+KS
Sbjct: 1 MDDFVKQLAEEKLSKGTKEHDRLSSAVQS----LEDERKLRKRSESLYRKLAQELSEVKS 56
Query: 169 SFTSALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLH 228
+ ++ +K++E+ +K+LE LCDEFA+GI YE+E+H LKQK +K+W + D ++L
Sbjct: 57 TLSNCVKEMERGTESKKILERLCDEFAKGIKSYEREIHGLKQKLDKNWKGWDEQDHMILC 116
Query: 229 ISESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSL 288
I+ESWLDER+Q + NG S ++KL EIETF+K QN+ S N++ R+RR SL
Sbjct: 117 IAESWLDERIQ-----SGNG-----SALEKLEFEIETFLKTNQNADS--NEIARNRRTSL 164
Query: 289 ESVPLNDAVSAP-QAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKS 347
ESVP N A+SAP + ++++DS GS S+CFEL K AK + DET K
Sbjct: 165 ESVPFN-AMSAPIWEVDREEEEDSGGSGSNCFELKKHGSDVAKPPRG-------DETEKP 216
Query: 348 NVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISE-EPEHFEISESG 406
+ K ++ PS+ K + A +++N + + TR +E EPE +E
Sbjct: 217 ELIKVGVSERPQRRSQSPSSLQVKFEDQMA-WAMSSNEK--KKTRANEMEPE----TEKC 269
Query: 407 GFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQAS 466
G E N + E+ T + L+SE+ EASCS R + S
Sbjct: 270 GKETNNVVGEMIRTHRR-----------LSSET---------REIDEASCSYPS-RRRES 308
Query: 467 PVRQWMASLDISEASKVHSGSKDNTLKAKLLEARSKGQRSRLRTLK 512
P+RQW + ++ G KDNTLK KL EAR+ R R+R K
Sbjct: 309 PIRQW-NTRTVTPDLGAPRGVKDNTLKTKLSEARTTSSRPRVRLFK 353
>UniRef100_Q9CAW9 Hypothetical protein F24K9.26 [Arabidopsis thaliana]
Length = 622
Score = 197 bits (501), Expect = 8e-49
Identities = 113/311 (36%), Positives = 193/311 (61%), Gaps = 8/311 (2%)
Query: 19 PASYGSSMEMTPYNPAATPN-SSLDFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQHSSN 77
P + GS M++ + TP S++ K R+ + +L TS ELLK++NR+W +++ SS+
Sbjct: 194 PINSGSFMDIETRSRVETPTGSTVGVKTRLKDCSNALTTSKELLKIINRMWGQDDRPSSS 253
Query: 78 ISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQS 137
+SL+ AL +EL+R R++V +L+ + + + +++ LMK+ AE+K V KS EQ+ + AA++S
Sbjct: 254 MSLVSALHSELERARLQVNQLIHEHKPENNDISYLMKRFAEEKAVWKSNEQEVVEAAIES 313
Query: 138 VRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARG 197
V ELE ERKLR+R ES+++KL +EL+E KS+ A+K++E E+ R ++E +CDE AR
Sbjct: 314 VAGELEVERKLRRRFESLNKKLGKELAETKSALMKAVKEIENEKRARVMVEKVCDELARD 373
Query: 198 INEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVD 257
I+E + EV LK++S K + + +R +L ++++ +ER+QM+L A++ +KN+ VD
Sbjct: 374 ISEDKAEVEELKRESFK-VKEEVEKEREMLQLADALREERVQMKLSEAKHQLEEKNAAVD 432
Query: 258 KLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGDD------DDDDS 311
KL +++T++KAK+ N LN +S +D +++ S
Sbjct: 433 KLRNQLQTYLKAKRCKEKTREPPQTQLHNEEAGDYLNHHISFGSYNIEDGEVENGNEEGS 492
Query: 312 VGSDSHCFELN 322
SD H ELN
Sbjct: 493 GESDLHSIELN 503
>UniRef100_Q945L6 AT3g11590/F24K9_26 [Arabidopsis thaliana]
Length = 622
Score = 196 bits (497), Expect = 2e-48
Identities = 112/311 (36%), Positives = 192/311 (61%), Gaps = 8/311 (2%)
Query: 19 PASYGSSMEMTPYNPAATPN-SSLDFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQHSSN 77
P + GS M++ + TP S++ K R+ + +L TS ELLK++NR+W +++ SS+
Sbjct: 194 PINSGSFMDIETRSRVETPTGSTVGVKTRLKDCSNALTTSKELLKIINRMWGQDDRPSSS 253
Query: 78 ISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQS 137
+SL+ AL +EL+R R++V +L+ + + + +++ LMK+ AE+K V KS EQ+ + AA++S
Sbjct: 254 MSLVSALHSELERARLQVNQLIHEHKPENNDISYLMKRFAEEKAVWKSNEQEVVEAAIES 313
Query: 138 VRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARG 197
V ELE ERKLR+R ES+++KL +EL+E KS+ A+K++E E+ R ++E +CDE AR
Sbjct: 314 VAGELEVERKLRRRFESLNKKLGKELAETKSALMKAVKEIENEKRARVMVEKVCDELARD 373
Query: 198 INEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVD 257
I+E + EV LK++S K + + +R +L ++++ +ER+QM+L A++ +KN+ VD
Sbjct: 374 ISEDKAEVEELKRESFK-VKEEVEKEREMLQLADALREERVQMKLSEAKHQLEEKNAAVD 432
Query: 258 KLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGDD------DDDDS 311
KL +++T++K K+ N LN +S +D +++ S
Sbjct: 433 KLRNQLQTYLKTKRCKEKTREPPQTQLHNEEAGDYLNHHISFGSYNIEDGEVENGNEEGS 492
Query: 312 VGSDSHCFELN 322
SD H ELN
Sbjct: 493 GESDLHSIELN 503
>UniRef100_O82344 Hypothetical protein At2g46250 [Arabidopsis thaliana]
Length = 468
Score = 183 bits (465), Expect = 1e-44
Identities = 124/365 (33%), Positives = 193/365 (51%), Gaps = 47/365 (12%)
Query: 9 RNNHALQPLSPASYGSS---MEMTPYNPAATPNSSLDFKGRIGEPHYSLKTSTELLKVLN 65
RN LQP+SPAS SS +++ PA + S K Y L +ST+LLKVLN
Sbjct: 115 RNGDLLQPISPASCSSSSSSLQVVVRKPAFSQTGSSAVKSA----SYGLGSSTKLLKVLN 170
Query: 66 RIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKS 125
RIWSLEEQ+++N+SL++ALK ELD R ++KE+ + ++ D+ +RK
Sbjct: 171 RIWSLEEQNTANMSLVRALKMELDECRAEIKEVQQRKKLS-------------DRPLRKK 217
Query: 126 KEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRK 185
KE++ + +S++ EL+DERK+RK SE++HRKL REL E K + ALKDLE+E R
Sbjct: 218 KEEEEVKDVFRSIKRELDDERKVRKESETLHRKLTRELCEAKHCLSKALKDLEKETQERV 277
Query: 186 LLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAA 245
++E+LCDEFA+ + +YE +V + +KS D++++ I+E W D+R+QM+LE
Sbjct: 278 VVENLCDEFAKAVKDYEDKVRRIGKKSPVS-------DKVIVQIAEVWSDQRLQMKLEED 330
Query: 246 QNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGD 305
F+ ++AK+ SRS ++ R +SV ++ V +
Sbjct: 331 DKTFL----------------LRAKEKSRSQSSKGSGLRAKPDDSVSMHQRVCLKEL--- 371
Query: 306 DDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKS-VPGEGFKNHI 364
++ + + +L K S + DK E+S NV +S F +
Sbjct: 372 EEGLEKRTRRDNKLQLKKSSGQVLNLSMSSEGDKIHPESSPRNVDDHESQEKSRTFSKNH 431
Query: 365 PSNSN 369
P N N
Sbjct: 432 PRNVN 436
>UniRef100_Q7XVC5 OSJNBa0072D21.15 protein [Oryza sativa]
Length = 601
Score = 159 bits (401), Expect = 3e-37
Identities = 91/278 (32%), Positives = 166/278 (58%), Gaps = 9/278 (3%)
Query: 51 HYSLKTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVD 110
H SL S EL++VL IW E + S SLI AL++E+D R V++L+++++++ ++
Sbjct: 223 HNSLIASKELVRVLAHIWGPGELNPSTTSLISALRSEIDLARSHVRKLIKEQKSEG--IE 280
Query: 111 DLMKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSF 170
L KQ+ ++ KSK+++++ A+Q + EL+ E+K R+R+E I++KL L+ ++S
Sbjct: 281 SLKKQLVQEMESWKSKQKEKVANALQYIVSELDSEKKSRRRAERINKKLGMALANTEASL 340
Query: 171 TSALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHIS 230
+A K+LE+ER + +E +C E RGI E + EV LK+++EK + +R +L ++
Sbjct: 341 QAATKELERERKSKGRVEKICTELIRGIGEDKAEVEALKKETEKA-QEELQKEREMLQLA 399
Query: 231 ESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVI-----RDRR 285
+ W ++R+QM+L A+ F +KN+ +++L E++ ++ K+ + +Q+ + R
Sbjct: 400 DEWREQRVQMKLLEARLQFEEKNAAINQLHDELQAYLDTKKEHGQSNDQMTLLRASENGR 459
Query: 286 NSLESVPLNDAVSAPQAMGDDDDDD-SVGSDSHCFELN 322
+++ N + DDDDD S GSD H ELN
Sbjct: 460 EIADNIQKNSGECDDEDEDDDDDDSASEGSDMHSIELN 497
>UniRef100_Q6L4U8 Hypothetical protein P0010D04.11 [Oryza sativa]
Length = 677
Score = 132 bits (333), Expect = 3e-29
Identities = 126/473 (26%), Positives = 212/473 (44%), Gaps = 53/473 (11%)
Query: 81 IKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSVRD 140
I ALK EL + ++ EL + ++ + ++D L++ +AE+K +SKE D++ + +V++
Sbjct: 207 ISALKAELLQAHNRIHELEAESRSAKKKLDHLVRNLAEEKASWRSKENDKVRNILDAVKE 266
Query: 141 ELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARGINE 200
EL ERK R+R+E ++ KL ELSE+KS+ L+D E+ER R+L+E++CDE A+ I E
Sbjct: 267 ELNRERKNRQRAEIMNSKLVSELSELKSAAKRYLQDYEKERKARELMEEVCDELAKEIAE 326
Query: 201 YEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVDKLS 260
+ EV LK +S K + ++ +L ++E W +ER+QM+L A+ K S + KL
Sbjct: 327 DKAEVEALKSESMK-MRDEVEEEKKMLQMAEVWREERVQMKLVDAKLTLDSKYSQLSKLQ 385
Query: 261 LEIETFIKAKQNSRSAENQVIRDRRNSLESV-----------------PLND--AVSAPQ 301
++E+F+ Q + + +RD E++ P D AV
Sbjct: 386 SDLESFLSFHQGN-GVNKEALRDGERLREAICSMKFHDIKEFSYKPPPPSEDIFAVFEEL 444
Query: 302 AMGDDDDDDSVGSDSHCFELNKPSHMGAKVH-----KEEILDKNLDETSKSNVKKKKSVP 356
DD ++ +G + P K+H + L+K L++ S + +
Sbjct: 445 RERDDANEKEIGQCNG----GTPKRHATKIHTVSPETDIFLEKPLNKYSNQLCDRNEEED 500
Query: 357 GEGFKNHIPSNSNKKSQSVDAEEGLTT-----NIRLVEGTRISEEPEHFEISE-----SG 406
G++ + S S D E N V GT + + EISE +
Sbjct: 501 DSGWETVSHVDEQGSSNSPDGSEPSVNGFCGGNDASVSGTDWDDNRSNSEISEVCSTTAE 560
Query: 407 GFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQAS 466
+ +K + S N +LL +G+ + N S N +
Sbjct: 561 KYRKKGSSFGRLWRSSNGDGHKKTGSELL---NGRLSSGRMSNAALSPSLKNGEVCTVSP 617
Query: 467 PVRQWMASLDISEASKVHSG--------SKDNT--LKAKLLEARSKGQRSRLR 509
V +W L + G K NT LK+KLLEA+ G + +LR
Sbjct: 618 SVGEWSPDLLNPHVVRAMKGCIEWPRGAQKQNTHNLKSKLLEAKLDGHKVQLR 670
>UniRef100_Q9LPT5 F11F12.2 protein [Arabidopsis thaliana]
Length = 725
Score = 130 bits (326), Expect = 2e-28
Identities = 113/516 (21%), Positives = 242/516 (46%), Gaps = 70/516 (13%)
Query: 54 LKTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLM 113
L T E+ ++ + + +++Q ++ +SL+ +L+ EL+ ++++L ++++ + +++ +
Sbjct: 213 LDTMEEVHQIYSNMKRIDQQVNA-VSLVSSLEAELEEAHARIEDLESEKRSHKKKLEQFL 271
Query: 114 KQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSA 173
++++E++ +S+E +++ A + ++ ++ E+K R+R E ++ KL EL++ K +
Sbjct: 272 RKVSEERAAWRSREHEKVRAIIDDMKTDMNREKKTRQRLEIVNHKLVNELADSKLAVKRY 331
Query: 174 LKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESW 233
++D E+ER R+L+E++CDE A+ I E + E+ LK++S + D +R +L ++E W
Sbjct: 332 MQDYEKERKARELIEEVCDELAKEIGEDKAEIEALKRES-MSLREEVDDERRMLQMAEVW 390
Query: 234 LDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSA----ENQVIRDRRNSLE 289
+ER+QM+L A+ ++ S ++KL ++E+F++++ E +++R+ S+
Sbjct: 391 REERVQMKLIDAKVALEERYSQMNKLVGDLESFLRSRDIVTDVKEVREAELLRETAASVN 450
Query: 290 S--------VPLND----AVSAPQAMGDDDDDD----------SVGSDSHCFELN----- 322
VP N AV +G+ D + S S H L+
Sbjct: 451 IQEIKEFTYVPANPDDIYAVFEEMNLGEAHDREMEKSVAYSPISHDSKVHTVSLDANMMN 510
Query: 323 -KPSHMGAKVHKEEILDKNLD--------ETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQ 373
K H A H+ ++++ E S+ S+P KNH +SN S
Sbjct: 511 KKGRHSDAYTHQNGDIEEDDSGWETVSHLEEQGSSYSPDGSIPSVNNKNHNHRHSNASSG 570
Query: 374 SVDA-----EEGLTTNIRLVEGTRISEEPEHFEISESGGFERKNNLSELHSTSKNHIIDN 428
++ ++ +T + E I ++S R S S +I
Sbjct: 571 GTESLGKVWDDTMTPTTEISEVCSIPRRSSK-KVSSIAKLWRSTGASNGDRDSNYKVIS- 628
Query: 429 LIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASP----VRQWMASLDISEASKVH 484
+ +G + ++ G S + SP V QW +S + + V+
Sbjct: 629 ------MEGMNGGRVSNGRKSSAGMVSPDRVSSKGGFSPMMDLVGQWNSSPESANHPHVN 682
Query: 485 SG-----------SKDNTLKAKLLEARSKGQRSRLR 509
G ++ ++LK+KL+EAR + Q+ +L+
Sbjct: 683 RGGMKGCIEWPRGAQKSSLKSKLIEARIESQKVQLK 718
>UniRef100_Q9LTQ8 Gb|AAF00675.1 [Arabidopsis thaliana]
Length = 495
Score = 127 bits (319), Expect = 1e-27
Identities = 113/481 (23%), Positives = 219/481 (45%), Gaps = 66/481 (13%)
Query: 73 QHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLH 132
Q +++SL +++ +L R +K+L ++++ + +++ +K+++E++ +S+E +++
Sbjct: 30 QQVNDVSLASSIELKLQEARACIKDLESEKRSQKKKLEQFLKKVSEERAAWRSREHEKVR 89
Query: 133 AAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCD 192
A + ++ ++ E+K R+R E ++ KL EL++ K + + D +QER R+L+E++CD
Sbjct: 90 AIIDDMKADMNQEKKTRQRLEIVNSKLVNELADSKLAVKRYMHDYQQERKARELIEEVCD 149
Query: 193 EFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDK 252
E A+ I E + E+ LK +S + + D +R +L ++E W +ER+QM+L A+ +K
Sbjct: 150 ELAKEIEEDKAEIEALKSES-MNLREEVDDERRMLQMAEVWREERVQMKLIDAKVTLEEK 208
Query: 253 NSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESV----PLNDAVSAP-------- 300
S ++KL ++E F+ ++ + E +V R + SV + + P
Sbjct: 209 YSQMNKLVGDMEAFLSSRNTTGVKEVRVAELLRETAASVDNIQEIKEFTYEPAKPDDILM 268
Query: 301 ----QAMGDDDDDDSV----------GSDSHCFE-----LNKPSHMGAKVHKE---EILD 338
MG++ D +S S +H +NK H A + E D
Sbjct: 269 LFEQMNMGENQDRESEQYVAYSPVSHASKAHTVSPDVNLINKGRHSNAFTDQNGEFEEDD 328
Query: 339 KNLDETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPE 398
+ S S P E N ++ + S++ E T +R
Sbjct: 329 SGWETVSHSEEHGSSYSPDESIPNISNTHHRNSNVSMNGTEYEKTLLR------------ 376
Query: 399 HFEISESGGFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSN 458
EI E R+ + +L S +K + G++ N + + + SN
Sbjct: 377 --EIKEVCSVPRRQS-KKLPSMAKLWSSLEGMNGRV------SNARKSTVEMVSPETGSN 427
Query: 459 AGWRNQASPVRQWMASLDISEASKVHSGSK----------DNTLKAKLLEARSKGQRSRL 508
G N V QW +S D + A+ G K N+LK KL+EA+ + Q+ +L
Sbjct: 428 KGGFNTLDLVGQWSSSPDSANANLNRGGRKGCIEWPRGAHKNSLKTKLIEAQIESQKVQL 487
Query: 509 R 509
+
Sbjct: 488 K 488
>UniRef100_Q6YZ90 Hypothetical protein B1099H05.52 [Oryza sativa]
Length = 675
Score = 124 bits (311), Expect = 9e-27
Identities = 119/495 (24%), Positives = 222/495 (44%), Gaps = 64/495 (12%)
Query: 70 LEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQD 129
+EEQ N + L+ EL +TR +V +L +R + + ++D L K++ E+K + +E
Sbjct: 195 IEEQKGENY--VSNLQVELQQTRDRVGKLEAERISAKKQLDHLFKKLTEEKAAWRKREHK 252
Query: 130 RLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLED 189
++ A ++ ++ +LE E+K R++ E I+ KL EL E+K + + L++ + ER R+L E+
Sbjct: 253 KVQAILEDMKADLEHEKKNRRQLEKINIKLVDELKEVKMAANNLLQEYDNERKTRELTEE 312
Query: 190 LCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGF 249
+C + R + E++ E+ LKQ S K + D DR +L ++E W +ER+QM+L A+
Sbjct: 313 VCTKLVRELEEHKAEIEGLKQDSLKLRAE-VDEDRKLLQMAEVWREERVQMKLVDAKLTL 371
Query: 250 MDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESV---------------PLN 294
K + KL L++E I A + ++ +++ +N L+S+ P +
Sbjct: 372 EAKYEELSKLQLDVEAII-ASFSDTKGDDTIVQTAKNILQSIESTREQEIKFTYEPPPAS 430
Query: 295 DAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNL------------- 341
D + A ++ C + N P H + D L
Sbjct: 431 DDILAIIEELRPSEELETRETEPCHKHNSPVHESENQQDSPMTDIFLENPTKLYSNRSHY 490
Query: 342 ------DETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSV------DAEEGLTTNI--RL 387
D +S + ++ +N + NK + D+E G N+ L
Sbjct: 491 NESDMGDSSSWETISNEEMQGSSSSRNGSEPSVNKICDKISWTSGDDSEAGQNDNLSGEL 550
Query: 388 VEGTRISEEPEHFEISESGGFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNKHAE 447
+ +P + S F + + L K +++ + R ++E N H+
Sbjct: 551 SKAYFADRKPSKKKESAISKFWKSSPLKNC-EIFKIDVVEMMNRRS--SNERLSNGMHSS 607
Query: 448 HNNYGEASCSNAGWRNQASPVRQWMASLDISEASKVHSGSK-------DNTLKAKLLEAR 500
+ E + +AG + + + QW S S S+++ G + +LKAKLLEAR
Sbjct: 608 N----EGANQDAGLSSPS--IGQW--SSPDSMNSQLNRGFRGCMELVQKQSLKAKLLEAR 659
Query: 501 SKGQRSRLRTLKGPK 515
+ Q+ +LR + K
Sbjct: 660 MESQKIQLRHVLNQK 674
>UniRef100_Q7XPQ1 OSJNBa0053K19.22 protein [Oryza sativa]
Length = 696
Score = 124 bits (311), Expect = 9e-27
Identities = 118/486 (24%), Positives = 227/486 (46%), Gaps = 54/486 (11%)
Query: 70 LEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQD 129
L EQ + + + L+ EL + R +V EL +R+A + ++D L K++AE+K +S+E +
Sbjct: 212 LNEQQDA--TYVANLQMELQQARDRVSELETERRAAKKKLDHLFKKLAEEKAAWRSREHE 269
Query: 130 RLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLED 189
++ A ++ ++ +L+ E+K R+R E I+ KL EL E K S L++ + ER R+L E+
Sbjct: 270 KVRAILEDMKADLDHEKKNRRRLEMINLKLVNELKEAKMSAKQLLQEYDNERKARELTEE 329
Query: 190 LCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGF 249
+C+E AR + E + E+ LK S K + D +R +L ++E W +ER+QM+L A+
Sbjct: 330 VCNELAREVEEDKAEIEALKHDSLK-LREEVDEERKMLQMAEVWREERVQMKLVDAKLTL 388
Query: 250 MDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVS-----APQAMG 304
K + + KL ++E FI A +R + V+ + N ++++ A P A
Sbjct: 389 DAKYTQLSKLQQDVEAFIAACSCAR-GDIMVVEEAENIIQAIKSVRAQDIEFRYEPPAQS 447
Query: 305 DD----------DDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDE-----TSKSNV 349
+D ++ + C++ N + D L++ +SKS
Sbjct: 448 EDIFSIFEELRPSEEPVIKEIEPCYKNNSAMCESEIQEASPMTDIFLEKPTKVYSSKSPQ 507
Query: 350 KKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPEHFEISESGGFE 409
+ + G ++ + S S D E ++ +G+ FE E+ +
Sbjct: 508 NESDTEDGSSWETISHEDMQASSGSPDGSEPSVN--KICDGSISWTSRNDFEYKEAE--K 563
Query: 410 RKNNLSELHSTSKNH-IIDNLIRGQLLASESGKNNKHAEHN--------------NYGEA 454
K++ ++++ T+ N +L S KNN + + + G
Sbjct: 564 LKDDSTDIYLTNMNQPKKKESALSKLWKSSRPKNNDVCKKDAVETINGRSSNVRLSVGTH 623
Query: 455 SCSNAGWRN---QASPVRQWMASLDIS-EASKVHSG-------SKDNTLKAKLLEARSKG 503
S ++G + + + QW + +S + ++ G S+ ++LK+KL+EAR +
Sbjct: 624 STIDSGIQEIGLSSPSIGQWSSPDSMSMQFNRGFKGCMEYPRTSQKHSLKSKLMEARMES 683
Query: 504 QRSRLR 509
Q+ +LR
Sbjct: 684 QKVQLR 689
>UniRef100_Q6K6L0 Hypothetical protein P0047E05.7 [Oryza sativa]
Length = 403
Score = 122 bits (305), Expect = 5e-26
Identities = 96/364 (26%), Positives = 177/364 (48%), Gaps = 38/364 (10%)
Query: 53 SLKTSTELLKVLNRIWSLEE---QHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEV 109
SL S EL+K L IW + S+ SL+ AL+ ELD R + L ++ + E
Sbjct: 11 SLAASKELVKALAGIWGPGDGALNPSTASSLLSALRAELDLARAHARRLAKEDRRRGDEA 70
Query: 110 DDLMKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSS 169
++AED +++++ AAV+ EL+ ER+ R+R+E ++ KL R L++ +
Sbjct: 71 ARARARLAEDAREWGRRQREKAAAAVRVAAAELDGERRSRRRAERVNAKLGRALADAERE 130
Query: 170 FTSALKDLEQERTRRKLLEDLCDEFAR--------GINEYEQEVHTLKQKSEKDWVQRAD 221
++ ++LE+ER R+ LE +CDE R G E+EV +++++E+ + +
Sbjct: 131 LAASRRELERERRSRERLEKVCDELVRGGLVAAGGGGRGGEEEVEEMRREAERA-QEELE 189
Query: 222 HDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVI 281
+R +L +++ +ER+QM+L A+ F +KN++V++L E+E F+ +K++ + E
Sbjct: 190 KEREMLRLADELREERVQMKLLEARLQFEEKNAVVEQLRDELEAFLGSKKDRQQQEEPPP 249
Query: 282 RD----RRNSLESVPLNDAVSAPQAMGDDDDD------------------DSVGSDSHCF 319
D RR + + A GD +DD DS GS+ H
Sbjct: 250 PDADDHRRRRPDGHQFQSILVAVNKNGDHEDDNDGDEEDDGGGRGECVAEDSDGSEMHSI 309
Query: 320 ELNKPSHMGAKVHKEEILDKNLDETSKSN---VKKKKSVPGEGFKNHIPSNS-NKKSQSV 375
ELN + K++ T++S ++S G G ++ +++S+ +
Sbjct: 310 ELNVDGNSKDYSWSYTTASKDMTTTARSKNAAAIDRRSQEGAGEEDRWDDGGCSERSKDL 369
Query: 376 DAEE 379
D E+
Sbjct: 370 DEED 373
>UniRef100_Q9FMS1 Gb|AAF00675.1 [Arabidopsis thaliana]
Length = 450
Score = 117 bits (293), Expect = 1e-24
Identities = 89/314 (28%), Positives = 154/314 (48%), Gaps = 62/314 (19%)
Query: 24 SSMEMTPYNPAATPNSSLDFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQH-SSNISLIK 82
SS++ P + S + K R L TS EL+KVL RI L + H +++ LI
Sbjct: 125 SSIDFPPRSSDPISRLSSEVKTRFKNVSDGLTTSKELVKVLKRIGELGDDHKTASNRLIS 184
Query: 83 ALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSVRDEL 142
AL ELDR R +K L+ E+D+ E++ ++S+++E
Sbjct: 185 ALLCELDRARSSLKHLMS-------ELDE---------------EEEEKRRLIESLQEEA 222
Query: 143 EDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARGINEYE 202
ERKLR+R+E ++R+L REL+E K + +++++E+ + +LE++CDE +GI + +
Sbjct: 223 MVERKLRRRTEKMNRRLGRELTEAKETERKMKEEMKREKRAKDVLEEVCDELTKGIGDDK 282
Query: 203 QEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVDKLSLE 262
+E+ + +R ++HI++ +ER+QM+L A+ F DK + V++L E
Sbjct: 283 KEM---------------EKEREMMHIADVLREERVQMKLTEAKFEFEDKYAAVERLKKE 327
Query: 263 IETFIKAKQNSRSAENQVI----------RDRRNSLESVPLN--------------DAVS 298
+ + ++ S+E + I D + L+S+ LN D
Sbjct: 328 LRRVLDGEEGKGSSEIRRILEVIDGSGSDDDEESDLKSIELNMESGSKWGYVDSLKDRRR 387
Query: 299 APQAMGDDDDDDSV 312
+ GDDDDDD V
Sbjct: 388 FDGSGGDDDDDDPV 401
>UniRef100_Q93ZM2 AT5g22310/MWD9_9 [Arabidopsis thaliana]
Length = 481
Score = 113 bits (282), Expect = 2e-23
Identities = 83/284 (29%), Positives = 144/284 (50%), Gaps = 62/284 (21%)
Query: 54 LKTSTELLKVLNRIWSLEEQH-SSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDL 112
L TS EL+KVL RI L + H +++ LI AL ELDR R +K L+ E+D+
Sbjct: 186 LTTSKELVKVLKRIGELGDDHKTASNRLISALLCELDRARSSLKHLMS-------ELDE- 237
Query: 113 MKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTS 172
E++ ++S+++E ERKLR+R+E ++R+L REL+E K +
Sbjct: 238 --------------EEEEKRRLIESLQEEAMVERKLRRRTEKMNRRLGRELTEAKETERK 283
Query: 173 ALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISES 232
+++++E+ + +LE++CDE +GI + ++E+ + +R ++HI++
Sbjct: 284 MKEEMKREKRAKDVLEEVCDELTKGIGDDKKEM---------------EKEREMMHIADV 328
Query: 233 WLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVI----------R 282
+ER+QM+L A+ F DK + V++L E+ + ++ S+E + I
Sbjct: 329 LREERVQMKLTEAKFEFEDKYAAVERLKKELRRVLDGEEGKGSSEIRRILEVIDGSGSDD 388
Query: 283 DRRNSLESVPLN--------------DAVSAPQAMGDDDDDDSV 312
D + L+S+ LN D + GDDDDDD V
Sbjct: 389 DEESDLKSIELNMESGSKWGYVDSLKDRRRFDGSGGDDDDDDPV 432
>UniRef100_Q84P68 Hypothetical protein [Oryza sativa]
Length = 179
Score = 89.7 bits (221), Expect = 2e-16
Identities = 51/164 (31%), Positives = 94/164 (57%), Gaps = 2/164 (1%)
Query: 126 KEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRK 185
+E +++ A ++ ++ +L+ E+K R+R E I+ K EL E + S L++ + ER R+
Sbjct: 18 REHEKVRAILEDIKADLDHEKKNRRRPEMINLKHVNELKEAQMSAKQLLQEYDNERKARE 77
Query: 186 LLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAA 245
L E++C+E AR + E + E+ LK S K + D +R +L ++E W +ER+QM+L A
Sbjct: 78 LTEEVCNELAREVEEDKAEIEALKHDSLK-LREEVDEERKMLQMAEVWREERVQMKLVDA 136
Query: 246 QNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLE 289
+ K + + KL ++E FI A +R + V+ + N ++
Sbjct: 137 KLTLDAKYTQLSKLQQDVEAFIAACSCAR-GDIMVVEEAENIIQ 179
>UniRef100_Q5Z9M3 Intracellular protein transport protein USO1-like [Oryza sativa]
Length = 294
Score = 84.3 bits (207), Expect = 1e-14
Identities = 81/295 (27%), Positives = 137/295 (45%), Gaps = 36/295 (12%)
Query: 238 MQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSR--------SAENQVIRDRRNSLE 289
MQMQ + ++ SI ++LS EI +F+ +++S+ S E Q R SLE
Sbjct: 1 MQMQNADPKATLAERISITERLSSEIHSFLNTRRSSKPKDDKLYISNEKQDASLCRQSLE 60
Query: 290 SVPLNDAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETS---- 345
SV L+ A SAP+ + +DD+D+SV SD HCFEL+ H + +++ TS
Sbjct: 61 SVHLHGATSAPR-LAEDDNDNSVASDLHCFELSMHGHT---IQNNDLIGTRQRVTSCMYS 116
Query: 346 -KSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPEHFEISE 404
++ +P EG + S + K ++ G+ + + PE ++
Sbjct: 117 PMRRLEFSNGIPVEGSRISTMSPCSMKDKA--RPNGIREQLN-------ASTPEISPCND 167
Query: 405 SGGFERKNNLSELHSTSKNHIIDNLIRGQLLASESG-----KNNKHAEHNNYGEASCSNA 459
+ R + + + D+L++ + A + N+ H+ + + ++
Sbjct: 168 AKNAPRCAQDETVMTQVSQRLHDDLLKIKSEAPQHAYLGQKSNDHHSRAGQFRDQCTTSG 227
Query: 460 GWRNQASPVRQW--MASLD--ISEASKVHS-GSKDNTLKAKLLEARSKGQRSRLR 509
+ SP RQ +SLD I+EAS H K TLKAKLL+AR +GQ +R+R
Sbjct: 228 NVYDLCSPARQLNQRSSLDHEITEASPTHPLEGKSTTLKAKLLQARLEGQHARMR 282
>UniRef100_Q8H382 Hypothetical protein OSJNBa0061L20.108 [Oryza sativa]
Length = 291
Score = 84.0 bits (206), Expect = 1e-14
Identities = 47/146 (32%), Positives = 85/146 (58%), Gaps = 1/146 (0%)
Query: 53 SLKTSTELLKVLNRIWSLEEQ-HSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDD 111
SL TS EL+K L IW ++ + S SL+ AL ELD R V+ L + + E
Sbjct: 62 SLATSKELVKALVGIWGPDDGLNPSTASLLSALCAELDLARAHVRHLATEDRRHGDETAR 121
Query: 112 LMKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFT 171
+ Q+ E+ +S++++++ A V+ EL+ E++ R+R+E ++ KL + L++ +
Sbjct: 122 MRTQLVEEAREWRSQQREKVAAMVRVAAAELDGEQRSRRRAERVNAKLGKALADAERELA 181
Query: 172 SALKDLEQERTRRKLLEDLCDEFARG 197
++ ++LE+ER R+ LE +CD+ RG
Sbjct: 182 ASRRELERERRSRERLEKVCDKLVRG 207
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,045,250,611
Number of Sequences: 2790947
Number of extensions: 43134598
Number of successful extensions: 255748
Number of sequences better than 10.0: 6774
Number of HSP's better than 10.0 without gapping: 386
Number of HSP's successfully gapped in prelim test: 6699
Number of HSP's that attempted gapping in prelim test: 230735
Number of HSP's gapped (non-prelim): 20950
length of query: 676
length of database: 848,049,833
effective HSP length: 134
effective length of query: 542
effective length of database: 474,062,935
effective search space: 256942110770
effective search space used: 256942110770
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC140913.13


