

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.1 - phase: 0 /pseudo
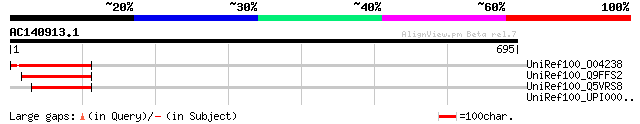
(695 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O04238 Transcription factor [Vicia faba] 169 3e-40
UniRef100_Q9FFS2 Transcription factor-like protein [Arabidopsis ... 81 9e-14
UniRef100_Q5VRS8 Putative transcription factor [Oryza sativa] 62 8e-08
UniRef100_UPI000030C5A4 UPI000030C5A4 UniRef100 entry 38 0.89
>UniRef100_O04238 Transcription factor [Vicia faba]
Length = 828
Score = 169 bits (428), Expect = 3e-40
Identities = 89/112 (79%), Positives = 95/112 (84%), Gaps = 2/112 (1%)
Query: 1 METNTSSPLSTLPESGAMTATANPVSPSLVNLYRITKVLERLATHFVPGNRSDAFEFFNL 60
ME+NTSS + P+ A T NPVSPSLVNLYRITKVL+RLA HF PGNRSD+FEFFNL
Sbjct: 1 MESNTSSSMP--PDLAATNGTTNPVSPSLVNLYRITKVLDRLAFHFQPGNRSDSFEFFNL 58
Query: 61 CLSLSRGIDYALANGEVPLKANELPILMKQMYQRKTDDHSQAAVMVLMISVK 112
CLSLSRGIDYALANGE P KANELP LMKQMYQRKTD+ S AAVMVLMISVK
Sbjct: 59 CLSLSRGIDYALANGEPPPKANELPTLMKQMYQRKTDELSLAAVMVLMISVK 110
>UniRef100_Q9FFS2 Transcription factor-like protein [Arabidopsis thaliana]
Length = 719
Score = 81.3 bits (199), Expect = 9e-14
Identities = 41/96 (42%), Positives = 60/96 (61%)
Query: 17 AMTATANPVSPSLVNLYRITKVLERLATHFVPGNRSDAFEFFNLCLSLSRGIDYALANGE 76
A T + SLVN +R+ V +RL H G + D EF C+S ++GID+A+AN +
Sbjct: 11 AGTGLREKTAASLVNSFRLASVTQRLRYHIQDGAKVDPKEFQICCISFAKGIDFAIANND 70
Query: 77 VPLKANELPILMKQMYQRKTDDHSQAAVMVLMISVK 112
+P K E P L+KQ+ + TD +++ A+MVLMISVK
Sbjct: 71 IPKKVEEFPWLLKQLCRHGTDVYTKTALMVLMISVK 106
>UniRef100_Q5VRS8 Putative transcription factor [Oryza sativa]
Length = 872
Score = 61.6 bits (148), Expect = 8e-08
Identities = 33/85 (38%), Positives = 55/85 (63%), Gaps = 2/85 (2%)
Query: 30 VNLYRITKVLERLATHFVPGNRS--DAFEFFNLCLSLSRGIDYALANGEVPLKANELPIL 87
+N R+ + +RL THF G + + + +L + +RGID+AL++G+VP A+E+P +
Sbjct: 34 MNARRLVMIGDRLRTHFRGGGGTVLEPPDLAHLVYAFARGIDFALSSGDVPTVASEIPSI 93
Query: 88 MKQMYQRKTDDHSQAAVMVLMISVK 112
+K++Y D Q++VMVLMIS K
Sbjct: 94 LKKVYLVGKDQFLQSSVMVLMISCK 118
>UniRef100_UPI000030C5A4 UPI000030C5A4 UniRef100 entry
Length = 255
Score = 38.1 bits (87), Expect = 0.89
Identities = 22/72 (30%), Positives = 43/72 (59%), Gaps = 3/72 (4%)
Query: 232 FIQGPTFEWILDHRCPPV*LVSLNLGQIFFKLWVSSMGIILYLLPI*VLDPCLRIQFFHQ 291
FI +++IL+ + + ++SL+L I LW + GI+ +L+ I ++ +I F H+
Sbjct: 113 FISLIIYDYILEKKFTDLFIISLSLNLI---LWTKAEGIVYFLIIISCINLIKKINFSHR 169
Query: 292 IMFSLLYLLMQI 303
I+F++L L + I
Sbjct: 170 IIFNILCLSLII 181
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.357 0.160 0.557
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 949,168,530
Number of Sequences: 2790947
Number of extensions: 33250341
Number of successful extensions: 142958
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 142953
Number of HSP's gapped (non-prelim): 5
length of query: 695
length of database: 848,049,833
effective HSP length: 134
effective length of query: 561
effective length of database: 474,062,935
effective search space: 265949306535
effective search space used: 265949306535
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 78 (34.7 bits)
Medicago: description of AC140913.1


