

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140848.2 + phase: 0 /pseudo
(573 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
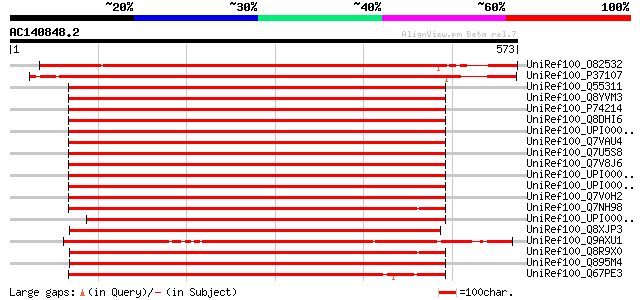
Sequences producing significant alignments: (bits) Value
UniRef100_O82532 Signal recognition particle 54 kDa subunit [Pis... 801 0.0
UniRef100_P37107 Signal recognition particle 54 kDa protein, chl... 760 0.0
UniRef100_Q55311 Signal sequence binding protein [Synechococcus ... 476 e-133
UniRef100_Q8YVM3 Signal recognition particle protein [Anabaena sp.] 474 e-132
UniRef100_P74214 Signal recognition particle protein [Synechocys... 473 e-132
UniRef100_Q8DHI6 Signal recognition particle protein [Synechococ... 470 e-131
UniRef100_UPI0000286CB5 UPI0000286CB5 UniRef100 entry 451 e-125
UniRef100_Q7VAU4 Signal recognition particle GTPase [Prochloroco... 447 e-124
UniRef100_Q7U5S8 Signal recognition particle protein [Synechococ... 445 e-123
UniRef100_Q7V8J6 Signal recognition particle protein [Prochloroc... 439 e-121
UniRef100_UPI0000305F06 UPI0000305F06 UniRef100 entry 435 e-120
UniRef100_UPI000031BAD7 UPI000031BAD7 UniRef100 entry 434 e-120
UniRef100_Q7V0H2 Signal recognition particle protein [Prochloroc... 430 e-119
UniRef100_Q7NH98 Signal recognition particle protein SRP54 [Gloe... 429 e-119
UniRef100_UPI0000330C1A UPI0000330C1A UniRef100 entry 410 e-113
UniRef100_Q8XJP3 Signal recognition particle protein [Clostridiu... 409 e-113
UniRef100_Q9AXU1 Chloroplast SRP54 [Chlamydomonas reinhardtii] 409 e-112
UniRef100_Q8R9X0 Signal recognition particle GTPase [Thermoanaer... 409 e-112
UniRef100_Q895M4 Signal recognition particle, subunit ffh/srp54 ... 407 e-112
UniRef100_Q67PE3 Signal recognition particle [Symbiobacterium th... 400 e-110
>UniRef100_O82532 Signal recognition particle 54 kDa subunit [Pisum sativum]
Length = 525
Score = 801 bits (2069), Expect = 0.0
Identities = 439/550 (79%), Positives = 465/550 (83%), Gaps = 40/550 (7%)
Query: 34 RNSFAKEIWGFVQSKSVTMRRMDMIRPVVVRAEMFGQLTTGLESAWNKLKGEEVLTKDNI 93
RNSFA+EIWG VQSKS+T RR DMIRPVVVRAEMFGQLT+GLESAWNKLK EEVLTK+NI
Sbjct: 5 RNSFAREIWGLVQSKSLTSRRTDMIRPVVVRAEMFGQLTSGLESAWNKLKREEVLTKENI 64
Query: 94 VEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVKIVHDELVQLMG 153
VEPMRDIRRA ADVSLPVVRRFVQSVTD+AVGVGVTRGVKPDQQLVKIVHDELVQLMG
Sbjct: 65 VEPMRDIRRA--RADVSLPVVRRFVQSVTDKAVGVGVTRGVKPDQQLVKIVHDELVQLMG 122
Query: 154 GEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGDVYRPAAIDQL 213
GEVSELTFA +GPTVILLAGLQGVGKTTVCAKLA YFKKQGKSCMLVAGDVYRPAAID L
Sbjct: 123 GEVSELTFANSGPTVILLAGLQGVGKTTVCAKLATYFKKQGKSCMLVAGDVYRPAAIDXL 182
Query: 214 TILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRLQIDKAMMDELKDV 273
TILGKQVDVPVY AGTDVKPS IAKQGFEEAKKKKIDVVIVDTAG LQIDKAMMDELK+V
Sbjct: 183 TILGKQVDVPVYAAGTDVKPSVIAKQGFEEAKKKKIDVVIVDTAGSLQIDKAMMDELKEV 242
Query: 274 KRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGK 333
K+VLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGK
Sbjct: 243 KQVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALSVKEVSGK 302
Query: 334 PIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEDLQKKIMSAKF 393
PIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVM +EDAED+QKKIMSAKF
Sbjct: 303 PIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMLEEDAEDMQKKIMSAKF 362
Query: 394 DFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTPAQIREAERNLEIMEVIIKAMTPEEREK 453
DFND LKQTR VA+MGSVSRV+GMIPGMAKVTPAQIREAE+NL+ MEVII+AMTPEER K
Sbjct: 363 DFNDLLKQTRTVAKMGSVSRVLGMIPGMAKVTPAQIREAEKNLQFMEVIIEAMTPEERGK 422
Query: 454 PELLAESPVRRKRVAQDSGKTEQQTDAILS-------NFEKYTTIMSGKPTCSSTISNAC 506
PELLAE PVRRK VAQ+SGKTEQQ +++ +K +M G T+SN
Sbjct: 423 PELLAEFPVRRKSVAQESGKTEQQVSQLVAQLFQMRVRMKKLMGVMEG--GSMPTLSNL- 479
Query: 507 SDEEPNGCNGRWIDANT**S*GST*NRGKVCLYQAPPGTARRRKKAESRRLF--ADSTLR 564
EE N + APPGTARR+KK +RLF S +
Sbjct: 480 --EEALKANKK-----------------------APPGTARRKKKGGLKRLFKGRSSKIS 514
Query: 565 QP-PRGFGSK 573
P PRGFGSK
Sbjct: 515 LPAPRGFGSK 524
>UniRef100_P37107 Signal recognition particle 54 kDa protein, chloroplast precursor
[Arabidopsis thaliana]
Length = 564
Score = 760 bits (1962), Expect = 0.0
Identities = 410/559 (73%), Positives = 457/559 (81%), Gaps = 43/559 (7%)
Query: 23 SSSKLCSSLSYRNSFAKEIWGFVQSKSVTMRRMDMIRPVVVRAEMFGQLTTGLESAWNKL 82
+S+ L SS RN +EIW +V+SK+V R VRAEMFGQLT GLE+AW+KL
Sbjct: 38 NSASLSSS---RNLSTREIWSWVKSKTVVGH--GRYRRSQVRAEMFGQLTGGLEAAWSKL 92
Query: 83 KGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVK 142
KGEEVLTKDNI EPMRDIRRALLEADVSLPVVRRFVQSV+DQAVG+GV RGVKPDQQLVK
Sbjct: 93 KGEEVLTKDNIAEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGMGVIRGVKPDQQLVK 152
Query: 143 IVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAG 202
IVHDELV+LMGGEVSEL FAK+GPTVILLAGLQGVGKTTVCAKLA Y KKQGKSCML+AG
Sbjct: 153 IVHDELVKLMGGEVSELQFAKSGPTVILLAGLQGVGKTTVCAKLACYLKKQGKSCMLIAG 212
Query: 203 DVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRLQI 262
DVYRPAAIDQL ILG+QV VPVYTAGTDVKP+DIAKQG +EAKK +DVVI+DTAGRLQI
Sbjct: 213 DVYRPAAIDQLVILGEQVGVPVYTAGTDVKPADIAKQGLKEAKKNNVDVVIMDTAGRLQI 272
Query: 263 DKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGG 322
DK MMDELKDVK+ LNPTEVLLVVDAMTGQEAAALVTTFN+EIGITGAILTKLDGDSRGG
Sbjct: 273 DKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAAALVTTFNVEIGITGAILTKLDGDSRGG 332
Query: 323 AALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAE 382
AALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKA EVM+QEDAE
Sbjct: 333 AALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKATEVMRQEDAE 392
Query: 383 DLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTPAQIREAERNLEIMEVI 442
DLQKKIMSAKFDFNDFLKQTRAVA+MGS++RV+GMIPGM KV+PAQIREAE+NL +ME +
Sbjct: 393 DLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVLGMIPGMGKVSPAQIREAEKNLLVMEAM 452
Query: 443 IKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFEKYTTIM--------SG 494
I+ MTPEERE+PELLAESP RRKR+A+DSGKTEQQ A+++ + M G
Sbjct: 453 IEVMTPEERERPELLAESPERRKRIAKDSGKTEQQVSALVAQIFQMRVKMKNLMGVMEGG 512
Query: 495 KPTCSSTISNACSDEEPNGCNGRWIDANT**S*GST*NRGKVCLYQAPPGTARRRKKAES 554
S + +A E+ +APPGTARR++KA+S
Sbjct: 513 SIPALSGLEDALKAEQ-----------------------------KAPPGTARRKRKADS 543
Query: 555 RRLFADSTLRQP-PRGFGS 572
R+ F +S +P PRGFGS
Sbjct: 544 RKKFVESASSKPGPRGFGS 562
>UniRef100_Q55311 Signal sequence binding protein [Synechococcus sp.]
Length = 485
Score = 476 bits (1226), Expect = e-133
Identities = 241/427 (56%), Positives = 321/427 (74%), Gaps = 1/427 (0%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF L LE AW KL+G++ +++ N+ E ++++RRALLEADV+L VV+ F+ V ++AV
Sbjct: 1 MFDALAERLEQAWTKLRGQDKISESNVQEALKEVRRALLEADVNLGVVKEFIAQVQEKAV 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V G++PDQQ +K+V+DELVQ+MG L A PTVIL+AGLQG GKTT AKL
Sbjct: 61 GTQVISGIRPDQQFIKVVYDELVQVMGETHVPLAQAAKAPTVILMAGLQGAGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
A + +K+G+S +LVA DVYRPAAIDQL LGKQ+DVPV+ G+D P +IA+QG E+A++
Sbjct: 121 ALHLRKEGRSTLLVATDVYRPAAIDQLITLGKQIDVPVFELGSDANPVEIARQGVEKARQ 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+ ID VIVDTAGRLQID MM+EL VK + P EVLLVVD+M GQEAA+L +F+ IG
Sbjct: 181 EGIDTVIVDTAGRLQIDTEMMEELAQVKEAIAPHEVLLVVDSMIGQEAASLTRSFHERIG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGAILTKLDGDSRGGAALS++ VSG+PIK VG GE++E L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAILTKLDGDSRGGAALSIRRVSGQPIKFVGLGEKVEALQPFHPERMASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGM-AKVT 425
+ VEKAQE + D E +Q+KI++AKFDF DFLKQ R + MGS++ I +IPG+ K+
Sbjct: 301 TLVEKAQEEIDLADVEKMQEKILAAKFDFTDFLKQMRLLKNMGSLAGFIKLIPGLGGKIN 360
Query: 426 PAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNF 485
Q+R+ ER L+ +E +I +MTP+ER P+LL+ SP RR R+A+ G+TE + I+ F
Sbjct: 361 DEQLRQGERQLKRVEAMINSMTPQERRDPDLLSNSPSRRHRIAKGCGQTEAEIRQIIQQF 420
Query: 486 EKYTTIM 492
++ T+M
Sbjct: 421 QQMRTMM 427
>UniRef100_Q8YVM3 Signal recognition particle protein [Anabaena sp.]
Length = 490
Score = 474 bits (1219), Expect = e-132
Identities = 240/426 (56%), Positives = 317/426 (74%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF L LESAW KL+G++ +++ NI + +R++RRALLEADV+L VV+ F+ V +A
Sbjct: 1 MFDALADRLESAWKKLRGQDKISESNIQDALREVRRALLEADVNLQVVKDFISEVEAKAQ 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V GV+PDQQ +KIV+DELVQ+MG E L + PT++L+AGLQG GKTT AKL
Sbjct: 61 GAEVISGVRPDQQFIKIVYDELVQVMGEENVPLAEVEQKPTIVLMAGLQGTGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
A + +K +SC+LVA DVYRPAAIDQL LGKQ+DVPV+ G+D P +IA+QG E AK
Sbjct: 121 ALHLRKLERSCLLVATDVYRPAAIDQLLTLGKQIDVPVFELGSDADPVEIARQGVERAKA 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+ ++ VI+DTAGRLQID+ MM EL +K + P E LLVVD+MTGQEAA L TF+ +IG
Sbjct: 181 EGVNTVIIDTAGRLQIDEDMMAELARIKETVQPHETLLVVDSMTGQEAANLTRTFHEQIG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGAILTK+DGDSRGGAALSV+++SG PIK VG GE++E L+PFYPDRMA RILGMGDVL
Sbjct: 241 ITGAILTKMDGDSRGGAALSVRQISGAPIKFVGVGEKVEALQPFYPDRMASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQE DAE +Q+KI+SAKFDF DF+KQ R + MGS+ ++ MIPGM K++
Sbjct: 301 TLVEKAQEEFDLADAEKMQEKILSAKFDFTDFVKQLRMLKNMGSLGGLLKMIPGMGKLSD 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
Q+++ E L+ E +I +MT +ER P+LLA SP RR+R+A SG E + ++++F+
Sbjct: 361 DQLKQGETQLKRCEAMINSMTMQERRDPDLLASSPNRRRRIASGSGYKEADVNKLVADFQ 420
Query: 487 KYTTIM 492
K ++M
Sbjct: 421 KMRSLM 426
>UniRef100_P74214 Signal recognition particle protein [Synechocystis sp.]
Length = 482
Score = 473 bits (1216), Expect = e-132
Identities = 239/426 (56%), Positives = 316/426 (74%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF L LE AW KL+G++ +++ NI E ++++RRALL ADV+L VV+ F++ V +A+
Sbjct: 1 MFDALADRLEDAWKKLRGQDKISESNIKEALQEVRRALLAADVNLQVVKGFIKDVEQKAL 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V GV P QQ +KIV+DELV LMG L A+ PTVIL+AGLQG GKTT AKL
Sbjct: 61 GADVISGVNPGQQFIKIVYDELVNLMGESNVPLAQAEQAPTVILMAGLQGTGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
A Y +KQ +S ++VA DVYRPAAIDQL LG+Q+DVPV+ G+D P +IA+QG E+AK+
Sbjct: 121 ALYLRKQKRSALMVATDVYRPAAIDQLKTLGQQIDVPVFDLGSDANPVEIARQGVEKAKE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+D V++DTAGRLQID MM EL ++K+V+ P + LLVVDAMTGQEAA L TF+ +IG
Sbjct: 181 LGVDTVLIDTAGRLQIDPQMMAELAEIKQVVKPDDTLLVVDAMTGQEAANLTHTFHEQIG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGAILTKLDGD+RGGAALSV+++SG+PIK VG GE++E L+PFYPDR+A RIL MGDVL
Sbjct: 241 ITGAILTKLDGDTRGGAALSVRQISGQPIKFVGVGEKVEALQPFYPDRLASRILNMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQE + D E LQ KI+ A FDF+DF+KQ R + MGS+ ++ MIPGM K++
Sbjct: 301 TLVEKAQEAIDVGDVEKLQNKILEATFDFDDFIKQMRFMKNMGSLGGLLKMIPGMNKLSS 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
I + E+ L+ E +I +MT EER+ P+LLA+SP RR+R+AQ SG +E +++NF
Sbjct: 361 GDIEKGEKELKRTEAMISSMTTEERKNPDLLAKSPSRRRRIAQGSGHSETDVSKLITNFT 420
Query: 487 KYTTIM 492
K T+M
Sbjct: 421 KMRTMM 426
>UniRef100_Q8DHI6 Signal recognition particle protein [Synechococcus elongatus]
Length = 469
Score = 470 bits (1209), Expect = e-131
Identities = 237/426 (55%), Positives = 319/426 (74%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF L LESAW L+G++ +T++NI E +R++RRALLEADV+L V + F+++V +A+
Sbjct: 1 MFDALAERLESAWKTLRGQDKITENNIQEALREVRRALLEADVNLQVAKDFIENVRQRAI 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V GV+PDQQ +KIV+D LV++MG S L + PT+IL+AGLQG GKTT AKL
Sbjct: 61 GAEVIAGVRPDQQFIKIVYDALVEVMGESHSPLAHVEPPPTIILMAGLQGTGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
A + +K+G+S +LVA DVYRPAAIDQL LGKQ+ VPV+ GT+V P +IA+QG +AK+
Sbjct: 121 ALHLRKEGRSTLLVATDVYRPAAIDQLMTLGKQIGVPVFEMGTEVSPVEIARQGVAKAKE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+D VI+DTAGRLQID MM EL +K + P E LLVVDAMTGQEAA L F+ ++G
Sbjct: 181 LGVDTVIIDTAGRLQIDAEMMAELAAIKEGVQPHETLLVVDAMTGQEAANLTRAFHEQVG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGAILTKLDGD+RGGAALSV++VSG+PIK +G GE++E L+PFYPDR+A RILGMGDVL
Sbjct: 241 ITGAILTKLDGDTRGGAALSVRQVSGQPIKFIGVGEKVEALQPFYPDRLASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQE + DAE + +KI+ A+FDF+DFLKQ R + MGS++ +I +IPGM K++
Sbjct: 301 TLVEKAQEEVDLADAEKMSRKILEAQFDFDDFLKQLRLLKNMGSLAGIIKLIPGMNKIST 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
Q+++ E+ L+ E +I +MT EER PELLA SP RR+RVA+ SG ++S+F+
Sbjct: 361 EQLQQGEQQLKRAEAMINSMTREERRNPELLASSPSRRERVAKGSGYQVADVTKLVSDFQ 420
Query: 487 KYTTIM 492
+ ++M
Sbjct: 421 RMRSLM 426
>UniRef100_UPI0000286CB5 UPI0000286CB5 UniRef100 entry
Length = 463
Score = 451 bits (1160), Expect = e-125
Identities = 220/426 (51%), Positives = 310/426 (72%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L+ E A L+G++ +++ N+ ++D+RRALLEADVSLPVV+ FV V D+AV
Sbjct: 1 MFDELSARFEDAVKGLRGQDSISETNVDVALKDVRRALLEADVSLPVVKEFVAEVRDKAV 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV PDQ+ +++VH++LV++MGG+ + L A PTV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVSPDQKFIQVVHEQLVEVMGGDNAPLAKAAEAPTVVLMAGLQGAGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
+ K QG+ ++V DVYRPAAI+QL LG Q+DV V++ G + KP DIA G +AK+
Sbjct: 121 GLHLKDQGRRALMVGADVYRPAAIEQLKTLGAQIDVEVFSLGAEAKPEDIAAAGLAKAKE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+ D ++VDTAGRLQID MM+E+ ++ + P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRTAVQPDEVLLVVDSMIGQEAAELTRAFHDQVG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE++E L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQ+ ++ D E +QKK+ A FDF+DF+KQ R + +MGS+ ++ MIPGM K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEATFDFSDFVKQMRLIKRMGSLGGLMKMIPGMNKIDD 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
+++ E+ L+ +E +I +MT +ERE P+LLA P RR+R+A SG D +L++F+
Sbjct: 361 GMLKQGEQQLKRIEAMIGSMTQQERENPDLLASQPSRRRRIASGSGHQAADVDKVLADFQ 420
Query: 487 KYTTIM 492
K M
Sbjct: 421 KMRGFM 426
>UniRef100_Q7VAU4 Signal recognition particle GTPase [Prochlorococcus marinus]
Length = 485
Score = 447 bits (1150), Expect = e-124
Identities = 225/426 (52%), Positives = 312/426 (72%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L+ E A L+GE +++DN+ ++ +RRALLEADVSL VV++FVQ V D+A+
Sbjct: 1 MFDELSARFEDAVKGLRGENKISEDNVDIALKQVRRALLEADVSLSVVKQFVQEVRDKAI 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV P Q+ +++VH ELV++MGG S L + P+VIL+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVNPGQKFIEVVHQELVEVMGGANSPLADSTKKPSVILMAGLQGAGKTTAAAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
Y K +G ++VA DVYRPAAIDQL LGKQ++V V++ G + KP DIA G ++A++
Sbjct: 121 GLYLKDKGLKPLMVAADVYRPAAIDQLNTLGKQINVEVFSLGKESKPEDIAASGLKKAQE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+ D +IVDTAGRLQID+ MM+E+ ++ ++P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EDFDTLIVDTAGRLQIDEEMMNEMVRIRSAVDPDEVLLVVDSMIGQEAADLTRAFHEKVG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE++E L+PF+P+RMAGRILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMAGRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQ+ ++ DAE +Q+K A FDF+DF+KQ R + +MGS+ ++ MIPGM K+
Sbjct: 301 TLVEKAQKEVEIADAEQMQRKFQEATFDFSDFVKQMRLIKRMGSLGGLMKMIPGMNKIDD 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
++ E L+ +E +I +MT E+ +PELLA P RRKRVA SG T + D +L++F+
Sbjct: 361 GMLKSGEDQLKRIEAMIGSMTNAEQTQPELLAAQPSRRKRVAFGSGHTPVEVDKVLADFQ 420
Query: 487 KYTTIM 492
K +M
Sbjct: 421 KMRGLM 426
>UniRef100_Q7U5S8 Signal recognition particle protein [Synechococcus sp.]
Length = 487
Score = 445 bits (1144), Expect = e-123
Identities = 219/426 (51%), Positives = 310/426 (72%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L+ E A LKG++ ++ N+ ++D+RRALLEADVSLPVV+ FV V ++AV
Sbjct: 1 MFDELSARFEDAVKGLKGQDKISDTNVEVALKDVRRALLEADVSLPVVKDFVADVREKAV 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV PDQ+ +++VH++LV++MGG+ + L A PTV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVSPDQKFIQVVHEQLVEVMGGDNAPLAKAAEAPTVVLMAGLQGAGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
+ K QG+ ++V DVYRPAAI+QL LG Q+ V V++ GT+ KP +IA G +AK+
Sbjct: 121 GLHLKDQGRRALMVGADVYRPAAIEQLKTLGGQIGVEVFSLGTEAKPEEIAAAGLAKAKE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+ D ++VDTAGRLQID MM+E+ ++ + P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRTAVQPDEVLLVVDSMIGQEAAELTRAFHDQVG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITGA+LTKLDGDSRGGAALS+++VSG+PIK +G GE++E L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGAVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQ+ ++ D E +QKK+ A FDF+DF++Q R + +MGS+ ++ MIPGM K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEATFDFSDFVQQMRLIKRMGSLGGLMKMIPGMNKLDD 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
+++ E+ L+ +E +I +MTP+ERE P+LLA P RR+R+A SG D +L++F+
Sbjct: 361 GILKQGEQQLKRIEAMIGSMTPKERENPDLLAGQPSRRRRIAAGSGHQPADVDKVLADFQ 420
Query: 487 KYTTIM 492
K M
Sbjct: 421 KMRGFM 426
>UniRef100_Q7V8J6 Signal recognition particle protein [Prochlorococcus marinus]
Length = 497
Score = 439 bits (1128), Expect = e-121
Identities = 217/426 (50%), Positives = 309/426 (71%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L+ E A L+G+ ++ N+ + ++ +RRALL ADVSL VVR FV+ V +AV
Sbjct: 1 MFEELSARFEDAVKGLRGQAKISDTNVEDALKQVRRALLGADVSLEVVREFVEEVRQKAV 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV PDQ+ V++VH +LV++MGG+ + + A+ PTV+L+AGLQG GKTT AKL
Sbjct: 61 GAEVVRGVTPDQKFVQVVHQQLVEVMGGDNAPMAEAEDSPTVVLMAGLQGAGKTTATAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
+ K QG+ ++VA DVYRPAAIDQL LG+Q+ V V++ G DVKP +IA G +A++
Sbjct: 121 GLHLKDQGQRPLMVAADVYRPAAIDQLRTLGEQIGVDVFSLGDDVKPEEIAAAGLAKARE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
+ D ++VDTAGRLQID MM+E+ ++ + P EVLLVVD+M GQEAA L F+ ++G
Sbjct: 181 EGFDTLLVDTAGRLQIDTEMMEEMVRIRSAVEPDEVLLVVDSMIGQEAAELTRAFHDKVG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
ITG++LTKLDGDSRGGAALS+++VSG+PIK +G GE++E L+PF+P+RMA RILGMGDVL
Sbjct: 241 ITGSVLTKLDGDSRGGAALSIRKVSGQPIKFIGTGEKVEALQPFHPERMASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ VEKAQ+ ++ D E +QKK+ A FDF+DFL+Q R + +MGS+ +I M+PGM K+
Sbjct: 301 TLVEKAQKEVELADVEKMQKKLQEASFDFSDFLQQMRLIKRMGSLGGLIKMMPGMNKLDD 360
Query: 427 AQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFE 486
+++ E+ L+ +E +I +MT +ER +PELLA P RR+R+A SG + D +L++F+
Sbjct: 361 GMLKQGEQQLKRIEAMIGSMTADERNQPELLASQPSRRRRIAGGSGHSPADVDKVLADFQ 420
Query: 487 KYTTIM 492
K M
Sbjct: 421 KMRGFM 426
>UniRef100_UPI0000305F06 UPI0000305F06 UniRef100 entry
Length = 498
Score = 435 bits (1118), Expect = e-120
Identities = 218/427 (51%), Positives = 310/427 (72%), Gaps = 1/427 (0%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L++ E A L+GE ++++NI + +++++RALL+ADVSL VV+ F+ V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNINDALKEVKRALLDADVSLSVVKEFISDVKDKAI 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV P Q+ +++V+ EL+ +MG E S L K PTVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNENKNSPTVILMAGLQGAGKTTATGKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGT-DVKPSDIAKQGFEEAK 245
Y K++ K +LVA D+YRPAA++QL LG Q D+ V++A + KP +IAKQ A
Sbjct: 121 GLYLKEKDKKVLLVAADIYRPAAVEQLKTLGSQYDLEVFSAKEKNSKPEEIAKQALNFAS 180
Query: 246 KKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEI 305
+ + +I+DTAGRLQID +MM+E+ +K V NP EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ENDFNTIIIDTAGRLQIDDSMMNEMVRIKEVSNPDEVLLVVDSMIGQEAAELTKSFHEKV 240
Query: 306 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDV 365
GI+GAILTKLDGDSRGGAALS++++SGKPIK +G GE++E L+PF+P+RMA RILGMGDV
Sbjct: 241 GISGAILTKLDGDSRGGAALSIRKISGKPIKFIGVGEKIEALQPFHPERMASRILGMGDV 300
Query: 366 LSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVT 425
L+ VEKAQ+ ++ DAE +QKK+ A FDFNDF+KQ R + +MGS+ +I +IPGM K+
Sbjct: 301 LTLVEKAQKEVELADAEAMQKKLQEATFDFNDFVKQMRLIKRMGSLGGLIKLIPGMNKID 360
Query: 426 PAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNF 485
I++ E L+ +E +I +MT EE++KPE+LA P RR+R+AQ SG + D +L++F
Sbjct: 361 DGMIKDGEDQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYEAKDVDKVLADF 420
Query: 486 EKYTTIM 492
++ M
Sbjct: 421 QRMRGFM 427
>UniRef100_UPI000031BAD7 UPI000031BAD7 UniRef100 entry
Length = 489
Score = 434 bits (1115), Expect = e-120
Identities = 217/427 (50%), Positives = 309/427 (71%), Gaps = 1/427 (0%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L++ E A L+GE ++++NI + +++++RALL+ADVSL VV+ F+ V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNINDALKEVKRALLDADVSLSVVKEFISDVKDKAI 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV P Q+ +++V+ EL+ +MG E S L + PTVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNENENSPTVILMAGLQGAGKTTATGKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTA-GTDVKPSDIAKQGFEEAK 245
Y K++ K +LVA D+YRPAA++QL LG Q D+ V++A G + KP +IAK A
Sbjct: 121 GLYLKEKDKKVLLVAADIYRPAAVEQLKTLGSQYDLEVFSAKGKNSKPEEIAKDALNYAS 180
Query: 246 KKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEI 305
+ + +I+DTAGRLQID +MM E+ +K V NP EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ENNFNSIIIDTAGRLQIDDSMMSEMVRIKEVSNPDEVLLVVDSMIGQEAADLTKSFHEKV 240
Query: 306 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDV 365
GI+GAILTKLDGDSRGGAALS++++SGKPIK +G GE++E L+PF+P+RMA RILGMGDV
Sbjct: 241 GISGAILTKLDGDSRGGAALSIRKISGKPIKFIGVGEKIEALQPFHPERMASRILGMGDV 300
Query: 366 LSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVT 425
L+ VEKAQ+ ++ DAE +QKK+ A FDFNDF+KQ R + +MGS+ +I +IPGM K+
Sbjct: 301 LTLVEKAQKEVELADAEAMQKKLQEATFDFNDFVKQMRLIKRMGSLGGLIKLIPGMNKID 360
Query: 426 PAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNF 485
I++ E L+ +E +I +MT EE++KPE+LA P RR+R+AQ SG + D +L++F
Sbjct: 361 DGMIKDGEDQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYEAKDVDKVLADF 420
Query: 486 EKYTTIM 492
++ M
Sbjct: 421 QRMRGFM 427
>UniRef100_Q7V0H2 Signal recognition particle protein [Prochlorococcus marinus subsp.
pastoris]
Length = 498
Score = 430 bits (1105), Expect = e-119
Identities = 217/427 (50%), Positives = 308/427 (71%), Gaps = 1/427 (0%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF +L++ E A L+GE ++++NI + ++ +++ALL+ADVSL VV+ FV V D+A+
Sbjct: 1 MFDELSSRFEDAVKGLRGEAKISENNIDDALKQVKKALLDADVSLSVVKEFVSDVKDKAI 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G V RGV P Q+ +++V+ EL+ +MG E S L +T PTVIL+AGLQG GKTT KL
Sbjct: 61 GEEVVRGVNPGQKFIEVVNKELINIMGNENSPLNKKETTPTVILMAGLQGAGKTTATGKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGT-DVKPSDIAKQGFEEAK 245
Y K++ + +LVA D+YRPAA++QL +G Q D+ V++A + KP +IA+ A
Sbjct: 121 GLYLKEKEEKVLLVAADIYRPAAVEQLKTIGNQYDLEVFSAKEKNSKPEEIARDALSYAN 180
Query: 246 KKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEI 305
ID +IVDTAGRLQID++MM E+ +K+V P EVLLVVD+M GQEAA L +F+ ++
Sbjct: 181 ANNIDTLIVDTAGRLQIDESMMGEMVRIKQVTKPDEVLLVVDSMIGQEAADLTKSFHEKV 240
Query: 306 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDV 365
GITGAILTKLDGDSRGGAALS++++SGKPIK +G GE++E L+PF+P+RMA RILGMGDV
Sbjct: 241 GITGAILTKLDGDSRGGAALSIRKISGKPIKFIGTGEKIEALQPFHPERMASRILGMGDV 300
Query: 366 LSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVT 425
L+ VEKAQ+ ++ DAE +QKK+ A FDFNDF+KQ R + +MGS+ +I +IPGM K+
Sbjct: 301 LTLVEKAQKEVELADAEVMQKKLQEATFDFNDFVKQMRLMKRMGSLGGLIKLIPGMNKID 360
Query: 426 PAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNF 485
I+ E L+ +E +I +MT EE++KPE+LA P RR+R+AQ SG + D +L++F
Sbjct: 361 DGMIKNGEEQLKKIESMISSMTLEEKQKPEVLAAQPSRRQRIAQGSGYQAKDVDKVLADF 420
Query: 486 EKYTTIM 492
++ M
Sbjct: 421 QRMRGFM 427
>UniRef100_Q7NH98 Signal recognition particle protein SRP54 [Gloeobacter violaceus]
Length = 497
Score = 429 bits (1104), Expect = e-119
Identities = 226/428 (52%), Positives = 302/428 (69%), Gaps = 4/428 (0%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF L+ LE AW KL+G++ +T+ NI E +R++RRALLEADV+ V + FV V D A+
Sbjct: 1 MFESLSEKLEGAWRKLRGQDRITEGNIDEALREVRRALLEADVNFQVAKEFVADVRDDAL 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTF--AKTGPTVILLAGLQGVGKTTVCA 184
G V GV PDQQ +KIVHD+LV LMG + L AK P V+L+AGLQG GKTT A
Sbjct: 61 GAEVVSGVTPDQQFIKIVHDQLVALMGEQNVPLAEPRAKGRPAVVLMAGLQGTGKTTASA 120
Query: 185 KLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEA 244
KLA Y +K G+ +L A DVYRPAAIDQL LG Q+ VPV+T G + P DIA+ E A
Sbjct: 121 KLALYLQKNGQKPLLAAADVYRPAAIDQLQTLGGQIQVPVFTLGKEADPVDIARASLERA 180
Query: 245 KKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLE 304
+ DV+IVDTAGRL ID AMM EL+ +K ++P E+LLVVDAMTGQEAA L F+
Sbjct: 181 IADRHDVLIVDTAGRLSIDDAMMAELERIKAAIDPEEILLVVDAMTGQEAANLTRAFHDR 240
Query: 305 IGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGD 364
+GITGAILTKLDGD+RGGAALSV+++SG+PIK VG GE++E L+PFYP+RMA RILGMGD
Sbjct: 241 LGITGAILTKLDGDTRGGAALSVRKISGQPIKFVGVGEKVEALQPFYPERMASRILGMGD 300
Query: 365 VLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKV 424
VL+ VEKAQE + DA +++++++ +FDF F+KQ R + MG + V+ MIPGM K+
Sbjct: 301 VLTLVEKAQEEIDFADAAKMEQQLLTGQFDFESFIKQMRMIKNMGPLGGVLKMIPGMNKI 360
Query: 425 TPAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSN 484
+ QIR+ E L+ E +I +MT +ER P+L+ S R++R+++ SG + + +L++
Sbjct: 361 SDDQIRQGEVQLKKAEAMIGSMTTQERRNPDLINLS--RKRRISKGSGVSLEDVGKLLND 418
Query: 485 FEKYTTIM 492
F K +M
Sbjct: 419 FGKMRQMM 426
>UniRef100_UPI0000330C1A UPI0000330C1A UniRef100 entry
Length = 478
Score = 410 bits (1054), Expect = e-113
Identities = 205/406 (50%), Positives = 296/406 (72%), Gaps = 1/406 (0%)
Query: 88 LTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVKIVHDE 147
++++NI + ++++++ALL ADVSL VV+ F+ V +A+G V RGV P Q+ +++V+ E
Sbjct: 2 ISENNINDALKEVKKALLNADVSLSVVKDFISDVKTKAIGEEVVRGVNPGQKFIEVVNKE 61
Query: 148 LVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGDVYRP 207
L+ +MG E S L + PTVIL+AGLQG GKTT KL Y K++ K +LVA D+YRP
Sbjct: 62 LINIMGNENSPLIEKEEKPTVILMAGLQGAGKTTATGKLGLYLKEKEKKVLLVAADIYRP 121
Query: 208 AAIDQLTILGKQVDVPVYTAG-TDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRLQIDKAM 266
AA++QL +G Q D+ V++A + +P +IAK A + +D +IVDTAGRLQID++M
Sbjct: 122 AAVEQLKTIGNQYDLEVFSAKENNNRPEEIAKDALNYATENDVDTLIVDTAGRLQIDESM 181
Query: 267 MDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGGAALS 326
M+E+ +K V NP EVLLVVD+M GQEAA L +F+ ++GI+GAILTKLDGDSRGGAALS
Sbjct: 182 MNEMVRIKEVTNPDEVLLVVDSMIGQEAADLTKSFHEKVGISGAILTKLDGDSRGGAALS 241
Query: 327 VKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAEDLQK 386
++++SGKPIK +G GE++E L+PF+P+RMA RILGMGDVL+ VEKAQ+ ++ DAE +QK
Sbjct: 242 IRKISGKPIKFIGIGEKIEALQPFHPERMASRILGMGDVLTLVEKAQKEVELADAEVMQK 301
Query: 387 KIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTPAQIREAERNLEIMEVIIKAM 446
K+ A FDFNDF+KQ R + +MGS+ +I +IPGM K+ I+ E L+ +E +I +M
Sbjct: 302 KLQEATFDFNDFVKQMRLMKRMGSLGGLIKLIPGMNKIDDGMIKNGEEQLKKIESMISSM 361
Query: 447 TPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFEKYTTIM 492
T EE++KPE+LA P RR+R+A+ SG + D +L++F++ M
Sbjct: 362 TLEEKQKPEVLAAQPSRRQRIAKGSGYQAKDVDKVLADFQRMRGFM 407
>UniRef100_Q8XJP3 Signal recognition particle protein [Clostridium perfringens]
Length = 452
Score = 409 bits (1052), Expect = e-113
Identities = 204/421 (48%), Positives = 301/421 (71%), Gaps = 2/421 (0%)
Query: 68 FGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVG 127
F L + L+ KLKG+ LT+ +I E MR+++ ALLEADV+ VV++F+ +V D+ VG
Sbjct: 3 FDGLASKLQDTLKKLKGKGKLTEKDIKEAMREVKLALLEADVNFKVVKKFISNVKDKCVG 62
Query: 128 VGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLA 187
V + P QQ++KIV+DEL LMG S+L ++ GPTV +L GLQG GKTT+ KLA
Sbjct: 63 EEVLNSLTPGQQVIKIVNDELTTLMGETESKLKYSDNGPTVFMLVGLQGAGKTTMAGKLA 122
Query: 188 NYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKK 247
+ +K+ K +LVA D+YRPAAI QL ++GKQ+D+PV++ G VK DIAK E AK
Sbjct: 123 LHLRKKNKKPLLVACDIYRPAAIKQLQVVGKQIDIPVFSMGDKVKAVDIAKAAIEHAKDN 182
Query: 248 KIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGI 307
+VVI+DTAGRL ID+ +M ELKDVK V NP+E+LLVVDAMTGQ+A + TFN + +
Sbjct: 183 GNNVVIIDTAGRLHIDEDLMQELKDVKEVSNPSEILLVVDAMTGQDAVNVAETFNNSLDL 242
Query: 308 TGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLS 367
+G ILTKLDGD+RGGAALS+++++GKPIK VG GE+M D+E F+PDRMA RILGMGDVLS
Sbjct: 243 SGIILTKLDGDTRGGAALSIRDITGKPIKFVGVGEKMSDIEVFHPDRMASRILGMGDVLS 302
Query: 368 FVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGM--AKVT 425
+EKAQ+ + Q++A L +K+++ +F+F+D+L + ++G ++++I MIPG+ ++
Sbjct: 303 LIEKAQQAIDQDEASKLSEKMLNQEFNFDDYLSAMDQMKKLGPINKLIEMIPGVNTKELE 362
Query: 426 PAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNF 485
+ E+ + ++ II++MT +ER++P L+ + R++R+A+ SG T Q+ + +L +
Sbjct: 363 GIDFSQGEKQMATVKAIIQSMTAKERKQPSLVIGNGSRKRRIAKGSGTTVQEVNKVLKGY 422
Query: 486 E 486
E
Sbjct: 423 E 423
>UniRef100_Q9AXU1 Chloroplast SRP54 [Chlamydomonas reinhardtii]
Length = 549
Score = 409 bits (1051), Expect = e-112
Identities = 236/510 (46%), Positives = 335/510 (65%), Gaps = 26/510 (5%)
Query: 61 VVVRAEMFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQS 120
+V+R+ MF L+ +E A + LT +N+ EP++++RRALLEADVSLPVVRRF++
Sbjct: 60 LVIRSAMFDSLSRSIEKAQRLIGKSGTLTAENMKEPLKEVRRALLEADVSLPVVRRFIKK 119
Query: 121 VTDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKT 180
V ++A+G V GV PD Q +K V +EL+ LMGG T+++ +IL+AGLQGVG+
Sbjct: 120 VEERALGTKVLEGVTPDVQFIKTVSNELIDLMGGGNRGKTWSRASRKIILMAGLQGVGRH 179
Query: 181 TVCA-KLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQ 239
CA KLA Y + + +L+ DVY AA QL+ G +DVPV+ GTDV P IAK+
Sbjct: 180 --CAGKLALYLRARE---LLLLHDVY--AAHRQLSA-GAAIDVPVFEDGTDVSPVRIAKK 231
Query: 240 GFEEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVT 299
G EEA++ +D VI+DTAGRLQ+D+ MM EL+DVK + P++ LLVVDAMTGQEAA LV
Sbjct: 232 GVEEARRLGVDAVIIDTAGRLQVDEGMMAELRDVKSAVRPSDTLLVVDAMTGQEAANLVR 291
Query: 300 TFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRI 359
+FN + I+GAILTK+DGDSRGGAALSV+EVSGKPIK VG GE+ME LEPFYP+RMA RI
Sbjct: 292 SFNEAVDISGAILTKMDGDSRGGAALSVREVSGKPIKFVGVGEKMEALEPFYPERMASRI 351
Query: 360 LGMGDVLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIP 419
LGMGDVL+ EKA+ +++EDA+ +++M KFDFNDFL Q +A+ MG + +++ M+P
Sbjct: 352 LGMGDVLTLYEKAEAAIKEEDAQKTMERLMEEKFDFNDFLNQWKAMNNMGGL-QMLKMMP 410
Query: 420 GMAKVTPAQIREAERNLEIMEVIIKAMTPEEREKPELLAES-PVRRKRVAQDSGKTEQQT 478
G K++ + EAE+ + E II AM EER PE+L ++ +RR+RVAQDSGK+E +
Sbjct: 411 GFNKISEKHLYEAEKQFGVYEAIIGAMDEEERSNPEVLIKNLALRRRRVAQDSGKSEAEV 470
Query: 479 DAILSNFEKYTTIMSGKPTCSSTISNACSDEEPNGCNGRWIDANT**S*GST*NRGKVCL 538
+++ YT++ + S + + +P N + + GK
Sbjct: 471 TKLMA---AYTSMKAQVGGMSKLLKLQKAGADPQKANSLLQELVA--------SAGK--- 516
Query: 539 YQAPPGTARRRKKAESRRLFADSTLRQPPR 568
+ PG RR+K+ ++ + ++ +P R
Sbjct: 517 -KVAPGKVRRKKEKDALSQGSRASAHRPNR 545
>UniRef100_Q8R9X0 Signal recognition particle GTPase [Thermoanaerobacter
tengcongensis]
Length = 447
Score = 409 bits (1050), Expect = e-112
Identities = 209/425 (49%), Positives = 297/425 (69%), Gaps = 2/425 (0%)
Query: 68 FGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVG 127
F L+ L+ + KL+G+ LT+ +I E MR+++ ALLEADV+ VV+ F+ SVT++A+G
Sbjct: 4 FESLSERLQGVFKKLRGKGKLTEKDIKEAMREVKVALLEADVNFKVVKDFINSVTEKALG 63
Query: 128 VGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLA 187
V + P QQ++KIVH+EL++LMG S L P VI++ GLQG GKTT C KLA
Sbjct: 64 QEVMESLTPAQQVIKIVHEELIKLMGSVESRLNLGSKVPAVIMMVGLQGSGKTTACGKLA 123
Query: 188 NYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKK 247
N KKQGK+ +LVA D RPAAI QL +LG ++VPV+T G V +DIAK + AK
Sbjct: 124 NLLKKQGKNPLLVACDTVRPAAIKQLQVLGANINVPVFTMGDKVDTADIAKASIDYAKSH 183
Query: 248 KIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGI 307
+DVVI+DTAGRL ID +M+EL +K ++P E+LLV DAMTGQ+A + ++FN + I
Sbjct: 184 NVDVVIIDTAGRLHIDDELMEELVRIKEAVHPDEILLVADAMTGQDAVNVASSFNERLDI 243
Query: 308 TGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLS 367
TG ILTKLDGD+RGGAALS+K V+ KPIK VG GE++ D+EPFYPDRMA RILGMGDVL+
Sbjct: 244 TGVILTKLDGDTRGGAALSIKAVTQKPIKYVGIGEKLGDIEPFYPDRMASRILGMGDVLT 303
Query: 368 FVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTPA 427
+EKAQ + ++ A ++ +KI+S +F DFL+Q R++ MG + +++ MIPG+ K
Sbjct: 304 LIEKAQAAIDEKKALEMGQKILSKQFTLEDFLEQLRSLKNMGPLDQLLAMIPGVNKSVLK 363
Query: 428 QIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFEK 487
+ +E++L+ +E II +MT EER+ P ++ S R++R+A+ SG T Q+ + +L FE+
Sbjct: 364 NVEVSEKDLKRIEAIILSMTKEERQNPSIINGS--RKRRIARGSGTTIQEVNNLLKQFEE 421
Query: 488 YTTIM 492
+M
Sbjct: 422 TKKMM 426
>UniRef100_Q895M4 Signal recognition particle, subunit ffh/srp54 [Clostridium tetani]
Length = 451
Score = 407 bits (1046), Expect = e-112
Identities = 204/427 (47%), Positives = 302/427 (69%), Gaps = 2/427 (0%)
Query: 68 FGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVG 127
F L + L+ + KL+G+ L++ +I E MR++R ALLEADV+ +V+ FV+SV+++ G
Sbjct: 3 FDGLASKLQETFKKLRGKGKLSEKDIKEAMREVRLALLEADVNYKIVKDFVKSVSEKCNG 62
Query: 128 VGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLA 187
V + P QQ+VKIV+DEL++LMG + SE+ F PTVI+L GLQG GKTT+ KLA
Sbjct: 63 KEVLESLTPGQQVVKIVNDELIELMGSKESEINFEANKPTVIMLVGLQGAGKTTMAGKLA 122
Query: 188 NYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKK 247
+K+ K +LVA D+YRPAAI QL ++G Q+DVPV+T G P DI+K + AK+
Sbjct: 123 LQLRKKNKKPLLVACDIYRPAAIKQLQVVGSQIDVPVFTLGDKTNPVDISKAAIKHAKEN 182
Query: 248 KIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGI 307
++ +I+DTAGRL ID+ +MDELK +K + P EVLLVVD+MTGQ+A + +FN ++ I
Sbjct: 183 NLNTIIIDTAGRLHIDENLMDELKSIKENIEPNEVLLVVDSMTGQDAINVAESFNEKLNI 242
Query: 308 TGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLS 367
+G ILTKLDGD+RGGAALS+K ++GKPIK +G GE+M DLE FYP+RMA RILGMGDVLS
Sbjct: 243 SGVILTKLDGDTRGGAALSIKAMTGKPIKFIGIGEKMNDLEVFYPERMASRILGMGDVLS 302
Query: 368 FVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPG--MAKVT 425
+EKA++ + +++AE + +IM+ +F+ DFL + + ++G +++++ M+PG ++
Sbjct: 303 LIEKAEQEIDKDEAEKIGSRIMNQEFNLEDFLSAMQQMKKLGPLNKLVEMLPGANSKELK 362
Query: 426 PAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNF 485
+ E++L+ +E II +MT +ER+ P L++ SP R+KR+A SG T QQ + IL +F
Sbjct: 363 GIDFDKGEKDLKRIESIISSMTIKERKNPSLVSGSPTRKKRIANGSGTTVQQVNKILKDF 422
Query: 486 EKYTTIM 492
E +M
Sbjct: 423 ENMRKMM 429
>UniRef100_Q67PE3 Signal recognition particle [Symbiobacterium thermophilum]
Length = 454
Score = 400 bits (1028), Expect = e-110
Identities = 207/429 (48%), Positives = 301/429 (69%), Gaps = 9/429 (2%)
Query: 67 MFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAV 126
MF L L+ A+ +L+G+ LT+ ++ E +R++R ALLEADV+ VV+ F+ V ++AV
Sbjct: 1 MFESLQERLQEAFKRLRGKGKLTESDVNEALREVRLALLEADVNFKVVKSFIARVKERAV 60
Query: 127 GVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKL 186
G + + P Q ++KIV++ELV LMGGE L A PT+I+L GLQG GKTT AKL
Sbjct: 61 GQEILESLNPAQMVIKIVYEELVALMGGESVGLNMADRPPTIIMLCGLQGAGKTTHAAKL 120
Query: 187 ANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKK 246
A KK+GK +LVA DVYRPAAI QL +LG+QV VPVY+ G P DIA+ A +
Sbjct: 121 ALKLKKEGKKPLLVAADVYRPAAIKQLEVLGEQVGVPVYSEGDQASPVDIAQHAVAYAAE 180
Query: 247 KKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG 306
++ DV+IVDTAGRL ID+ +M+EL+ +K + P + LLV+DAM GQ+A F+ ++G
Sbjct: 181 ERRDVIIVDTAGRLTIDEELMEELQQIKARIQPHDTLLVLDAMIGQDAVTTAEHFHTKLG 240
Query: 307 ITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVL 366
I G ILTKLDGD+RGGAALSV+EV+G+PIK VG GE+++ LEPFYPDR+A RILGMGDVL
Sbjct: 241 IDGVILTKLDGDTRGGAALSVREVTGRPIKFVGVGEKLDALEPFYPDRLASRILGMGDVL 300
Query: 367 SFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTP 426
+ +EKAQE + ++ AE++ +K++ A+FDF DFL+Q RA+ +MG + ++ M+PG+
Sbjct: 301 TLIEKAQEQVDRKQAEEMARKMLKAQFDFEDFLEQLRALQRMGPLQDILKMLPGVG---- 356
Query: 427 AQIREA---ERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILS 483
AQ+++ E+ L+ +E II +MTP+ER P+++ + RR+R+A+ SG+ + ++
Sbjct: 357 AQLKDINIDEKELKHIEAIILSMTPKERRNPDII--NVRRRERIARGSGRPIAEVHRLIK 414
Query: 484 NFEKYTTIM 492
FE+ +M
Sbjct: 415 QFEQTRKLM 423
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 866,792,652
Number of Sequences: 2790947
Number of extensions: 34900952
Number of successful extensions: 125813
Number of sequences better than 10.0: 1819
Number of HSP's better than 10.0 without gapping: 1564
Number of HSP's successfully gapped in prelim test: 255
Number of HSP's that attempted gapping in prelim test: 121732
Number of HSP's gapped (non-prelim): 1996
length of query: 573
length of database: 848,049,833
effective HSP length: 133
effective length of query: 440
effective length of database: 476,853,882
effective search space: 209815708080
effective search space used: 209815708080
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 78 (34.7 bits)
Medicago: description of AC140848.2


