

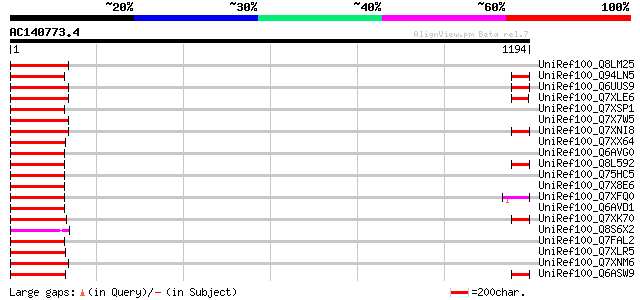
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140773.4 + phase: 0 /pseudo
(1194 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8LM25 Putative retroelement [Oryza sativa] 106 5e-21
UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sa... 105 1e-20
UniRef100_Q6UUS9 Putative gag-pol [Oryza sativa] 104 1e-20
UniRef100_Q7XLE6 OSJNBa0013A04.23 protein [Oryza sativa] 104 2e-20
UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa] 104 2e-20
UniRef100_Q7X7W5 OSJNBb0046P18.13 protein [Oryza sativa] 103 2e-20
UniRef100_Q7XNI8 OSJNBb0032D24.5 protein [Oryza sativa] 103 2e-20
UniRef100_Q7XX64 OSJNBa0052P16.15 protein [Oryza sativa] 103 3e-20
UniRef100_Q6AVG0 Putative retrotransposon gag protein [Oryza sat... 103 3e-20
UniRef100_Q8L592 Putative gag-pol [Oryza sativa] 102 5e-20
UniRef100_Q75HC5 Putative reverse transcriptase [Oryza sativa] 102 5e-20
UniRef100_Q7X8E6 OSJNBb0012A12.6 protein [Oryza sativa] 102 5e-20
UniRef100_Q7XFQ0 Putative retroelement [Oryza sativa] 102 5e-20
UniRef100_Q6AVD1 Putative polyprotein [Oryza sativa] 102 5e-20
UniRef100_Q7XK70 OSJNBa0028M15.17 protein [Oryza sativa] 102 7e-20
UniRef100_Q8S6X2 Putative pol polyprotein [Oryza sativa] 102 7e-20
UniRef100_Q7FAL2 OSJNBa0042N22.17 protein [Oryza sativa] 102 9e-20
UniRef100_Q7XLR5 OSJNBb0093G06.6 protein [Oryza sativa] 101 2e-19
UniRef100_Q7XNM6 OSJNBb0068N06.4 protein [Oryza sativa] 100 2e-19
UniRef100_Q6ASW9 Reverse transcriptase (RNA-dependent DNA polyme... 100 2e-19
>UniRef100_Q8LM25 Putative retroelement [Oryza sativa]
Length = 858
Score = 106 bits (264), Expect = 5e-21
Identities = 52/134 (38%), Positives = 85/134 (62%), Gaps = 1/134 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG+ ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 365 PLYINGYVNGMPLSKIMVDGGAVVNLMPYATFRKLGRNSEDLIKTNMVLKDFGGNPSETK 424
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D ++
Sbjct: 425 GVLNVELTVGSKTVPTTFFVIDRKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIK 483
Query: 121 NVEADQSYFLAEVN 134
V AD+S +A +
Sbjct: 484 IVPADRSVNVASAD 497
>UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 105 bits (261), Expect = 1e-20
Identities = 51/125 (40%), Positives = 80/125 (63%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ ++
Sbjct: 363 PLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPSETM 422
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 423 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTIHQCLIQWQGD-KIE 481
Query: 121 NVEAD 125
V AD
Sbjct: 482 IVPAD 486
Score = 63.5 bits (153), Expect = 4e-08
Identities = 28/41 (68%), Positives = 33/41 (80%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EAIPLK V + I F+Q+ IIYR GIP+TITTDQG
Sbjct: 1315 FTKWVEAIPLKKVDSGDAIQFVQEHIIYRVGIPQTITTDQG 1355
>UniRef100_Q6UUS9 Putative gag-pol [Oryza sativa]
Length = 1672
Score = 104 bits (260), Expect = 1e-20
Identities = 54/133 (40%), Positives = 83/133 (61%), Gaps = 1/133 (0%)
Query: 2 LFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLG 61
L+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ + G
Sbjct: 656 LYINGYVNGKPMSKMMVDGGAAVNLIPYAKFRKLGRNAEDLIKTNMVLKDFGGNPSETKG 715
Query: 62 AIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVEN 121
+ +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D L E
Sbjct: 716 VLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGDKL-EI 774
Query: 122 VEADQSYFLAEVN 134
V AD+S +A N
Sbjct: 775 VPADRSVNVASAN 787
Score = 61.6 bits (148), Expect = 1e-07
Identities = 27/41 (65%), Positives = 32/41 (77%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EAIPLK V + I F Q+ +IYRF IP+TITTDQG
Sbjct: 1407 FTKWLEAIPLKKVDSGDAIQFFQEHVIYRFDIPQTITTDQG 1447
>UniRef100_Q7XLE6 OSJNBa0013A04.23 protein [Oryza sativa]
Length = 1235
Score = 104 bits (259), Expect = 2e-20
Identities = 51/134 (38%), Positives = 82/134 (61%), Gaps = 1/134 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+++ +NG ++K++VDGGA VNL+P + K+G DL NV+L ++ G+ +
Sbjct: 205 PLYVKGFVNGKPMSKMMVDGGAAVNLMPYATFRKLGKNADDLIKTNVVLNDFGGNPSENK 264
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL +GS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 265 GVLNVELTIGSKTDPTTFFVIDGKGSYSMLLGRDWIHANCCIPSTMHQSLIQWQGD-KIE 323
Query: 121 NVEADQSYFLAEVN 134
V AD + N
Sbjct: 324 IVLADSQLKMESPN 337
Score = 58.2 bits (139), Expect = 2e-06
Identities = 22/40 (55%), Positives = 32/40 (80%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQ 1193
FTKW+EA+PLK V + I ++++I+YRFG+P+ ITTDQ
Sbjct: 970 FTKWVEAVPLKKVDSGDAIQLVKEYIVYRFGVPQIITTDQ 1009
>UniRef100_Q7XSP1 OSJNBa0070M12.15 protein [Oryza sativa]
Length = 1685
Score = 104 bits (259), Expect = 2e-20
Identities = 52/125 (41%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 739 PLYINGYVNGKPMSKMMVDGGAAVNLMPYTTFRKLGRTTEDLMKTNMVLKDFGGNPSETK 798
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D VE
Sbjct: 799 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KVE 857
Query: 121 NVEAD 125
V AD
Sbjct: 858 IVPAD 862
>UniRef100_Q7X7W5 OSJNBb0046P18.13 protein [Oryza sativa]
Length = 766
Score = 103 bits (258), Expect = 2e-20
Identities = 53/134 (39%), Positives = 83/134 (61%), Gaps = 1/134 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N+ L ++ G+ +
Sbjct: 357 PLYINGYVNGKPMSKMMVDGGAVVNLMPYATFRKLGRNAEDLIKMNMALKDFGGNPSETK 416
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 417 GVLNVELSVGSKTVPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 475
Query: 121 NVEADQSYFLAEVN 134
V AD+S +A +
Sbjct: 476 IVPADRSVNVASAD 489
>UniRef100_Q7XNI8 OSJNBb0032D24.5 protein [Oryza sativa]
Length = 1355
Score = 103 bits (258), Expect = 2e-20
Identities = 52/134 (38%), Positives = 83/134 (61%), Gaps = 1/134 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 352 PLYINGYVNGKPLSKMIVDGGAAVNLMPYATFRKLGRSSDDLIKTNMVLKDFGGNQSETK 411
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+ Q + W+ D +E
Sbjct: 412 GVLNVELTVGSKTVPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMQQCLIQWQGD-KIE 470
Query: 121 NVEADQSYFLAEVN 134
V AD+S +A +
Sbjct: 471 IVPADRSVNIASAD 484
Score = 65.1 bits (157), Expect = 1e-08
Identities = 28/41 (68%), Positives = 34/41 (82%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW++AIPLK V + I F+Q+ IIYRFGIP+TITTDQG
Sbjct: 1241 FTKWVDAIPLKKVDSGDAIQFVQEHIIYRFGIPQTITTDQG 1281
>UniRef100_Q7XX64 OSJNBa0052P16.15 protein [Oryza sativa]
Length = 858
Score = 103 bits (257), Expect = 3e-20
Identities = 52/127 (40%), Positives = 79/127 (61%), Gaps = 1/127 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++ ++VDGGATVNL+P + K+G DL N++L ++ G+ +
Sbjct: 567 PLYINGFVNGKPMSNMMVDGGATVNLMPYATFRKLGRNAEDLSKTNMVLKDFGGNPSETK 626
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K +++LLGR+WIH +PST+HQ + W E +E
Sbjct: 627 GVLNVELTVGSKTIPTTFFVIDGKGCYSLLLGRDWIHANCCIPSTMHQCLIQW-EGNKIE 685
Query: 121 NVEADQS 127
V AD+S
Sbjct: 686 IVPADRS 692
>UniRef100_Q6AVG0 Putative retrotransposon gag protein [Oryza sativa]
Length = 1251
Score = 103 bits (257), Expect = 3e-20
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 700 PLYINGYVNGKSMSKMMVDGGAAVNLMPYATFKKLGRNAEDLIKTNMVLKDFGGNPSETR 759
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 760 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 818
Query: 121 NVEAD 125
V AD
Sbjct: 819 IVPAD 823
>UniRef100_Q8L592 Putative gag-pol [Oryza sativa]
Length = 1383
Score = 102 bits (255), Expect = 5e-20
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 649 PLYINGYVNGKPMSKMIVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPSETK 708
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 709 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 767
Query: 121 NVEAD 125
V AD
Sbjct: 768 IVPAD 772
Score = 57.8 bits (138), Expect = 2e-06
Identities = 25/41 (60%), Positives = 30/41 (72%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EAIPLK + I F+Q+ IIYRFGI + I TDQG
Sbjct: 1241 FTKWVEAIPLKKANSGDAIQFVQEHIIYRFGISQNIMTDQG 1281
>UniRef100_Q75HC5 Putative reverse transcriptase [Oryza sativa]
Length = 1149
Score = 102 bits (255), Expect = 5e-20
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 662 PLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLEDFGGNPSETK 721
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 722 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 780
Query: 121 NVEAD 125
V AD
Sbjct: 781 IVPAD 785
>UniRef100_Q7X8E6 OSJNBb0012A12.6 protein [Oryza sativa]
Length = 243
Score = 102 bits (255), Expect = 5e-20
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 48 PLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNSYDLIKTNMVLKDFGGNPSETK 107
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 108 GVLNVELTVGSKTVPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 166
Query: 121 NVEAD 125
V AD
Sbjct: 167 IVPAD 171
>UniRef100_Q7XFQ0 Putative retroelement [Oryza sativa]
Length = 1025
Score = 102 bits (255), Expect = 5e-20
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 120 PLYINGYVNGKPLSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPSETK 179
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 180 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 238
Query: 121 NVEAD 125
V AD
Sbjct: 239 IVPAD 243
Score = 64.3 bits (155), Expect = 2e-08
Identities = 35/69 (50%), Positives = 41/69 (58%), Gaps = 8/69 (11%)
Query: 1134 RGD*TYFIEMI--------QIYLSRDRLFTKWIEAIPLKDVTQEEVISFIQKFIIYRFGI 1185
RG Y I MI Q L FTKW+EAIPLK V + I F+ + IIYRFG+
Sbjct: 903 RGWGIYMIGMINPPSSKGHQFILVATDYFTKWVEAIPLKKVDSGDAIQFVHEHIIYRFGL 962
Query: 1186 PETITTDQG 1194
P+TITTDQG
Sbjct: 963 PQTITTDQG 971
>UniRef100_Q6AVD1 Putative polyprotein [Oryza sativa]
Length = 1418
Score = 102 bits (255), Expect = 5e-20
Identities = 51/125 (40%), Positives = 79/125 (62%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 856 PLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPSETK 915
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D +E
Sbjct: 916 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIE 974
Query: 121 NVEAD 125
V AD
Sbjct: 975 IVPAD 979
>UniRef100_Q7XK70 OSJNBa0028M15.17 protein [Oryza sativa]
Length = 1218
Score = 102 bits (254), Expect = 7e-20
Identities = 50/129 (38%), Positives = 79/129 (60%), Gaps = 1/129 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 339 PLYINGYVNGKPMSKMMVDGGAVVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPSETK 398
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VG+ TTF V+ K ++++LLGR+WIH +PS +HQ + W+ D +E
Sbjct: 399 GGLNVELTVGNKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSAMHQCLIKWQGD-KIE 457
Query: 121 NVEADQSYF 129
V D YF
Sbjct: 458 IVPTDSYYF 466
Score = 62.4 bits (150), Expect = 8e-08
Identities = 28/41 (68%), Positives = 33/41 (80%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EAIPLK V + I F+Q+ IIYRFGI +TITTDQG
Sbjct: 953 FTKWVEAIPLKKVDSGDAIQFVQEHIIYRFGILQTITTDQG 993
>UniRef100_Q8S6X2 Putative pol polyprotein [Oryza sativa]
Length = 505
Score = 102 bits (254), Expect = 7e-20
Identities = 55/136 (40%), Positives = 82/136 (59%), Gaps = 1/136 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+++ I+G V+++LVD GA VNL+P S ++G D +LK N+IL + G +
Sbjct: 321 PLYLKGHIDGKPVSRMLVDRGAAVNLMPYSLFKRLGRGDDELKKTNMILNGFNGEPTEAK 380
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G +EL +G+ TTF +V + ++NV+LGR WIH VPST+HQ + W D VE
Sbjct: 381 GIFSVELTMGNKTLPTTFFIVDVQGSYNVILGRCWIHANCCVPSTLHQCLIQWDGDD-VE 439
Query: 121 NVEADQSYFLAEVNTI 136
V+AD S +A + I
Sbjct: 440 IVQADTSTEVAMADAI 455
>UniRef100_Q7FAL2 OSJNBa0042N22.17 protein [Oryza sativa]
Length = 1436
Score = 102 bits (253), Expect = 9e-20
Identities = 52/125 (41%), Positives = 80/125 (63%), Gaps = 1/125 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+S +
Sbjct: 888 PLYINGYVNGKPMSKMMVDGGAAVNLMPYTTFRKLGRNAEDLIRMNMVLKDFGGNSSETK 947
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS T+F V+ K ++++LLGR+WIH +PST+HQ + W+ D VE
Sbjct: 948 GVLNVELTVGSKTIPTSFFVIDGKGSYSLLLGRDWIHVNCCIPSTMHQCLIQWQGD-KVE 1006
Query: 121 NVEAD 125
V AD
Sbjct: 1007 IVPAD 1011
>UniRef100_Q7XLR5 OSJNBb0093G06.6 protein [Oryza sativa]
Length = 1165
Score = 101 bits (251), Expect = 2e-19
Identities = 49/127 (38%), Positives = 79/127 (61%), Gaps = 1/127 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL++ +NG ++K++V+GGA VNL+P + K+G DL N++L N+ G+ +
Sbjct: 279 PLYVNGYVNGKPMSKMMVNGGAAVNLMPYATFRKLGRNAEDLIKTNMVLKNFGGNPSETK 338
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL GS TTF V+ K ++N+LLG +WIH +PST+HQ + W+ D +E
Sbjct: 339 GVLNVELTAGSKTIPTTFFVIDGKGSYNLLLGSDWIHANCCIPSTMHQCLIQWQGD-KIE 397
Query: 121 NVEADQS 127
V AD++
Sbjct: 398 IVPADRT 404
>UniRef100_Q7XNM6 OSJNBb0068N06.4 protein [Oryza sativa]
Length = 959
Score = 100 bits (250), Expect = 2e-19
Identities = 50/134 (37%), Positives = 83/134 (61%), Gaps = 1/134 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
P ++ +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 527 PFYVNGYVNGKPMSKMMVDGGAVVNLMPYATFRKLGRNAEDLIKTNMVLKDFGGNPSETK 586
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VGS TTF V+ K ++++LLGR+WIH +PST++Q + W+ D +E
Sbjct: 587 GVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMNQCLIQWQVD-KIE 645
Query: 121 NVEADQSYFLAEVN 134
V AD+S +A +
Sbjct: 646 IVPADRSVNVASAD 659
>UniRef100_Q6ASW9 Reverse transcriptase (RNA-dependent DNA polymerase) domain
containing protein [Oryza sativa]
Length = 1372
Score = 100 bits (250), Expect = 2e-19
Identities = 49/127 (38%), Positives = 81/127 (63%), Gaps = 1/127 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL+I +NG ++K++VDGGA VNL+P + K+G DL N++L ++ G+ +
Sbjct: 436 PLYINGYVNGKPMSKMMVDGGAAVNLMPYATFRKLGRNVEDLIKTNMVLKDFGGNPSETK 495
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G + +EL VG+ TTF V+ K ++++LLGR+WIH +PST+HQ + W+ D ++
Sbjct: 496 GVLNVELTVGNKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQCLIQWQGD-KIQ 554
Query: 121 NVEADQS 127
V AD++
Sbjct: 555 IVPADRA 561
Score = 65.1 bits (157), Expect = 1e-08
Identities = 29/41 (70%), Positives = 33/41 (79%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EAIPLK V + I F Q+ IIYRFGIP+TITTDQG
Sbjct: 1107 FTKWVEAIPLKKVDSGDAIQFFQEHIIYRFGIPQTITTDQG 1147
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.348 0.153 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,770,698,925
Number of Sequences: 2790947
Number of extensions: 68312604
Number of successful extensions: 229231
Number of sequences better than 10.0: 736
Number of HSP's better than 10.0 without gapping: 345
Number of HSP's successfully gapped in prelim test: 391
Number of HSP's that attempted gapping in prelim test: 228157
Number of HSP's gapped (non-prelim): 1220
length of query: 1194
length of database: 848,049,833
effective HSP length: 139
effective length of query: 1055
effective length of database: 460,108,200
effective search space: 485414151000
effective search space used: 485414151000
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 81 (35.8 bits)
Medicago: description of AC140773.4


