

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140722.2 + phase: 0
(555 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
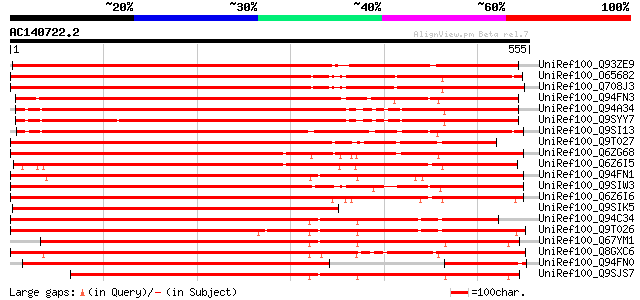
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93ZE9 At2g21540/F2G1.19 [Arabidopsis thaliana] 683 0.0
UniRef100_O65682 Hypothetical protein T4L20.160 [Arabidopsis tha... 636 0.0
UniRef100_Q708J3 Can of worms 1 [Arabidopsis thaliana] 636 0.0
UniRef100_Q94FN3 Phosphatidylinositol transfer-like protein II [... 635 e-180
UniRef100_Q94A34 C7A10_870/C7A10_870 [Arabidopsis thaliana] 627 e-178
UniRef100_Q9SYY7 Hypothetical protein C7A10.870 [Arabidopsis tha... 621 e-176
UniRef100_Q9SI13 Putative phosphatidylinositol/phophatidylcholin... 613 e-174
UniRef100_Q9T027 SEC14-like protein [Arabidopsis thaliana] 607 e-172
UniRef100_Q6ZG68 Putative SEC14 cytosolic factor [Oryza sativa] 598 e-169
UniRef100_Q6Z6I5 Putative phosphatidylinositol/ phophatidylcholi... 568 e-160
UniRef100_Q94FN1 Phosphatidylinositol transfer-like protein III ... 565 e-159
UniRef100_Q9SIW3 Putative phosphatidylinositol/phosphatidylcholi... 563 e-159
UniRef100_Q6Z6I6 Putative phosphatidylinositol/ phophatidylcholi... 558 e-157
UniRef100_Q9SIK5 Putative phosphatidylinositol/phophatidylcholin... 556 e-157
UniRef100_Q94C34 AT4g39170/T22F8_70 [Arabidopsis thaliana] 553 e-156
UniRef100_Q9T026 SEC14-like protein [Arabidopsis thaliana] 534 e-150
UniRef100_Q67YM1 Putative phosphatidylinositol/ phosphatidylchol... 528 e-148
UniRef100_Q8GXC6 Putative sec14 cytosolic factor [Arabidopsis th... 507 e-142
UniRef100_Q94FN0 Phosphatidylinositol transfer-like protein IV [... 501 e-140
UniRef100_Q9SJS7 Putative phosphatidylinositol/phosphatidylcholi... 497 e-139
>UniRef100_Q93ZE9 At2g21540/F2G1.19 [Arabidopsis thaliana]
Length = 548
Score = 683 bits (1762), Expect = 0.0
Identities = 346/543 (63%), Positives = 417/543 (76%), Gaps = 20/543 (3%)
Query: 4 SEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDALRQT 63
SEDEKK K+ S+KK A++AS+KFK+SFTK+ R++SRVMS+ I D D EELQAVDA RQ
Sbjct: 20 SEDEKKTKLCSLKKKAINASNKFKHSFTKRTRRNSRVMSVSIVDDIDLEELQAVDAFRQA 79
Query: 64 LILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELD 123
LIL+ELLPSKHDD HMMLRFLRARK+D+EK KQMWTDM+ WRKEFG DTIMEDF+F+E+D
Sbjct: 80 LILDELLPSKHDDHHMMLRFLRARKFDLEKAKQMWTDMIHWRKEFGVDTIMEDFDFKEID 139
Query: 124 EVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPA 183
EVLK YPQG+HGVDKDGRPVYIERLGQVD KL+QVT+++RY+KYHVREFE+ F +KLPA
Sbjct: 140 EVLKYYPQGYHGVDKDGRPVYIERLGQVDATKLMQVTTIDRYVKYHVREFEKTFNIKLPA 199
Query: 184 CSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSG 243
CSIAAKKHIDQSTTILDVQGVGL+S +KAARDLLQR+QKID DNYPE+LNRMFIINAGSG
Sbjct: 200 CSIAAKKHIDQSTTILDVQGVGLKSFSKAARDLLQRIQKIDSDNYPETLNRMFIINAGSG 259
Query: 244 FRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSD 303
FRLLW+TVKSFLDPKTT+KIHVLGNKYQSKLLE+ID++ELPEFLGG CTCADKGGCM SD
Sbjct: 260 FRLLWSTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDSNELPEFLGGNCTCADKGGCMRSD 319
Query: 304 KGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVHCLDQP 363
KGPWNDP+I KM QNG G+ K LS +EEKTI +E K DSF
Sbjct: 320 KGPWNDPDIFKMVQNGEGKCPRKTLSNIEEKTISVDENTTMK--SDSF-----------A 366
Query: 364 KSYGVYQYDSFVPVLDKAVD-SSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIM 422
K+ + F+P++DK V+ S+W + Y +D +S G GG+M
Sbjct: 367 KNKFDAENTKFIPMIDKTVNASTWPTNLHKSNYPEPEDLYSAVKPSQRRGGEGYLFGGVM 426
Query: 423 ALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIKAAAISNNEYMAMMKRMAELEE 482
+LVMG++T++R+T +MPRK+TEAA+YGG + + +SN EYM+M+KRMAELEE
Sbjct: 427 SLVMGLMTVVRLTKNMPRKLTEAAIYGG-----EVDKAETTMVSNQEYMSMVKRMAELEE 481
Query: 483 KVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKK 541
K L +P PEKE++L AL+RV LE +L TKK LE+ + Q + IDKKKK
Sbjct: 482 KCRSLDNQPAAFSPEKEQILTAALSRVDELELQLAQTKKTLEETMATQHVIMAYIDKKKK 541
Query: 542 KKK 544
KKK
Sbjct: 542 KKK 544
>UniRef100_O65682 Hypothetical protein T4L20.160 [Arabidopsis thaliana]
Length = 560
Score = 636 bits (1641), Expect = 0.0
Identities = 340/558 (60%), Positives = 423/558 (74%), Gaps = 24/558 (4%)
Query: 1 MEHSEDEKK-KKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAVD 58
+E SE+E+K K+ S+KK A++AS++FKNSF KKGR+ S RVMS+ IED DAE+LQA+D
Sbjct: 8 IEMSEEERKIVKISSLKKKAINASNRFKNSFKKKGRRSSSRVMSVPIEDDIDAEDLQALD 67
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ LIL+ELLPSK DD HMMLRFLRARK+DIEK KQMW+DM++WRK+FGADTI+EDF+
Sbjct: 68 AFRQALILDELLPSKLDDLHMMLRFLRARKFDIEKAKQMWSDMIQWRKDFGADTIIEDFD 127
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
FEE+DEV+K YPQG+HGVDK+GRPVYIERLGQ+D NKLLQVT+++RY+KYHV+EFE+ F
Sbjct: 128 FEEIDEVMKHYPQGYHGVDKEGRPVYIERLGQIDANKLLQVTTMDRYVKYHVKEFEKTFK 187
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
VK P+CS+AA KHIDQSTTILDVQGVGL++ +K+AR+LLQRL KID +NYPE+LNRMFII
Sbjct: 188 VKFPSCSVAANKHIDQSTTILDVQGVGLKNFSKSARELLQRLCKIDNENYPETLNRMFII 247
Query: 239 NAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGG 298
NAGSGFRLLW+TVKSFLDPKTT+KIHVLGNKY SKLLEVIDASELPEF GG CTC DKGG
Sbjct: 248 NAGSGFRLLWSTVKSFLDPKTTAKIHVLGNKYHSKLLEVIDASELPEFFGGACTCEDKGG 307
Query: 299 CMLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVH 358
CM SDKGPWNDPE+LK+A N + +S E K + Q + GF E+L+
Sbjct: 308 CMRSDKGPWNDPEVLKIAINREAK--CSPISEDEHKHVDQGRST--SGF-----ESLE-- 356
Query: 359 CLDQPKSYGVYQYDSFVPVLDKAVDSSW-KKTIQNDKYALSKDCFSNNNGMNSSGFSKQF 417
+ K+ Y+ + +DK++D +W KT + + + +SK G G
Sbjct: 357 -RIKKKTDEDNVYEKQIATIDKSMDMAWLAKTQKAENFPISKGITKLMKGAPKKG-DGLL 414
Query: 418 VGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIK------AAAISNNEYM 471
VGG+MA VMGIV ++R++ +PRK+TEAALYG + Y + + K AA +S++EYM
Sbjct: 415 VGGVMAFVMGIVAMVRLSKDVPRKLTEAALYGNSVCYEESTKSKQNQGQFAAPVSSSEYM 474
Query: 472 AMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQV 530
M+KRMAELE+K L +KP + EKEE L AL RV LEQEL TKKALE+AL Q
Sbjct: 475 LMVKRMAELEDKCMFLDLKPAHVESEKEEKLQAALNRVQVLEQELTETKKALEEALVSQK 534
Query: 531 ELEGQIDKKKKKKKKLVR 548
E+ I+ KKKKKKKLVR
Sbjct: 535 EILAYIE-KKKKKKKLVR 551
>UniRef100_Q708J3 Can of worms 1 [Arabidopsis thaliana]
Length = 557
Score = 636 bits (1640), Expect = 0.0
Identities = 337/562 (59%), Positives = 424/562 (74%), Gaps = 26/562 (4%)
Query: 1 MEHSEDEKK-KKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAVD 58
+E SE+E+K K+ S+KK A++AS++FKNSF KKGR+ S RVMS+ IED DAE+LQA+D
Sbjct: 8 IEMSEEERKIVKISSLKKKAINASNRFKNSFKKKGRRSSSRVMSVPIEDDIDAEDLQALD 67
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ LIL+ELLPSK DD HMMLRFLRARK+DIEK KQMW+DM++WRK+FGADTI+EDF+
Sbjct: 68 AFRQALILDELLPSKLDDLHMMLRFLRARKFDIEKAKQMWSDMIQWRKDFGADTIIEDFD 127
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
FEE+DEV+K YPQG+HGVDK+GRPVYIERLGQ+D NKLLQVT+++RY+KYHV+EFE+ F
Sbjct: 128 FEEIDEVMKHYPQGYHGVDKEGRPVYIERLGQIDANKLLQVTTMDRYVKYHVKEFEKTFK 187
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
VK P+CS+AA KHIDQSTTILDVQGVGL++ +K+AR+LLQRL KID +NYPE+LNRMFII
Sbjct: 188 VKFPSCSVAANKHIDQSTTILDVQGVGLKNFSKSARELLQRLCKIDNENYPETLNRMFII 247
Query: 239 NAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGG 298
NAGSGFRLLW+TVKSFLDPKTT+KIHVLGNKY SKLLEVIDASELPEF GG CTC DKGG
Sbjct: 248 NAGSGFRLLWSTVKSFLDPKTTAKIHVLGNKYHSKLLEVIDASELPEFFGGACTCEDKGG 307
Query: 299 CMLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVH 358
CM SDKGPWNDPE+LK+A N + +S E K + Q + GF E+L+
Sbjct: 308 CMRSDKGPWNDPEVLKIAINREAK--CSPISEDEHKHVDQGRST--SGF-----ESLE-- 356
Query: 359 CLDQPKSYGVYQYDSFVPVLDKAVDSSW-KKTIQNDKYALSK--DCFSNNNGMNSSGFSK 415
+ K+ Y+ + +DK++D +W KT + + + +SK +C+
Sbjct: 357 -RIKKKTDEDNVYEKQIATIDKSMDMAWLAKTQKAENFPISKGLECYVRKGAPKKG--DG 413
Query: 416 QFVGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIK------AAAISNNE 469
VGG+MA VMGIV ++R++ +PRK+TEAALYG + Y + + K AA +S++E
Sbjct: 414 LLVGGVMAFVMGIVAMVRLSKDVPRKLTEAALYGNSVCYEESTKSKQNQGQFAAPVSSSE 473
Query: 470 YMAMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALEDALTR 528
YM M+KRMAELE+K L +KP + EKEE L AL RV LEQEL TKKALE+AL
Sbjct: 474 YMLMVKRMAELEDKCMFLDLKPAHVESEKEEKLQAALNRVQVLEQELTETKKALEEALVS 533
Query: 529 QVELEGQIDKKKKKKKKLVRYI 550
Q E+ I+KKKKKKK + R +
Sbjct: 534 QKEILAYIEKKKKKKKLVFRIL 555
>UniRef100_Q94FN3 Phosphatidylinositol transfer-like protein II [Lotus japonicus]
Length = 550
Score = 635 bits (1637), Expect = e-180
Identities = 331/546 (60%), Positives = 406/546 (73%), Gaps = 20/546 (3%)
Query: 7 EKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDALRQTLIL 66
+K + GS+KK A+S S K R S+VMS+ IED DAE+L+AVD RQ LIL
Sbjct: 12 DKSDRAGSLKKKAMSLRSSLSR---KSRRSSSKVMSVEIEDVRDAEDLKAVDEFRQALIL 68
Query: 67 EELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVL 126
+ELLP KHDD H +LRFL+ARK+DIEK+KQMW+DML+WRKEFGADTI+EDF+F+E+DEV+
Sbjct: 69 DELLPEKHDDYHQLLRFLKARKFDIEKSKQMWSDMLQWRKEFGADTIVEDFDFKEIDEVV 128
Query: 127 KCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSI 186
K YP GHHGVDKDGRPVYIE +GQVD KL+QVT+++RY+KYHV+EFER F +K ACSI
Sbjct: 129 KYYPHGHHGVDKDGRPVYIENIGQVDATKLMQVTTMDRYIKYHVKEFERTFDLKFAACSI 188
Query: 187 AAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRL 246
AAKKHIDQSTTILDVQGVGL++ NK AR+L+ RLQKIDGDNYPE+LNRMFIINAGSGFR+
Sbjct: 189 AAKKHIDQSTTILDVQGVGLKNFNKHARELITRLQKIDGDNYPETLNRMFIINAGSGFRM 248
Query: 247 LWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGP 306
LW+TVKSFLDPKTTSKIHVLGNKYQSKLLEVIDAS+LPEFLGGTCTCAD+GGCM SDKGP
Sbjct: 249 LWSTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASQLPEFLGGTCTCADQGGCMRSDKGP 308
Query: 307 WNDPEILKMAQNGVGRYTIKALSG-VEEKTIKQEETAYQKGFKDSFPETLDVHCLDQPKS 365
W DPE+++M QNG + + K S VEEK I +E T F + +
Sbjct: 309 WKDPELVRMVQNGEHKCSRKCESPVVEEKKISEETTKMGANFTSQLSSV-----FGEVPA 363
Query: 366 YGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNS----SGFSKQFVGGI 421
V Y+ FVP D+ V WKK +N+++ +SK + ++S + Q G+
Sbjct: 364 TKVCNYEDFVPAADETV---WKKVEENEQFQMSKAVVETFSMVDSCKIHEKVNSQIFTGV 420
Query: 422 MALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDG---SMIKAAAISNNEYMAMMKRMA 478
MA VMGIVT++RMT +MP+K+T+A Y NSVY G S +IS E+M +MKRMA
Sbjct: 421 MAFVMGIVTMVRMTRNMPKKLTDANFY-SNSVYSGGQNPSDQTNPSISAQEFMTVMKRMA 479
Query: 479 ELEEKVTVLSVKPVMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDK 538
ELEEK+ ++ MPPEKEEMLN A++R LEQEL +TKKALED+L +Q EL I+K
Sbjct: 480 ELEEKMGNMNYNTCMPPEKEEMLNAAISRADALEQELMSTKKALEDSLAKQEELSAYIEK 539
Query: 539 KKKKKK 544
KKKKKK
Sbjct: 540 KKKKKK 545
>UniRef100_Q94A34 C7A10_870/C7A10_870 [Arabidopsis thaliana]
Length = 543
Score = 627 bits (1616), Expect = e-178
Identities = 336/550 (61%), Positives = 414/550 (75%), Gaps = 31/550 (5%)
Query: 7 EKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSI-CIEDSFDAEELQAVDALRQTLI 65
E K ++GS KK S+S + S TK+ R+ S+VMS+ IED DAEEL+AVDA RQ+LI
Sbjct: 8 ELKPRMGSFKK--RSSSKNLRYSMTKR-RRSSKVMSVEIIEDVHDAEELKAVDAFRQSLI 64
Query: 66 LEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEV 125
L+ELLP KHDD HMMLRFL+ARK+D+EKTKQMWT+ML+WRKEFGADT+ME+F+F+E+DEV
Sbjct: 65 LDELLPEKHDDYHMMLRFLKARKFDLEKTKQMWTEMLRWRKEFGADTVMEEFDFKEIDEV 124
Query: 126 LKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACS 185
LK YPQGHHGVDK+GRPVYIERLG VD KL+QVT+++RY+ YHV EFER F VK PACS
Sbjct: 125 LKYYPQGHHGVDKEGRPVYIERLGLVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACS 184
Query: 186 IAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFR 245
IAAKKHIDQSTTILDVQGVGL++ NKAARDL+ RLQK+DGDNYPE+LNRMFIINAGSGFR
Sbjct: 185 IAAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFR 244
Query: 246 LLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKG 305
+LWNTVKSFLDPKTT+KIHVLGNKYQSKLLE+ID SELPEFLGG+CTCAD GGCM SDKG
Sbjct: 245 MLWNTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDESELPEFLGGSCTCADNGGCMRSDKG 304
Query: 306 PWNDPEILKMAQNGVGRYTI-KALSGVEEKTIKQEETAYQKGFKDSFPETLDVHCLDQPK 364
PW +PEI+K NG + + EKTI +E+ + + + + +V + P
Sbjct: 305 PWKNPEIMKRVHNGDHKCSKGSQAENSGEKTIPEEDDSTTEPASEEEKASKEVEIV--PA 362
Query: 365 SYGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMAL 424
++ + + +A S K + YA+ + C NN G S F G+MAL
Sbjct: 363 AHPAWN-------MPEAHKFSLSK---KEVYAIQEAC---NNATTEGGRSPIFT-GVMAL 408
Query: 425 VMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIKAA---------AISNNEYMAMMK 475
VMG+VT+I++T ++PRK+TE+ LY D SM K+A AIS ++MA+MK
Sbjct: 409 VMGVVTMIKVTKNVPRKLTESTLYSSPVYCDDASMNKSAMQSEKMTVPAISGEDFMAIMK 468
Query: 476 RMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEG 534
RMAELE+KVTVLS +P VMPP+KEEMLN A++R + LEQEL ATKKAL+D+L RQ EL
Sbjct: 469 RMAELEQKVTVLSAQPTVMPPDKEEMLNAAISRSNVLEQELAATKKALDDSLGRQEELVA 528
Query: 535 QIDKKKKKKK 544
I+KKKKKKK
Sbjct: 529 YIEKKKKKKK 538
>UniRef100_Q9SYY7 Hypothetical protein C7A10.870 [Arabidopsis thaliana]
Length = 558
Score = 621 bits (1602), Expect = e-176
Identities = 336/550 (61%), Positives = 413/550 (75%), Gaps = 32/550 (5%)
Query: 7 EKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSI-CIEDSFDAEELQAVDALRQTLI 65
E K ++GS KK S+S + S TK+ R+ S+VMS+ IED DAEEL+AVDA RQ+LI
Sbjct: 24 ELKPRMGSFKK--RSSSKNLRYSMTKR-RRSSKVMSVEIIEDVHDAEELKAVDAFRQSLI 80
Query: 66 LEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEV 125
L+ELLP KHDD HMMLRFL+ARK+D+EKTKQMWT+ML+WRKEFGADT+ME F+F+E+DEV
Sbjct: 81 LDELLPEKHDDYHMMLRFLKARKFDLEKTKQMWTEMLRWRKEFGADTVME-FDFKEIDEV 139
Query: 126 LKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACS 185
LK YPQGHHGVDK+GRPVYIERLG VD KL+QVT+++RY+ YHV EFER F VK PACS
Sbjct: 140 LKYYPQGHHGVDKEGRPVYIERLGLVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACS 199
Query: 186 IAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFR 245
IAAKKHIDQSTTILDVQGVGL++ NKAARDL+ RLQK+DGDNYPE+LNRMFIINAGSGFR
Sbjct: 200 IAAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFR 259
Query: 246 LLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKG 305
+LWNTVKSFLDPKTT+KIHVLGNKYQSKLLE+ID SELPEFLGG+CTCAD GGCM SDKG
Sbjct: 260 MLWNTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDESELPEFLGGSCTCADNGGCMRSDKG 319
Query: 306 PWNDPEILKMAQNGVGRYTI-KALSGVEEKTIKQEETAYQKGFKDSFPETLDVHCLDQPK 364
PW +PEI+K NG + + EKTI +E+ + + + + +V + P
Sbjct: 320 PWKNPEIMKRVHNGDHKCSKGSQAENSGEKTIPEEDDSTTEPASEEEKASKEVEIV--PA 377
Query: 365 SYGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMAL 424
++ + + +A S K + YA+ + C NN G S F G+MAL
Sbjct: 378 AHPAWN-------MPEAHKFSLSK---KEVYAIQEAC---NNATTEGGRSPIFT-GVMAL 423
Query: 425 VMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIKAA---------AISNNEYMAMMK 475
VMG+VT+I++T ++PRK+TE+ LY D SM K+A AIS ++MA+MK
Sbjct: 424 VMGVVTMIKVTKNVPRKLTESTLYSSPVYCDDASMNKSAMQSEKMTVPAISGEDFMAIMK 483
Query: 476 RMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEG 534
RMAELE+KVTVLS +P VMPP+KEEMLN A++R + LEQEL ATKKAL+D+L RQ EL
Sbjct: 484 RMAELEQKVTVLSAQPTVMPPDKEEMLNAAISRSNVLEQELAATKKALDDSLGRQEELVA 543
Query: 535 QIDKKKKKKK 544
I+KKKKKKK
Sbjct: 544 YIEKKKKKKK 553
>UniRef100_Q9SI13 Putative phosphatidylinositol/phophatidylcholine transfer protein
[Arabidopsis thaliana]
Length = 558
Score = 613 bits (1580), Expect = e-174
Identities = 331/556 (59%), Positives = 404/556 (72%), Gaps = 37/556 (6%)
Query: 8 KKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICI-EDSFDAEELQAVDALRQTLIL 66
++ V S KK + SK S TKK R+ S+VMS+ I ED DAEEL+ VDA RQ LIL
Sbjct: 13 ERPNVCSFKK---RSCSKLSCSLTKK-RRSSKVMSVEIFEDEHDAEELKVVDAFRQVLIL 68
Query: 67 EELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVL 126
+ELLP KHDD HMMLRFL+ARK+D+EKT QMW+DML+WRKEFGADT+MEDFEF+E+DEVL
Sbjct: 69 DELLPDKHDDYHMMLRFLKARKFDLEKTNQMWSDMLRWRKEFGADTVMEDFEFKEIDEVL 128
Query: 127 KCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSI 186
K YPQGHHGVDK+GRPVYIERLGQVD KL+QVT+++RY+ YHV EFER F VK PACSI
Sbjct: 129 KYYPQGHHGVDKEGRPVYIERLGQVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACSI 188
Query: 187 AAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRL 246
AAKKHIDQSTTILDVQGVGL++ NKAARDL+ RLQK+DGDNYPE+LNRMFIINAGSGFR+
Sbjct: 189 AAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFRM 248
Query: 247 LWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGP 306
LWNTVKSFLDPKTT+KIHVLGNKYQSKLLE+IDASELPEFLGG+CTCAD GGCM SDKGP
Sbjct: 249 LWNTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDASELPEFLGGSCTCADNGGCMRSDKGP 308
Query: 307 WNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDV-HCLDQPKS 365
WN+P+I+K NG + + E +G + E + P
Sbjct: 309 WNNPDIMKRVNNG------DHICSKRSQADNAGENIISQGNNSAVEEAPETDQSQPSPCQ 362
Query: 366 YGVYQYDSF-VPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMAL 424
V + ++ +P K S + D YA+ + C + N S F+ G+MA
Sbjct: 363 NVVVAHPAWNIPEAHKFSLS------KRDVYAIQEACKATNESGRSPIFT-----GVMAF 411
Query: 425 VMGIVTIIRMTSSMPRKITEAALYGGNSVYYD----------GSMIKAAAISNNEYMAMM 474
VMG+VT+IR+T ++PRK+TE+ +Y + VY D G + IS ++MA+M
Sbjct: 412 VMGVVTMIRVTKNVPRKLTESTIY-SSPVYCDENSMNKSSMHGKKMATTTISGEDFMAVM 470
Query: 475 KRMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELE 533
KRMAELE+KVT LS +P MPPEKEEMLN A++R LEQEL ATKKAL+D+LTRQ +L
Sbjct: 471 KRMAELEQKVTNLSAQPATMPPEKEEMLNAAISRADFLEQELAATKKALDDSLTRQEDLV 530
Query: 534 GQIDKKKKKKKKLVRY 549
++ +KKKKKKLVR+
Sbjct: 531 AYVE-RKKKKKKLVRF 545
>UniRef100_Q9T027 SEC14-like protein [Arabidopsis thaliana]
Length = 550
Score = 607 bits (1564), Expect = e-172
Identities = 312/525 (59%), Positives = 394/525 (74%), Gaps = 21/525 (4%)
Query: 1 MEHSEDEKK-KKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDA 59
+E SED+K+ K+ S+KK A++A++KFK+S TKKGR+HSRV + I D D EELQAVDA
Sbjct: 17 VEISEDDKRLTKLCSLKKKAINATNKFKHSMTKKGRRHSRVACVSIVDEIDTEELQAVDA 76
Query: 60 LRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEF 119
RQ LIL+ELLPSKHDD HMMLRFLRARK+D+EK KQMW+DML WRKE+GADTIMEDF+F
Sbjct: 77 FRQALILDELLPSKHDDHHMMLRFLRARKFDLEKAKQMWSDMLNWRKEYGADTIMEDFDF 136
Query: 120 EELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAV 179
+E++EV+K YPQG+HGVDK+GRP+YIERLGQVD KL++VT+++RY+KYHV+EFE+ F V
Sbjct: 137 KEIEEVVKYYPQGYHGVDKEGRPIYIERLGQVDATKLMKVTTIDRYVKYHVKEFEKTFNV 196
Query: 180 KLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIIN 239
K PACSIAAK+HIDQSTTILDVQGVGL + NKAA+DLLQ +QKID DNYPE+LNRMFIIN
Sbjct: 197 KFPACSIAAKRHIDQSTTILDVQGVGLSNFNKAAKDLLQSIQKIDNDNYPETLNRMFIIN 256
Query: 240 AGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGC 299
AG GFRLLWNTVKSFLDPKTT+KIHVLGNKYQ+KLLE+IDA+ELPEFLGG CTCADKGGC
Sbjct: 257 AGCGFRLLWNTVKSFLDPKTTAKIHVLGNKYQTKLLEIIDANELPEFLGGKCTCADKGGC 316
Query: 300 MLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVHC 359
M SDKGPWNDPEI K+ QNG GR ++LSG+EEKTI + +K K ET
Sbjct: 317 MRSDKGPWNDPEIFKLVQNGEGRCLRRSLSGIEEKTIFEYNNETKK--KCEPEETHKQSA 374
Query: 360 LDQPKSYGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALS--KDCFSNNNGMNSSGFSKQF 417
+ K + D+ V D A + W + + + KD +S N + G+
Sbjct: 375 AEMEKKF----IDTNV---DAAAAADWPTKLNKAEKNPTDLKDVYSAVNPLERKGY---L 424
Query: 418 VGGIMALVMGIVTIIRMTSSMPRKITEAALYG--GNSVYYDGSMIKAAAISNNEYMAMMK 475
G +MAL+MGIV ++R+T +MPR++TEA +Y G++VY DG +S EY+AM+K
Sbjct: 425 YGSVMALLMGIVGVMRLTKNMPRRLTEANVYSREGSAVYQDG----VTVMSKQEYIAMVK 480
Query: 476 RMAELEEKVTVLSVKPVMPPEKEEMLNNALTRVSTLEQELGATKK 520
++ +LEEK + + E+E+ L+ AL R+ LE +L T K
Sbjct: 481 KITDLEEKCKSMEAQAAFYMEREKTLDAALRRIDQLELQLSETNK 525
>UniRef100_Q6ZG68 Putative SEC14 cytosolic factor [Oryza sativa]
Length = 605
Score = 598 bits (1542), Expect = e-169
Identities = 326/585 (55%), Positives = 421/585 (71%), Gaps = 45/585 (7%)
Query: 1 MEHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDAL 60
+E+SEDEKK K+ S+KK A+SAS K ++S KKGR+ S+V+SI I D D EE+QAVDA
Sbjct: 30 VEYSEDEKKAKIISLKKKAMSASQKLRHSM-KKGRRSSKVISISIADERDPEEVQAVDAF 88
Query: 61 RQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFE 120
RQ L+LEELLPS HDD HMMLRFL+ARK+D+EK KQMW DML+WRKEF ADTI+EDFEFE
Sbjct: 89 RQLLVLEELLPSHHDDYHMMLRFLKARKFDVEKAKQMWVDMLQWRKEFAADTILEDFEFE 148
Query: 121 ELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVK 180
E D+V +CYPQG+HGVDK+GRPVYIERLGQ++ N+L+QVT+++R++K HVREFE+ FAVK
Sbjct: 149 EADKVAECYPQGYHGVDKEGRPVYIERLGQINVNRLMQVTTMDRFIKNHVREFEKNFAVK 208
Query: 181 LPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINA 240
PACSIAAK HIDQSTTILDVQGVG++ +KAARDL+ +LQKIDGDNYPE+L RMFIINA
Sbjct: 209 FPACSIAAKCHIDQSTTILDVQGVGMKQFSKAARDLIGQLQKIDGDNYPETLCRMFIINA 268
Query: 241 GSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCM 300
G GFRLLW+TVKSFLDPKTT+KIHVLGNKYQSKLLEVIDASELPEF GGTC C +GGCM
Sbjct: 269 GPGFRLLWSTVKSFLDPKTTAKIHVLGNKYQSKLLEVIDASELPEFFGGTCQC--EGGCM 326
Query: 301 LSDKGPWNDPEILKMAQNGVG---RYTIKALSGVEEKTIKQEETAYQKGFKDSFP----- 352
+DKGPW D EILKM Q+G G ++ L E+ I +++T + K ++SF
Sbjct: 327 KADKGPWKDAEILKMVQSGAGWCGNLSLNHLDAEEKMMICEDDTMHTKR-QESFKYEGCT 385
Query: 353 ------------ETLDVHCLDQPK-----SYGVY-----QYDSFVPVLDKAVDSSWK-KT 389
+L C + P SY + Y VP+++KA+D+ + K
Sbjct: 386 LSRKISCARIEHPSLSPVCEELPPTILPVSYALSLTLGSAYSCDVPMVEKAIDAICQSKG 445
Query: 390 IQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALYG 449
+ ++ ++K + +NG N + GGIMALVM I T++R++ +MP+K+ A L
Sbjct: 446 LPDENVTVTKAIVNASNGSNPPLY-----GGIMALVMSIATMLRVSRNMPKKVLGATLGA 500
Query: 450 GNSVYYDG---SMIKAAAISNNEYMAMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNAL 505
++ S I A+S EY++ KR++++EEKV + KP MP +KEEML A+
Sbjct: 501 QSTSKIQAQQLSEISVEAVSVAEYVSSTKRLSDIEEKVIAILTKPAEMPADKEEMLKTAV 560
Query: 506 TRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKK-KKLVRY 549
+RVS LE+EL ATKKAL++ L RQ E+ I+KKKKKK K+L R+
Sbjct: 561 SRVSALEEELAATKKALQETLERQEEIMAYIEKKKKKKSKRLFRW 605
>UniRef100_Q6Z6I5 Putative phosphatidylinositol/ phophatidylcholine transfer protein
[Oryza sativa]
Length = 632
Score = 568 bits (1465), Expect = e-160
Identities = 310/599 (51%), Positives = 409/599 (67%), Gaps = 66/599 (11%)
Query: 5 EDEKKKKV-----GSIKKVALSASSKFKN---SFTKKG----RKHSRVMSICIEDSFDAE 52
EDE++ + S+++ A+SASSK S + KG ++ S+VMS+ IED DAE
Sbjct: 32 EDERRARALSSSSSSLRQRAMSASSKLLRTSLSRSSKGAAARQRSSKVMSVSIEDVRDAE 91
Query: 53 ELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADT 112
E+++VDA RQTL+LEELLP++HDD HMMLRFLRARK+DI+K+KQMW+DML+WRKEFG+DT
Sbjct: 92 EMKSVDAFRQTLVLEELLPARHDDYHMMLRFLRARKFDIDKSKQMWSDMLQWRKEFGSDT 151
Query: 113 IMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVRE 172
I++DF+FEE+D+VL+ YPQGHHGVD+DGRPVYIE+LG +D KLLQVTS++RY+KYHVRE
Sbjct: 152 ILDDFQFEEMDQVLEHYPQGHHGVDRDGRPVYIEKLGAIDTAKLLQVTSMDRYVKYHVRE 211
Query: 173 FERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESL 232
FERAFAVK PACSIAAK+H+DQSTTILDV GVG ++ NKAARDL+ RLQK+DGDNYPE+L
Sbjct: 212 FERAFAVKFPACSIAAKRHVDQSTTILDVSGVGYKNFNKAARDLIGRLQKVDGDNYPETL 271
Query: 233 NRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCT 292
RMFIINAG GFRLLWNTVKSFLDPKTT+KIHVLGNKYQSKLLEVID SELPEFLGGTCT
Sbjct: 272 CRMFIINAGQGFRLLWNTVKSFLDPKTTAKIHVLGNKYQSKLLEVIDPSELPEFLGGTCT 331
Query: 293 CADKGGCMLSDKGPWNDPEILKMAQNGVGR--YTIKALSGVEEKTIKQEETAYQKGFKDS 350
C +GGCM SDKGPW DPEI+KM Q G+GR + + +EK I +++ ++S
Sbjct: 332 C--EGGCMRSDKGPWKDPEIIKMVQCGMGRCGFNSSGHTEADEKMITEDDIVAIPKKQES 389
Query: 351 F-----------------PETLDVHCLDQPKSYGV-------YQYDSFVPVLDKAVDSSW 386
P+ +H + +S +YD P+ DK +D +W
Sbjct: 390 IRRDSVDSPKIPREKIEHPQMSPLHEMSTSESKAPPGQEGSSSRYDDLFPMPDKNMDFNW 449
Query: 387 KKTIQNDKYALSKDCFSNNNGMNSSGFS--KQFVGGIMALVMGIVTIIRMTSSMPRKITE 444
+ +K AL++D +++ G + +Q V G MA VMG+V + R+ P++ +
Sbjct: 450 NGEVSAEKLALARDMYASLPDAYKHGDAGDRQVVTGFMAFVMGVVAMFRVGKIAPKRAMD 509
Query: 445 AALYGGNSVYYDGSMIK-------------------AAAISNNEYMAMMKRMAELEEKVT 485
AA+ + +M K +S +Y A++KR+ +LEEKV
Sbjct: 510 AAM----GIATMEAMAKNRKLMQQQQRQLEQLPGPDTVTVSTAQYEALIKRLGDLEEKVA 565
Query: 486 VLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKK 543
L+ +P MP +KE++L A+TRV LE EL +TKK LE + +Q E+ I+KKKKK+
Sbjct: 566 ALTSRPPEMPADKEDLLKAAVTRVEALETELESTKKLLETSSGQQEEVLAYIEKKKKKR 624
>UniRef100_Q94FN1 Phosphatidylinositol transfer-like protein III [Lotus japonicus]
Length = 625
Score = 565 bits (1456), Expect = e-159
Identities = 317/595 (53%), Positives = 396/595 (66%), Gaps = 48/595 (8%)
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHS---RVMSICIEDSFDAEELQAVD 58
E+SEDE++ ++GS+KK ALSAS+KFK+S KK + RV S+ IED D EELQAVD
Sbjct: 30 ENSEDERRTRIGSLKKKALSASTKFKHSLRKKSSRRKSDGRVSSVSIEDVRDVEELQAVD 89
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ+LI++ELLP DD HMMLRFL+ARK+DIEK K MW +ML+WRKEFGADTIM+DFE
Sbjct: 90 AFRQSLIMDELLPQAFDDYHMMLRFLKARKFDIEKAKHMWAEMLQWRKEFGADTIMQDFE 149
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
F+ELDEV++ YP GHHGVDK+GRPVYIERLG+VD NKL+QVT+++RY++YHV+EFE++FA
Sbjct: 150 FQELDEVVRYYPHGHHGVDKEGRPVYIERLGKVDPNKLMQVTTMDRYVRYHVQEFEKSFA 209
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
+K PAC+IAAK+HID STTILDVQGVGL++ K+AR+L+ RLQK+DGDNYPE+L +MFII
Sbjct: 210 IKFPACTIAAKRHIDSSTTILDVQGVGLKNFTKSARELITRLQKVDGDNYPETLCQMFII 269
Query: 239 NAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGG 298
NAG GFRLLWNTVKSFLDPKTTSKIHVLGNKY SKLLEVIDASELPEFLGG CTC D+GG
Sbjct: 270 NAGPGFRLLWNTVKSFLDPKTTSKIHVLGNKYHSKLLEVIDASELPEFLGGACTCEDQGG 329
Query: 299 CMLSDKGPWNDPEILKMAQNGV---GRYTIKALSGVEEKTIKQEETAYQ--KGFKDSFPE 353
C+ SDKGPW +PEILKM NG R +K L+ E K I + Y KG S E
Sbjct: 330 CLRSDKGPWKNPEILKMVLNGEPRRARQVVKVLNS-EGKVIAYAKPRYPTVKGSDTSTAE 388
Query: 354 T-LDVHCLDQPKSYGVYQ-------------------------YDSFVPVLDKAVDSSWK 387
+ + + PK+ Y YD +VP++DK VD+ K
Sbjct: 389 SGSEAEDIASPKAMKNYSHLRLTPVREEAKIVGKSSYTNNFSGYDEYVPMVDKPVDAVLK 448
Query: 388 KTIQNDKYALSKDCFSNNNGMNS-SGFSKQFVGGIMALVMGIVTII-----RMTSSMPR- 440
K + S+ S + G + + I A ++ I TI R+T +P
Sbjct: 449 KQASLQRSYTSQGAPSRPATQRTPEGIQARILVAITAFLLTIFTIFRQVACRVTKKLPAI 508
Query: 441 -----KITEAALYGGNSVYYDGSMIKAAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMP 494
+ T + V S A N +M+KR+ ELEEKV L KP MP
Sbjct: 509 SSNHDQSTSEPTFDTTVVEVIPSSSTPAHTEENLLPSMLKRLGELEEKVDTLQSKPSEMP 568
Query: 495 PEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKKKKLVRY 549
EKEE+LN A+ RV LE EL ATKKAL +AL RQ EL ID++ + K + R+
Sbjct: 569 YEKEELLNAAVCRVDALEAELIATKKALYEALMRQEELLAYIDRQAEAKLRKKRF 623
>UniRef100_Q9SIW3 Putative phosphatidylinositol/phosphatidylcholine transfer protein
[Arabidopsis thaliana]
Length = 547
Score = 563 bits (1450), Expect = e-159
Identities = 302/565 (53%), Positives = 393/565 (69%), Gaps = 41/565 (7%)
Query: 1 MEHSEDEKKK-KVGSIKKVALSASSKFKNSFTKKGRK-HSRVMSICIEDSFDAEELQAVD 58
ME+SED +K K+ S+K+ A+SAS++FKNSF KK R+ S+++S+ D + ++ +V+
Sbjct: 8 MENSEDGRKLVKMSSLKQKAISASNRFKNSFKKKTRRTSSKIVSVANTDDINGDDYLSVE 67
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ L+L++LLP KHDD HMMLRFLRARK+D EK KQMW+DML+WR +FG DTI+EDFE
Sbjct: 68 AFRQVLVLDDLLPPKHDDLHMMLRFLRARKFDKEKAKQMWSDMLQWRMDFGVDTIIEDFE 127
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
FEE+D+VLK YPQG+HGVDK+GRPVYIERLGQ+D NKLLQ T+++RY KYHV+EFE+ F
Sbjct: 128 FEEIDQVLKHYPQGYHGVDKEGRPVYIERLGQIDANKLLQATTMDRYEKYHVKEFEKMFK 187
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
+K P+CS AAKKHIDQSTTI DVQGVGL++ NK+AR+LLQRL KID DNYPE+LNRMFII
Sbjct: 188 IKFPSCSAAAKKHIDQSTTIFDVQGVGLKNFNKSARELLQRLLKIDNDNYPETLNRMFII 247
Query: 239 NAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGG 298
NAG GFRLLW +K FLDPKTTSKIHVLGNKYQ KLLE IDASELP F GG CTCADKGG
Sbjct: 248 NAGPGFRLLWAPIKKFLDPKTTSKIHVLGNKYQPKLLEAIDASELPYFFGGLCTCADKGG 307
Query: 299 CMLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVH 358
C+ SDKGPWNDPE+LK+A+N R+ + E+ + +E T+ F E L+
Sbjct: 308 CLRSDKGPWNDPELLKIARNPEARF---STISEEDYLLVEEGTSMSMVF-----EPLE-- 357
Query: 359 CLDQPKSYGVYQYDSFVPVLDKAVDSSWK-----KTIQNDKYALSKDCFSNNNGMNSSGF 413
++ K+ + + +DK + S KT++ K KD
Sbjct: 358 -RNKMKTIEENVSEKHIDAVDKFMALSLPPKPHLKTLRKGKEPQKKD------------- 403
Query: 414 SKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSM--------IKAAAI 465
VGG++A VMGIV ++R++ ++PRK+T+ AL NSVYY+ + + A +
Sbjct: 404 DSFLVGGVIAFVMGIVAMLRLSKAVPRKLTDVALL-TNSVYYEEAKMSKPNQDEVSAPPV 462
Query: 466 SNNEYMAMMKRMAELEEKVTVLSVKPVMPP-EKEEMLNNALTRVSTLEQELGATKKALED 524
S++EY+ M+KRMAELEEK L K EK++ L AL RV LE EL TKKAL++
Sbjct: 463 SSSEYVIMVKRMAELEEKYKSLDSKSADEALEKDDKLQAALNRVQVLEHELSETKKALDE 522
Query: 525 ALTRQVELEGQIDKKKKKKKKLVRY 549
+ Q + I+KK KKK+ R+
Sbjct: 523 TMVNQQGILAYIEKKNKKKRMFFRF 547
>UniRef100_Q6Z6I6 Putative phosphatidylinositol/ phophatidylcholine transfer protein
[Oryza sativa]
Length = 624
Score = 558 bits (1439), Expect = e-157
Identities = 308/599 (51%), Positives = 400/599 (66%), Gaps = 53/599 (8%)
Query: 1 MEHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKH-SRVMSICIEDSFDAEELQAVDA 59
+E+SED+K+ ++GS+KK A+ AS+K ++S KK R+ SRV+S+ IED D EELQAV+A
Sbjct: 29 VENSEDDKRTRMGSLKKKAIDASTKIRHSLKKKKRRSGSRVLSVSIEDVRDLEELQAVEA 88
Query: 60 LRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEF 119
RQ LIL+ELLP++HDD HMMLRFL+AR++DIEK KQMWTDMLKWRKE+G DTI+EDF++
Sbjct: 89 FRQALILDELLPARHDDYHMMLRFLKARRFDIEKAKQMWTDMLKWRKEYGTDTIVEDFDY 148
Query: 120 EELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAV 179
ELD VL+ YP G+HGVDKDGRPVYIERLG+VD NKL+ VT+++RY++YHV+EFER+F +
Sbjct: 149 NELDAVLQYYPHGYHGVDKDGRPVYIERLGKVDPNKLMHVTTMDRYVRYHVKEFERSFLI 208
Query: 180 KLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIIN 239
K PACS+AAK+HID STTILDVQGVGL++ +K AR+L+ RLQKID DNYPE+L +MFI+N
Sbjct: 209 KFPACSLAAKRHIDSSTTILDVQGVGLKNFSKTARELIVRLQKIDNDNYPETLYQMFIVN 268
Query: 240 AGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGC 299
AG GFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGG CTC + GGC
Sbjct: 269 AGPGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGACTCPEYGGC 328
Query: 300 MLSDKGPWNDPEILKMAQNGVGRYT--IKALSGVEEKTIKQEETAY------QKGFKDSF 351
+ ++KGPW D IL + +G + I +S EEK I ++ + +S
Sbjct: 329 LKAEKGPWKDQNILNIVLSGEAQCARQIVTVSNGEEKIISYAKSKHHTIRGSDTSTAESG 388
Query: 352 PETLDV-------HCLDQPK--------------SYGVYQYDSFVPVLDKAVDSSWKKTI 390
E DV + PK S+ + VPV+DKAVD++WK+ +
Sbjct: 389 SEAEDVTSPKVLRSYISHPKLTPVREEVKMVRATSFSTRMPEYDVPVVDKAVDATWKREV 448
Query: 391 QNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSS-----MPRKITEA 445
SKD + +S+G + V ++A+ M I+T++R +P K
Sbjct: 449 TRKTAFSSKDSSLTSTESSSNGSLDRIVAVLLAVFMAIITLVRSVKDLAAKRLPDKNESE 508
Query: 446 ALYGGNSVYYDGSMIK---------AAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMPP 495
Y S Y SM K + + ++++R+ +LEEK +L KP MP
Sbjct: 509 QKY---STLYPDSMPKEEFRPPSPTPGFVEAELFSSVLQRLGDLEEKFLMLQDKPSEMPC 565
Query: 496 EKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKK-----KKKKKKLVRY 549
EKEE+LN A+ RV LE EL TKKAL +AL RQ EL ID K +KKKK + Y
Sbjct: 566 EKEELLNAAVRRVDALEAELIVTKKALHEALIRQEELLAYIDSKEVAKAQKKKKAMFCY 624
>UniRef100_Q9SIK5 Putative phosphatidylinositol/phophatidylcholine transfer protein
[Arabidopsis thaliana]
Length = 371
Score = 556 bits (1432), Expect = e-157
Identities = 268/350 (76%), Positives = 309/350 (87%), Gaps = 2/350 (0%)
Query: 4 SEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDALRQT 63
SEDEKK K+ S+KK A++AS+KFK+SFTK+ R++SRVMS+ I D D EELQAVDA RQ
Sbjct: 20 SEDEKKTKLCSLKKKAINASNKFKHSFTKRTRRNSRVMSVSIVDDIDLEELQAVDAFRQA 79
Query: 64 LILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELD 123
LIL+ELLPSKHDD HMMLRFLRARK+D+EK KQMWTDM+ WRKEFG DTIMEDF+F+E+D
Sbjct: 80 LILDELLPSKHDDHHMMLRFLRARKFDLEKAKQMWTDMIHWRKEFGVDTIMEDFDFKEID 139
Query: 124 EVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPA 183
EVLK YPQG+HGVDKDGRPVYIERLGQVD KL+QVT+++RY+KYHVREFE+ F +KLPA
Sbjct: 140 EVLKYYPQGYHGVDKDGRPVYIERLGQVDATKLMQVTTIDRYVKYHVREFEKTFNIKLPA 199
Query: 184 CSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSG 243
CSIAAKKHIDQSTTILDVQGVGL+S +KAARDLLQR+QKID DNYPE+LNRMFIINAGSG
Sbjct: 200 CSIAAKKHIDQSTTILDVQGVGLKSFSKAARDLLQRIQKIDSDNYPETLNRMFIINAGSG 259
Query: 244 FRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSD 303
FRLLW+TVKSFLDPKTT+KIHVLGNKYQSKLLE+ID++ELPEFLGG CTCADKGGCM SD
Sbjct: 260 FRLLWSTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDSNELPEFLGGNCTCADKGGCMRSD 319
Query: 304 KGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEE--TAYQKGFKDSF 351
KGPWNDP+I KM QNG G+ K LS +EEKTI +E T + +K+ F
Sbjct: 320 KGPWNDPDIFKMVQNGEGKCPRKTLSNIEEKTISVDENTTMVKAKYKNIF 369
>UniRef100_Q94C34 AT4g39170/T22F8_70 [Arabidopsis thaliana]
Length = 583
Score = 553 bits (1426), Expect = e-156
Identities = 305/557 (54%), Positives = 388/557 (68%), Gaps = 42/557 (7%)
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAVDAL 60
E+SEDE++ ++GS+KK A++AS+KFK+S KK RK RV S+ IED D EELQAVD
Sbjct: 30 ENSEDERRTRIGSLKKKAINASTKFKHSLKKKRRKSDVRVSSVSIEDVRDVEELQAVDEF 89
Query: 61 RQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFE 120
RQ L++EELLP KHDD HMMLRFL+ARK+DIEK K MW DM++WRKEFG DTI++DF+FE
Sbjct: 90 RQALVMEELLPHKHDDYHMMLRFLKARKFDIEKAKHMWADMIQWRKEFGTDTIIQDFQFE 149
Query: 121 ELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVK 180
E+DEVLK YP G+H VDK+GRPVYIERLG+VD NKL+QVT+++RY++YHV+EFER+F +K
Sbjct: 150 EIDEVLKYYPHGYHSVDKEGRPVYIERLGKVDPNKLMQVTTLDRYIRYHVKEFERSFMLK 209
Query: 181 LPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINA 240
PAC+IAAKK+ID STTILDVQGVGL++ K+AR+L+ RLQKIDGDNYPE+L++MFIINA
Sbjct: 210 FPACTIAAKKYIDSSTTILDVQGVGLKNFTKSARELITRLQKIDGDNYPETLHQMFIINA 269
Query: 241 GSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCM 300
G GFRLLW+TVKSFLDPKTTSKIHVLG KYQSKLLE+ID+SELPEFLGG CTCAD+GGCM
Sbjct: 270 GPGFRLLWSTVKSFLDPKTTSKIHVLGCKYQSKLLEIIDSSELPEFLGGACTCADQGGCM 329
Query: 301 LSDKGPWNDPEILKMAQNG---VGRYTIKALSGVEEKTIKQEETAYQ--KGFKDSFPETL 355
LSDKGPW +PEI+KM +G ++ +K L+ + K I + +Y KG S E+
Sbjct: 330 LSDKGPWKNPEIVKMVLHGGAHRAKHVVKVLNS-DGKVIAYAKPSYPWIKGSDTSTAESG 388
Query: 356 D--VHCLDQPKSYGVYQ--------------------------YDSFVPVLDKAVDSSWK 387
+ PK+ Y YD +VP++DKAVD++WK
Sbjct: 389 SEAEDIVVSPKAVKSYSHLRLTPVREEAKVGSGETSFAGSFAGYDEYVPMVDKAVDATWK 448
Query: 388 KTIQNDKYALSKDC-FSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAA 446
A SK N + FS + + MA VM I+T R S+ R +T+
Sbjct: 449 VKPTAINRAPSKGAHMPPNVPKDHESFSARVLVTFMAFVMAILTFFRTVSN--RVVTKQL 506
Query: 447 LYGGNSVYYDGSMIKAAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNAL 505
+ +GS AAA + +++K++ ELEEK+ L KP MP EKEE+LN A+
Sbjct: 507 PPPPSQPQIEGS---AAAEEADLLNSVLKKLTELEEKIGALQSKPSEMPYEKEELLNAAV 563
Query: 506 TRVSTLEQELGATKKAL 522
RV LE EL ATKKAL
Sbjct: 564 CRVDALEAELIATKKAL 580
>UniRef100_Q9T026 SEC14-like protein [Arabidopsis thaliana]
Length = 617
Score = 534 bits (1376), Expect = e-150
Identities = 308/594 (51%), Positives = 396/594 (65%), Gaps = 51/594 (8%)
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAVDAL 60
E+SEDE++ ++GS+KK A++AS+KFK+S KK RK RV S+ IED D EELQAVD
Sbjct: 30 ENSEDERRTRIGSLKKKAINASTKFKHSLKKKRRKSDVRVSSVSIEDVRDVEELQAVDEF 89
Query: 61 RQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFE 120
RQ L++EELLP KHDD HMMLRFL+ARK+DIEK K MW DM++WRKEFG DTI++DF+FE
Sbjct: 90 RQALVMEELLPHKHDDYHMMLRFLKARKFDIEKAKHMWADMIQWRKEFGTDTIIQDFQFE 149
Query: 121 ELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVK 180
E+DEVLK YP G+H VDK+GRPVYIERLG+VD NKL+QVT+++RY++YHV+EFER+F +K
Sbjct: 150 EIDEVLKYYPHGYHSVDKEGRPVYIERLGKVDPNKLMQVTTLDRYIRYHVKEFERSFMLK 209
Query: 181 LPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINA 240
PAC+IAAKK+ID STTILDVQGVGL++ K+AR+L+ RLQKIDGDNYPE+L++MFIINA
Sbjct: 210 FPACTIAAKKYIDSSTTILDVQGVGLKNFTKSARELITRLQKIDGDNYPETLHQMFIINA 269
Query: 241 GSGFRLLWNTVKSFLDPKTTSKIH----VLGNKYQSKLLEVIDASELPEFLGGTCTCADK 296
G GFRLLW+TVKSFLDPKTTSKIH +L Y S + I SELPEFLGG CTCAD+
Sbjct: 270 GPGFRLLWSTVKSFLDPKTTSKIHNYSILLCFAYISD-VSFICFSELPEFLGGACTCADQ 328
Query: 297 GGCMLSDKGPWNDPEILKMAQNG---VGRYTIKALSGVEEKTIKQEETAYQ--KGFKDSF 351
GGCMLSDKGPW +PEI+KM +G + +K L+ + K I + +Y KG S
Sbjct: 329 GGCMLSDKGPWKNPEIVKMVLHGGAHRAKQVVKVLNS-DGKVIAYAKPSYPWIKGSDTST 387
Query: 352 PETLD--VHCLDQPKSYGVYQ--------------------------YDSFVPVLDKAVD 383
E+ + PK+ Y YD +VP++DKAVD
Sbjct: 388 AESGSEAEDIVVSPKAVKSYSHLRLTPVREEAKVGSGETSFAGSFAGYDEYVPMVDKAVD 447
Query: 384 SSWKKTIQNDKYALSKDC-FSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKI 442
++WK A SK N + FS + + MA VM I+T R S+ R +
Sbjct: 448 ATWKVKPTAINRAPSKGAHMPPNVPKDHESFSARVLVTFMAFVMAILTFFRTVSN--RVV 505
Query: 443 TEAALYGGNSVYYDGSMIKAAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMPPEKEEML 501
T+ + +GS AAA + +++K++ ELEEK+ L KP MP EKEE+L
Sbjct: 506 TKQLPPPPSQPQIEGS---AAAEEADLLNSVLKKLTELEEKIGALQSKPSEMPYEKEELL 562
Query: 502 NNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKK----KKKKKLVRYIC 551
N A+ RV LE EL ATKKAL +AL RQ EL ID+++ +KK K + C
Sbjct: 563 NAAVCRVDALEAELIATKKALYEALMRQEELLAYIDRQEAAQHQKKNKRKQMFC 616
>UniRef100_Q67YM1 Putative phosphatidylinositol/ phosphatidylcholine transfer protein
[Arabidopsis thaliana]
Length = 572
Score = 528 bits (1361), Expect = e-148
Identities = 295/567 (52%), Positives = 380/567 (66%), Gaps = 56/567 (9%)
Query: 34 GRKHS--RVMSICIEDSFDAEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDI 91
GR+ S RV S+ IED D EELQAVDA RQ+L+++ELLP +HDD HMMLRFL+ARK+D+
Sbjct: 2 GRRKSDGRVSSVSIEDVRDVEELQAVDAFRQSLLMDELLPDRHDDYHMMLRFLKARKFDV 61
Query: 92 EKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQV 151
EK KQMW DM++WRKEFG DTI++DF+FEE++EVLK YPQ +HGVDK+GRP+YIERLG+V
Sbjct: 62 EKAKQMWADMIQWRKEFGTDTIIQDFDFEEINEVLKHYPQCYHGVDKEGRPIYIERLGKV 121
Query: 152 DCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNK 211
D N+L+QVTS++RY++YHV+EFER+F +K P+C+I+AK+HID STTILDVQGVGL++ NK
Sbjct: 122 DPNRLMQVTSMDRYVRYHVKEFERSFMIKFPSCTISAKRHIDSSTTILDVQGVGLKNFNK 181
Query: 212 AARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQ 271
+ARDL+ RLQKIDGDNYPE+L++MFIINAG GFRLLWNTVKSFLDPKT++KIHVLG KY
Sbjct: 182 SARDLITRLQKIDGDNYPETLHQMFIINAGPGFRLLWNTVKSFLDPKTSAKIHVLGYKYL 241
Query: 272 SKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGPWNDPEILKMAQNG---VGRYTIKAL 328
SKLLEVID +ELPEFLGG CTCAD+GGCMLSDKGPW +PEI+KM +G R +K L
Sbjct: 242 SKLLEVIDVNELPEFLGGACTCADQGGCMLSDKGPWKNPEIVKMVLHGGAHRARQVVKVL 301
Query: 329 SGVEEKTIKQEETAYQ--KGFKDSFPET-LDVHCLDQPKSYGVYQ--------------- 370
+ E K I + +Y KG S E+ D + PK+ +
Sbjct: 302 NS-EGKVIAYAKPSYTWIKGSDTSTAESGSDAEDIGSPKAIKSFSHLRLTPVREEAKIAG 360
Query: 371 ----------YDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGG 420
YD +VP++DKAVD++WK + A S + G + +
Sbjct: 361 ETSLAGSFPGYDEYVPMVDKAVDATWKVKPAIQRVASRGALMSPTVPKDHEGIKARVLVM 420
Query: 421 IMALVMGIVTIIR-MTSSMPRKITEA-ALYGGNSVYY--DGSMIKAAA--------ISNN 468
MA +M + T R +T +P T + A GN++ +G +K ++
Sbjct: 421 FMAFLMAVFTFFRTVTKKLPATTTSSPAETQGNAIELGSNGEGVKEECRPPSPVPDLTET 480
Query: 469 EYM-AMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDAL 526
+ + + K++ ELE K+ L KP MP EKEE+LN A+ RV LE EL ATKKAL +AL
Sbjct: 481 DLLNCVTKKLTELEGKIGTLQSKPNEMPYEKEELLNAAVCRVDALEAELIATKKALYEAL 540
Query: 527 TRQVEL--------EGQIDKKKKKKKK 545
RQ EL E Q K KKKKKK
Sbjct: 541 MRQEELLAYIDRQEEAQFQKMKKKKKK 567
>UniRef100_Q8GXC6 Putative sec14 cytosolic factor [Arabidopsis thaliana]
Length = 612
Score = 507 bits (1306), Expect = e-142
Identities = 296/593 (49%), Positives = 379/593 (62%), Gaps = 54/593 (9%)
Query: 2 EHSEDEKKKKVGSI--KKVALSASSKFKNSFTKKG----RKHSRVMSICIEDSFDAEELQ 55
E SEDEKK ++G+ KK A ASSK ++S KKG R R S+ IED D EEL+
Sbjct: 30 EVSEDEKKTRIGNFNFKKKAAKASSKLRHSLKKKGSSRRRSSDRTFSLTIEDIHDVEELR 89
Query: 56 AVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIME 115
AVD R L+ E LLP DD H+MLRFL+ARK+DI KTK MW++M+KWRK+FG DTI E
Sbjct: 90 AVDEFRNLLVSENLLPPTLDDYHIMLRFLKARKFDIGKTKLMWSNMIKWRKDFGTDTIFE 149
Query: 116 DFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFER 175
DFEFEE DEVLK YP G+HGVDK+GRPVYIERLG VD KL+QVT+VER+++YHVREFE+
Sbjct: 150 DFEFEEFDEVLKYYPHGYHGVDKEGRPVYIERLGLVDPAKLMQVTTVERFIRYHVREFEK 209
Query: 176 AFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRM 235
+KLPAC IAAK+HID STTILDVQGVG ++ +K ARDL+ +LQKID DNYPE+L+RM
Sbjct: 210 TVNIKLPACCIAAKRHIDSSTTILDVQGVGFKNFSKPARDLIIQLQKIDNDNYPETLHRM 269
Query: 236 FIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
FIIN GSGF+L+W TVK FLDPKT +KIHV+GNKYQ+KLLE+IDAS+LP+FLGGTCTCAD
Sbjct: 270 FIINGGSGFKLVWATVKQFLDPKTVTKIHVIGNKYQNKLLEIIDASQLPDFLGGTCTCAD 329
Query: 296 KGGCMLSDKGPWNDPEILKMAQNG---------------VGRYTIKALSGVE-------E 333
+GGCM SDKGPWNDPEILKM Q+G V + SG++ E
Sbjct: 330 RGGCMRSDKGPWNDPEILKMLQSGGPLCRHNSALNSFSRVSSCDKPSFSGIKASDTSTAE 389
Query: 334 KTIKQEETAYQKGFKDSFPETLDVHCLDQPKSYGVY-----QYDSFVPVLDKAVDSSWKK 388
+ EE A K ++ L C D + Y +YDS P++DK VD +W
Sbjct: 390 SGSEVEEMASPKVNRELRVPKLTPVCEDIRGTAISYPTDSSEYDS--PMVDKVVDVAW-- 445
Query: 389 TIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALY 448
+ ++K SK + SG + L+M V + + S+ E
Sbjct: 446 -MAHEKPKASK----GSEDTPDSGKIRTVTYIWRWLMMFFVNLFTLLISLALPQREGHSQ 500
Query: 449 GGNSVYYDG---------SMIKAAAISNNEYMAMMKRMAELEEKV-TVLSVKPVMPPEKE 498
+SV DG S A N + +++ R+ +LE++V T+ S + MP EKE
Sbjct: 501 SESSV--DGPNARESRPPSPAFATIAERNVFSSVVNRLGDLEKQVETLHSKRHEMPREKE 558
Query: 499 EMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKKKKLVRYIC 551
E+LN A+ RV LE EL ATKKAL +AL RQ +L ID+++ +K + +C
Sbjct: 559 ELLNTAVYRVDALEAELIATKKALHEALMRQDDLLAYIDREEDEKYHKKKKVC 611
>UniRef100_Q94FN0 Phosphatidylinositol transfer-like protein IV [Lotus japonicus]
Length = 482
Score = 501 bits (1289), Expect = e-140
Identities = 238/330 (72%), Positives = 290/330 (87%), Gaps = 1/330 (0%)
Query: 14 SIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDALRQTLILEELLPSK 73
S KK A++AS+ +NS T+KGR+ S+VMS+ IED DAEEL+AV+ RQ LI ++LLP+K
Sbjct: 11 SFKKKAINASNMLRNSLTRKGRRSSKVMSVEIEDVHDAEELKAVEEFRQALISDDLLPAK 70
Query: 74 HDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGH 133
HDD HMMLRFL+ARK++I+K+KQMW+DMLKWRKEFGADTI+E+FEF+E+DEVLK YPQGH
Sbjct: 71 HDDYHMMLRFLKARKFEIDKSKQMWSDMLKWRKEFGADTIVEEFEFKEIDEVLKYYPQGH 130
Query: 134 HGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKHID 193
HGVDK+GRPVYIE+LGQVD KL+QVT+++RY+KYHV+EFE+ F +K ACSIAAKKHID
Sbjct: 131 HGVDKEGRPVYIEQLGQVDATKLMQVTTMDRYIKYHVKEFEKTFDLKFAACSIAAKKHID 190
Query: 194 QSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKS 253
QSTTILDVQGVGL+S NK AR+L+ R+QK+DGDNYPE+LNRMFIINAGSGFR+LWNTVKS
Sbjct: 191 QSTTILDVQGVGLKSFNKHARELVTRIQKVDGDNYPETLNRMFIINAGSGFRILWNTVKS 250
Query: 254 FLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGPWNDPEIL 313
FLDPKTT+KI+VLGNKY +KLLE+IDASELPEFLGGTCTC D+GGCM SDKGPW D EIL
Sbjct: 251 FLDPKTTAKINVLGNKYDTKLLEIIDASELPEFLGGTCTCTDQGGCMRSDKGPWKDEEIL 310
Query: 314 KMAQNGVGRYTIKALS-GVEEKTIKQEETA 342
+M QNG + + K S G EEK I +++T+
Sbjct: 311 RMVQNGAHKCSRKPESHGEEEKPISEDKTS 340
Score = 88.6 bits (218), Expect = 4e-16
Identities = 45/88 (51%), Positives = 63/88 (71%), Gaps = 4/88 (4%)
Query: 466 SNNEYMAMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALED 524
++ E+ +MKRMAELEEK+T ++ +P MPPEKEEMLN ++R LE++L TKKALED
Sbjct: 398 TSKEFTTVMKRMAELEEKMTTMNHQPATMPPEKEEMLNATISRADVLEKQLMDTKKALED 457
Query: 525 ALTRQVELEGQIDKKKKKKKKLVRYICC 552
+L +Q L ++KKK+KKK + CC
Sbjct: 458 SLAKQEVLSAYVEKKKQKKK---TFFCC 482
>UniRef100_Q9SJS7 Putative phosphatidylinositol/phosphatidylcholine transfer protein
[Arabidopsis thaliana]
Length = 531
Score = 497 bits (1280), Expect = e-139
Identities = 274/527 (51%), Positives = 355/527 (66%), Gaps = 48/527 (9%)
Query: 66 LEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEV 125
++ELLP +HDD HMMLRFL+ARK+D+EK KQMW DM++WRKEFG DTI++DF+FEE++EV
Sbjct: 1 MDELLPDRHDDYHMMLRFLKARKFDVEKAKQMWADMIQWRKEFGTDTIIQDFDFEEINEV 60
Query: 126 LKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACS 185
LK YPQ +HGVDK+GRP+YIERLG+VD N+L+QVTS++RY++YHV+EFER+F +K P+C+
Sbjct: 61 LKHYPQCYHGVDKEGRPIYIERLGKVDPNRLMQVTSMDRYVRYHVKEFERSFMIKFPSCT 120
Query: 186 IAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFR 245
I+AK+HID STTILDVQGVGL++ NK+ARDL+ RLQKIDGDNYPE+L++MFIINAG GFR
Sbjct: 121 ISAKRHIDSSTTILDVQGVGLKNFNKSARDLITRLQKIDGDNYPETLHQMFIINAGPGFR 180
Query: 246 LLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKG 305
LLWNTVKSFLDPKT++KIHVLG KY SKLLEVID +ELPEFLGG CTCAD+GGCMLSDKG
Sbjct: 181 LLWNTVKSFLDPKTSAKIHVLGYKYLSKLLEVIDVNELPEFLGGACTCADQGGCMLSDKG 240
Query: 306 PWNDPEILKMAQNG---VGRYTIKALSGVEEKTIKQEETAYQ--KGFKDSFPET-LDVHC 359
PW +PEI+KM +G R +K L+ E K I + +Y KG S E+ D
Sbjct: 241 PWKNPEIVKMVLHGGAHRARQVVKVLNS-EGKVIAYAKPSYTWIKGSDTSTAESGSDAED 299
Query: 360 LDQPKSYGVYQ-------------------YDSFVPVLDKAVDSSWKKTIQNDKYALSKD 400
+ PK+ + YD +VP++DKAVD++WK + A
Sbjct: 300 IGSPKAIKSFSHLRLTPVPGETSLAGSFPGYDEYVPMVDKAVDATWKVKPAIQRVASRGA 359
Query: 401 CFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIR-MTSSMPRKITEA-ALYGGNSVYY--D 456
S + G + + MA +M + T R +T +P T + A GN++ +
Sbjct: 360 LMSPTVPKDHEGIKARVLVMFMAFLMAVFTFFRTVTKKLPATTTSSPAETQGNAIELGSN 419
Query: 457 GSMIKAAA--------ISNNEYM-AMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNALT 506
G +K ++ + + + K++ ELE K+ L KP MP EKEE+LN A+
Sbjct: 420 GEGVKEECRPPSPVPDLTETDLLNCVTKKLTELEGKIGTLQSKPNEMPYEKEELLNAAVC 479
Query: 507 RVSTLEQELGATKKALEDALTRQVEL--------EGQIDKKKKKKKK 545
RV LE EL ATKKAL +AL RQ EL E Q K KKKKKK
Sbjct: 480 RVDALEAELIATKKALYEALMRQEELLAYIDRQEEAQFQKMKKKKKK 526
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 909,791,238
Number of Sequences: 2790947
Number of extensions: 38486439
Number of successful extensions: 142216
Number of sequences better than 10.0: 447
Number of HSP's better than 10.0 without gapping: 243
Number of HSP's successfully gapped in prelim test: 204
Number of HSP's that attempted gapping in prelim test: 141357
Number of HSP's gapped (non-prelim): 558
length of query: 555
length of database: 848,049,833
effective HSP length: 133
effective length of query: 422
effective length of database: 476,853,882
effective search space: 201232338204
effective search space used: 201232338204
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 77 (34.3 bits)
Medicago: description of AC140722.2


