

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.1 - phase: 0
(313 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
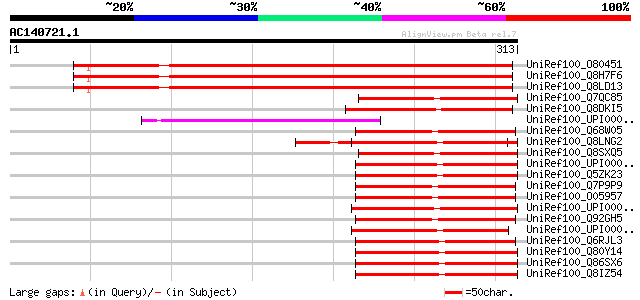
Sequences producing significant alignments: (bits) Value
UniRef100_O80451 Expressed protein [Arabidopsis thaliana] 364 1e-99
UniRef100_Q8H7F6 Hypothetical protein [Arabidopsis thaliana] 362 9e-99
UniRef100_Q8LD13 Hypothetical protein [Arabidopsis thaliana] 361 2e-98
UniRef100_Q7QC85 ENSANGP00000022155 [Anopheles gambiae str. PEST] 108 2e-22
UniRef100_Q8DKI5 Ycf64 protein [Synechococcus elongatus] 107 3e-22
UniRef100_UPI00002B0665 UPI00002B0665 UniRef100 entry 107 4e-22
UniRef100_Q68W05 Glutaredoxin 3 [Rickettsia typhi] 107 4e-22
UniRef100_Q8LNG2 Putative PKCq-interacting protein [Oryza sativa] 107 5e-22
UniRef100_Q8SXQ5 RH03087p [Drosophila melanogaster] 107 5e-22
UniRef100_UPI00003ADFB5 UPI00003ADFB5 UniRef100 entry 106 7e-22
UniRef100_Q5ZK23 Hypothetical protein [Gallus gallus] 106 7e-22
UniRef100_Q7P9P9 Glutaredoxin-like protein grla [Rickettsia sibi... 105 1e-21
UniRef100_O05957 Probable monothiol glutaredoxin grlA [Rickettsi... 105 2e-21
UniRef100_UPI000033F072 UPI000033F072 UniRef100 entry 105 2e-21
UniRef100_Q92GH5 Probable monothiol glutaredoxin grlA [Rickettsi... 105 2e-21
UniRef100_UPI00002A410B UPI00002A410B UniRef100 entry 104 3e-21
UniRef100_Q6RJL3 Putative glutaredoxin-like protein [uncultured ... 104 3e-21
UniRef100_Q80Y14 2900070E19Rik protein [Mus musculus] 103 5e-21
UniRef100_Q86SX6 Full-length cDNA clone CS0DL002YE04 of B cells ... 103 5e-21
UniRef100_Q8IZ54 Chromosome 14 open reading frame 87 [Homo sapiens] 103 5e-21
>UniRef100_O80451 Expressed protein [Arabidopsis thaliana]
Length = 293
Score = 364 bits (935), Expect = 1e-99
Identities = 176/273 (64%), Positives = 223/273 (81%), Gaps = 8/273 (2%)
Query: 40 QNTPTLSL--NFHPNSSTNLRPISLKPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGL 97
+NTP ++L F P+ S +L+ P + +S+ +A AVK+L +T + ++
Sbjct: 25 RNTPVITLYSRFTPSFSFPSLSFTLRDTAPSRRRSFFIASAVKSLTETELLPITE----- 79
Query: 98 TGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRES 157
+ +PS +GVYAVYDK+ ELQF+G+SRNIAA+V AH KSVPELCGS KVG+V+EPD+
Sbjct: 80 -ADSIPSASGVYAVYDKSDELQFVGISRNIAASVSAHLKSVPELCGSVKVGIVEEPDKAV 138
Query: 158 LTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVD 217
LTQAWK W+EEHIK+TGKVPPGN+SGN T+V+ P+KK+D+RLTPGRHV+LTVPLEEL+D
Sbjct: 139 LTQAWKLWIEEHIKVTGKVPPGNKSGNNTFVKQTPRKKSDIRLTPGRHVELTVPLEELID 198
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
+LVKE+KVVAFIKGSRSAP CGFSQ+V+GILE +GVDYE+VDVLD++YN+GLRETLK YS
Sbjct: 199 RLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGVDYETVDVLDDEYNHGLRETLKNYS 258
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
NWPTFPQIF+ GELVGGCDILTSM E GE+A +
Sbjct: 259 NWPTFPQIFVKGELVGGCDILTSMYENGELANI 291
>UniRef100_Q8H7F6 Hypothetical protein [Arabidopsis thaliana]
Length = 293
Score = 362 bits (928), Expect = 9e-99
Identities = 175/273 (64%), Positives = 222/273 (81%), Gaps = 8/273 (2%)
Query: 40 QNTPTLSL--NFHPNSSTNLRPISLKPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGL 97
+NTP ++L F P+ S +L+ P + +S+ +A AVK+L +T + ++
Sbjct: 25 RNTPVITLYSRFTPSFSFPSLSFTLRDTAPSRRRSFFIASAVKSLTETELLPITE----- 79
Query: 98 TGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRES 157
+ +PS +GVYAVYDK+ ELQF+G+SRNIAA+V AH KSVPELCGS KVG+V+EPD+
Sbjct: 80 -ADSIPSASGVYAVYDKSDELQFVGISRNIAASVSAHLKSVPELCGSVKVGIVEEPDKAV 138
Query: 158 LTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVD 217
LTQAWK W+EEHIK+TGKVPPGN+SGN T+V+ P+KK+D+RLTPGRHV+LTVPL EL+D
Sbjct: 139 LTQAWKLWIEEHIKVTGKVPPGNKSGNNTFVKQTPRKKSDIRLTPGRHVELTVPLGELID 198
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
+LVKE+KVVAFIKGSRSAP CGFSQ+V+GILE +GVDYE+VDVLD++YN+GLRETLK YS
Sbjct: 199 RLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGVDYETVDVLDDEYNHGLRETLKNYS 258
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
NWPTFPQIF+ GELVGGCDILTSM E GE+A +
Sbjct: 259 NWPTFPQIFVKGELVGGCDILTSMYENGELANI 291
>UniRef100_Q8LD13 Hypothetical protein [Arabidopsis thaliana]
Length = 293
Score = 361 bits (926), Expect = 2e-98
Identities = 175/273 (64%), Positives = 222/273 (81%), Gaps = 8/273 (2%)
Query: 40 QNTPTLSL--NFHPNSSTNLRPISLKPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGL 97
+NTP ++L F P+ S +L+ P + +S+ +A AVK+L +T + ++
Sbjct: 25 RNTPVITLYSRFTPSFSFPSLSFTLRDTAPSRRRSFFIASAVKSLTETELLPITE----- 79
Query: 98 TGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRES 157
+ +PS +GVYAVYDK+ ELQF+G+SRNIAA+V AH KSVPELCGS KVG+V+EPD+
Sbjct: 80 -ADSIPSASGVYAVYDKSDELQFVGISRNIAASVSAHLKSVPELCGSVKVGIVEEPDKAV 138
Query: 158 LTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVD 217
LTQAWK W+EEHIK+TGKVPPGN+SGN T+V+ P+KK+D+RLTPGRHV+LTV LEEL+D
Sbjct: 139 LTQAWKLWIEEHIKVTGKVPPGNKSGNNTFVKQTPRKKSDIRLTPGRHVELTVXLEELID 198
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
+LVKE+KVVAFIKGSRSAP CGFSQ+V+GILE +GVDYE+VDVLD++YN+GLRETLK YS
Sbjct: 199 RLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGVDYETVDVLDDEYNHGLRETLKNYS 258
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
NWPTFPQIF+ GELVGGCDILTSM E GE+A +
Sbjct: 259 NWPTFPQIFVKGELVGGCDILTSMYENGELANI 291
>UniRef100_Q7QC85 ENSANGP00000022155 [Anopheles gambiae str. PEST]
Length = 121
Score = 108 bits (270), Expect = 2e-22
Identities = 52/98 (53%), Positives = 66/98 (67%), Gaps = 3/98 (3%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
++KLV NKVV F+KG+ AP CGFS V+ IL V Y+S DVL N LR+ +K
Sbjct: 15 IEKLVSNNKVVVFMKGNPDAPRCGFSNAVVQILRMHSVKYDSHDVLQ---NEALRQGIKD 71
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+SNWPT PQ+F+NGE VGGCDIL M++ GE+ KK
Sbjct: 72 FSNWPTIPQVFINGEFVGGCDILLQMHQNGELIDELKK 109
>UniRef100_Q8DKI5 Ycf64 protein [Synechococcus elongatus]
Length = 121
Score = 107 bits (268), Expect = 3e-22
Identities = 49/103 (47%), Positives = 70/103 (67%), Gaps = 3/103 (2%)
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
+T L +D LVK NK++ F+KGS+ P CGFS + IL GV YE+VDVL++ +
Sbjct: 15 MTPELHAKIDNLVKSNKIIVFMKGSKLMPQCGFSNNAVQILNALGVPYETVDVLED---F 71
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
+R+ +K+YSNWPT PQ+F+NGE +GG DIL + + GE+ L
Sbjct: 72 EIRQGIKEYSNWPTIPQVFINGEFIGGSDILIELYQSGELQQL 114
>UniRef100_UPI00002B0665 UPI00002B0665 UniRef100 entry
Length = 150
Score = 107 bits (267), Expect = 4e-22
Identities = 57/150 (38%), Positives = 87/150 (58%), Gaps = 4/150 (2%)
Query: 82 LADTSPVTVSPENDGLTGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVLAHRKSVPEL 141
LAD V + NDG +P+ GVYA+YD G LQ++GLSR + A+V H +P+
Sbjct: 3 LADADAVLLV--NDGHADARIPTSPGVYAIYDARGTLQYVGLSRRVNASVKVHAFEMPQY 60
Query: 142 CGSFKVGVVDEPDRESLTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQPK-KKADLRL 200
C + + ++ + L AWK+W+ EH+ G +PPGN GN + + + KA +RL
Sbjct: 61 CHAVRCVGMEGASKADLQTAWKTWVMEHVAAEGGLPPGNAPGNTLFSERRARPSKACVRL 120
Query: 201 TPGRHVQLTV-PLEELVDKLVKENKVVAFI 229
+ G++ ++T L E +D VKE K+VAFI
Sbjct: 121 SDGKNPEMTAEALTEAIDACVKEYKIVAFI 150
>UniRef100_Q68W05 Glutaredoxin 3 [Rickettsia typhi]
Length = 111
Score = 107 bits (267), Expect = 4e-22
Identities = 47/99 (47%), Positives = 71/99 (71%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+ + +K+NKVV F+KG++ P CGFS V+ IL K GV++ ++VL ++ LRE L
Sbjct: 8 KFIQNAIKKNKVVLFMKGTKEMPACGFSGTVVAILNKLGVEFSDINVL---FDTSLREDL 64
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
KK+S+WPTFPQ+++NGELVGGCDI+ + + GE+ + K
Sbjct: 65 KKFSDWPTFPQLYINGELVGGCDIVKELYQNGELEKMLK 103
>UniRef100_Q8LNG2 Putative PKCq-interacting protein [Oryza sativa]
Length = 491
Score = 107 bits (266), Expect = 5e-22
Identities = 47/102 (46%), Positives = 76/102 (74%), Gaps = 3/102 (2%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
L + +++LV + V F+KG+ P CGFS+KV+ +L++EGV++ S D+L ++ +RE
Sbjct: 150 LNKRLEQLVNSHPVFLFMKGTPEQPRCGFSRKVVDVLKQEGVEFGSFDILTDN---DVRE 206
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+KK+SNWPTFPQ++ GEL+GGCDI+ +M+E GE+ +FK+
Sbjct: 207 GMKKFSNWPTFPQLYCKGELLGGCDIVIAMHESGELKDVFKE 248
Score = 97.8 bits (242), Expect = 3e-19
Identities = 50/131 (38%), Positives = 81/131 (61%), Gaps = 7/131 (5%)
Query: 177 PPGNESGNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAP 236
P G+++ A +P +K + LT +E ++ LV + V+AFIKG+ P
Sbjct: 255 PQGSKNEEAVKAKPDTEKSGAV----SEPALLTAAQKERLESLVNFSTVMAFIKGTPEEP 310
Query: 237 LCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCD 296
CGFS K++ IL++E + + S D+L +D +R+ LK SNWP++PQ+++NGELVGG D
Sbjct: 311 KCGFSGKLVHILKQEKIPFSSFDILTDDE---VRQGLKLLSNWPSYPQLYINGELVGGSD 367
Query: 297 ILTSMNEKGEV 307
I+ M++ GE+
Sbjct: 368 IVMEMHKSGEL 378
Score = 92.8 bits (229), Expect = 1e-17
Identities = 41/96 (42%), Positives = 64/96 (65%), Gaps = 3/96 (3%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
LE+ + L+ V+ F+KG+ AP CGFS KV+ L++ GV + + D+L ++ +R+
Sbjct: 393 LEDRLKALISSAPVMLFMKGTPDAPRCGFSSKVVNALKQAGVSFGAFDILSDEE---VRQ 449
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
LK YSNWPTFPQ++ EL+GGCDI+ + + GE+
Sbjct: 450 GLKTYSNWPTFPQLYYKSELIGGCDIVLELEKSGEL 485
>UniRef100_Q8SXQ5 RH03087p [Drosophila melanogaster]
Length = 159
Score = 107 bits (266), Expect = 5e-22
Identities = 50/98 (51%), Positives = 68/98 (69%), Gaps = 3/98 (3%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
+DKLV+ NKVV F+KG+ AP CGFS V+ I+ GV Y++ DVL N LR+ +K
Sbjct: 44 MDKLVRTNKVVVFMKGNPQAPRCGFSNAVVQIMRMHGVQYDAHDVLQ---NESLRQGVKD 100
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
Y++WPT PQ+F+NGE VGGCDIL M++ G++ KK
Sbjct: 101 YTDWPTIPQVFINGEFVGGCDILLQMHQSGDLIEELKK 138
>UniRef100_UPI00003ADFB5 UPI00003ADFB5 UniRef100 entry
Length = 135
Score = 106 bits (265), Expect = 7e-22
Identities = 53/101 (52%), Positives = 70/101 (68%), Gaps = 4/101 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV-DYESVDVLDEDYNYGLRET 272
E V++LV+E+ VV F+KGS + PLCGFS V+ IL GV DY + DVL + LR+
Sbjct: 21 EAVERLVREHPVVVFMKGSPAQPLCGFSNAVVQILRLHGVEDYRAHDVLQDP---DLRQG 77
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+K YSNWPT PQ++LNGE VGGCDIL M++ G++ KK
Sbjct: 78 IKNYSNWPTIPQVYLNGEFVGGCDILLQMHQNGDLVEELKK 118
>UniRef100_Q5ZK23 Hypothetical protein [Gallus gallus]
Length = 162
Score = 106 bits (265), Expect = 7e-22
Identities = 53/101 (52%), Positives = 70/101 (68%), Gaps = 4/101 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV-DYESVDVLDEDYNYGLRET 272
E V++LV+E+ VV F+KGS + PLCGFS V+ IL GV DY + DVL + LR+
Sbjct: 48 EAVERLVREHPVVVFMKGSPAQPLCGFSNAVVQILRLHGVEDYRAHDVLQDP---DLRQG 104
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+K YSNWPT PQ++LNGE VGGCDIL M++ G++ KK
Sbjct: 105 IKNYSNWPTIPQVYLNGEFVGGCDILLQMHQNGDLVEELKK 145
>UniRef100_Q7P9P9 Glutaredoxin-like protein grla [Rickettsia sibirica]
Length = 107
Score = 105 bits (263), Expect = 1e-21
Identities = 47/99 (47%), Positives = 71/99 (71%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+ ++ +K NKVV F+KG + +P CGFS V+ IL K GV++ ++VL ++ LRE L
Sbjct: 8 KFIENEIKNNKVVLFMKGIKKSPACGFSGTVVAILNKLGVEFRDINVL---FDAELREDL 64
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
KK+S+WPTFPQ+++NGELVGGCDI+ + + GE+ + K
Sbjct: 65 KKFSDWPTFPQLYINGELVGGCDIVRELYQSGELEKMLK 103
>UniRef100_O05957 Probable monothiol glutaredoxin grlA [Rickettsia prowazekii]
Length = 107
Score = 105 bits (262), Expect = 2e-21
Identities = 47/99 (47%), Positives = 69/99 (69%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E + +K+NKVV F+KG++ P CGFS V+ IL K GV++ ++VL ++ LRE L
Sbjct: 8 EFIQNAIKKNKVVLFMKGTKEMPACGFSGTVVAILNKLGVEFSDINVL---FDTALREDL 64
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
KK+S+WPTFPQ+++NG LVGGCDI + + GE+ + K
Sbjct: 65 KKFSDWPTFPQLYINGVLVGGCDIAKELYQNGELEKMLK 103
>UniRef100_UPI000033F072 UPI000033F072 UniRef100 entry
Length = 104
Score = 105 bits (261), Expect = 2e-21
Identities = 47/102 (46%), Positives = 71/102 (69%), Gaps = 3/102 (2%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
+EE + + +KENK++ ++KGS P CGFS K + L G ++ VD+LD N +R+
Sbjct: 3 IEEKIKEQIKENKILLYMKGSPYEPKCGFSAKTVQALIDCGAEFSYVDILD---NQDIRQ 59
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
TL S+WPTFPQ+F+ GEL+GGCDI+T M+E+GE+ + K+
Sbjct: 60 TLPSISDWPTFPQLFVKGELIGGCDIVTEMHEEGELQAIIKE 101
>UniRef100_Q92GH5 Probable monothiol glutaredoxin grlA [Rickettsia conorii]
Length = 107
Score = 105 bits (261), Expect = 2e-21
Identities = 47/99 (47%), Positives = 70/99 (70%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+ ++ +K NKVV F+KG + +P CGFS V+ IL K GV++ ++VL ++ LRE L
Sbjct: 8 KFIENEIKNNKVVLFMKGIKKSPACGFSGTVVAILNKLGVEFRDINVL---FDAELREDL 64
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
KK+S+WPTFPQ+++NGELVGGCDI + + GE+ + K
Sbjct: 65 KKFSDWPTFPQLYINGELVGGCDIARELYQSGELEKMLK 103
>UniRef100_UPI00002A410B UPI00002A410B UniRef100 entry
Length = 103
Score = 104 bits (260), Expect = 3e-21
Identities = 45/97 (46%), Positives = 69/97 (70%), Gaps = 3/97 (3%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
+++L+ + ENK+V F+KG+ P+CGFS +V+ IL ++Y++ DVL ++ GLR
Sbjct: 5 IKQLITSSISENKIVLFMKGTPEMPMCGFSSQVVQILNSFKIEYKTFDVLSDE---GLRS 61
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVA 308
+K++SNWPT PQ+++NGE VGGCDI M E GE+A
Sbjct: 62 GIKEFSNWPTLPQLYINGEFVGGCDICVEMFENGELA 98
>UniRef100_Q6RJL3 Putative glutaredoxin-like protein [uncultured bacterium]
Length = 110
Score = 104 bits (260), Expect = 3e-21
Identities = 47/99 (47%), Positives = 70/99 (70%), Gaps = 3/99 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E ++ VK+NKV+ F+KG+RS P CGFS + I ++ GV YE+ DVL + LR+ +
Sbjct: 6 EQIESAVKKNKVMIFMKGNRSFPQCGFSAATVAIFDQLGVPYETADVLSDPE---LRDGI 62
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
K+YSNWPT PQ++++G+ VGGCDI+ ++E GE+ L K
Sbjct: 63 KRYSNWPTIPQVYIDGKFVGGCDIIRELHETGELEPLVK 101
>UniRef100_Q80Y14 2900070E19Rik protein [Mus musculus]
Length = 162
Score = 103 bits (258), Expect = 5e-21
Identities = 52/101 (51%), Positives = 69/101 (67%), Gaps = 4/101 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV-DYESVDVLDEDYNYGLRET 272
E +D LVK++KVV F+KG+ P CGFS V+ IL GV DY + +VLD+ LR+
Sbjct: 49 EQLDALVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYAAYNVLDDPE---LRQG 105
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+K YSNWPT PQ++LNGE VGGCDIL M++ G++ KK
Sbjct: 106 IKDYSNWPTIPQVYLNGEFVGGCDILLQMHQNGDLVEELKK 146
>UniRef100_Q86SX6 Full-length cDNA clone CS0DL002YE04 of B cells (Ramos cell line) of
Homo sapiens [Homo sapiens]
Length = 176
Score = 103 bits (258), Expect = 5e-21
Identities = 52/101 (51%), Positives = 69/101 (67%), Gaps = 4/101 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV-DYESVDVLDEDYNYGLRET 272
E +D LVK++KVV F+KG+ P CGFS V+ IL GV DY + +VLD+ LR+
Sbjct: 62 EQLDALVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYAAYNVLDDPE---LRQG 118
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+K YSNWPT PQ++LNGE VGGCDIL M++ G++ KK
Sbjct: 119 IKDYSNWPTIPQVYLNGEFVGGCDILLQMHQNGDLVEELKK 159
>UniRef100_Q8IZ54 Chromosome 14 open reading frame 87 [Homo sapiens]
Length = 157
Score = 103 bits (258), Expect = 5e-21
Identities = 52/101 (51%), Positives = 69/101 (67%), Gaps = 4/101 (3%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGV-DYESVDVLDEDYNYGLRET 272
E +D LVK++KVV F+KG+ P CGFS V+ IL GV DY + +VLD+ LR+
Sbjct: 43 EQLDALVKKDKVVVFLKGTPEQPQCGFSNAVVQILRLHGVRDYAAYNVLDDPE---LRQG 99
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+K YSNWPT PQ++LNGE VGGCDIL M++ G++ KK
Sbjct: 100 IKDYSNWPTIPQVYLNGEFVGGCDILLQMHQNGDLVEELKK 140
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 581,165,940
Number of Sequences: 2790947
Number of extensions: 26604519
Number of successful extensions: 54049
Number of sequences better than 10.0: 1027
Number of HSP's better than 10.0 without gapping: 697
Number of HSP's successfully gapped in prelim test: 330
Number of HSP's that attempted gapping in prelim test: 52696
Number of HSP's gapped (non-prelim): 1054
length of query: 313
length of database: 848,049,833
effective HSP length: 127
effective length of query: 186
effective length of database: 493,599,564
effective search space: 91809518904
effective search space used: 91809518904
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140721.1


