

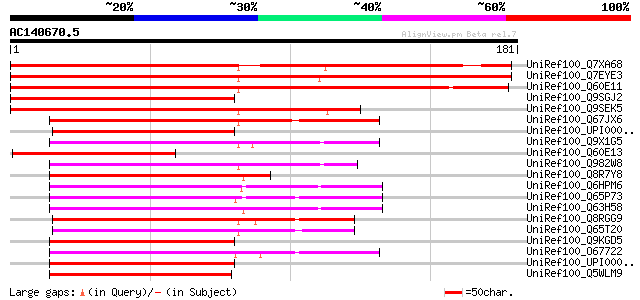
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.5 + phase: 0 /pseudo/partial
(181 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q7XA68 At1g01640 [Arabidopsis thaliana] 217 1e-55
UniRef100_Q7EYE3 Putative ribosomal protein I [Oryza sativa] 207 8e-53
UniRef100_Q60E11 Putative ribosomal protein I [Oryza sativa] 207 1e-52
UniRef100_Q9SGJ2 Putative ribosomal protein L13 [Arabidopsis tha... 139 3e-32
UniRef100_Q9SEK5 Ribosomal protein I [Ceratopteris richardii] 124 1e-27
UniRef100_Q67JX6 50S ribosomal protein L13 [Symbiobacterium ther... 93 4e-18
UniRef100_UPI000033130D UPI000033130D UniRef100 entry 91 2e-17
UniRef100_Q9X1G5 50S ribosomal protein L13 [Thermotoga maritima] 91 2e-17
UniRef100_Q60E13 Hypothetical protein OSJNBa0004B23.10 [Oryza sa... 89 4e-17
UniRef100_Q982W8 50S ribosomal protein L13 [Rhizobium loti] 88 9e-17
UniRef100_Q8R7Y8 Ribosomal protein L13 [Thermoanaerobacter tengc... 88 9e-17
UniRef100_Q6HPM6 Ribosomal protein L13 [Bacillus thuringiensis] 88 9e-17
UniRef100_Q65P73 50S ribosomal protein L13 [Bacillus licheniformis] 88 9e-17
UniRef100_Q63H58 Ribosomal protein L13 [Bacillus cereus] 88 9e-17
UniRef100_Q8RGG9 LSU ribosomal protein L13P [Fusobacterium nucle... 87 2e-16
UniRef100_Q65T20 50S ribosomal protein L13 [Mannheimia succinici... 87 2e-16
UniRef100_Q9KGD5 50S ribosomal protein L13 [Bacillus halodurans] 87 2e-16
UniRef100_O67722 50S ribosomal protein L13 [Aquifex aeolicus] 86 3e-16
UniRef100_UPI000027A8AF UPI000027A8AF UniRef100 entry 86 4e-16
UniRef100_Q5WLM9 50S ribosomal protein L13 [Bacillus clausii] 86 4e-16
>UniRef100_Q7XA68 At1g01640 [Arabidopsis thaliana]
Length = 205
Score = 217 bits (552), Expect = 1e-55
Identities = 119/194 (61%), Positives = 135/194 (69%), Gaps = 28/194 (14%)
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
+A AGIKRI LDGLRWRVFDA+GQ+LGRLASQI+TV+ KDKPTY PNRDDGD+CIVLNA
Sbjct: 17 KAVAGIKRINLDGLRWRVFDARGQVLGRLASQISTVLQAKDKPTYCPNRDDGDICIVLNA 76
Query: 61 KDIAVTGRKLTDKVYYWHTG-----KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVM---- 111
K+I TGRKLTDK Y WHTG KE ++L+ AK +K + M
Sbjct: 77 KEIGFTGRKLTDKFYRWHTGYIGHLKE-------RSLKDQMAKDPTEVIRKAVWRMLPSN 129
Query: 112 ------DRKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENA 165
DRKLRIF G EHPFGD+PLEP+VMPPR VREMRPRARRAMIRAQKKAEQ +
Sbjct: 130 NLRDDRDRKLRIFEGGEHPFGDKPLEPFVMPPRRVREMRPRARRAMIRAQKKAEQAE--- 186
Query: 166 EQRQNAEEMKNSKK 179
E+K KK
Sbjct: 187 ---NEGTEVKKGKK 197
>UniRef100_Q7EYE3 Putative ribosomal protein I [Oryza sativa]
Length = 229
Score = 207 bits (528), Expect = 8e-53
Identities = 107/187 (57%), Positives = 131/187 (69%), Gaps = 8/187 (4%)
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
+A AG++RI LDGLRWRVFDAKGQ+LGRLASQIA V+ GKDKPTY P+ ++GDMC+VLNA
Sbjct: 14 KALAGLRRINLDGLRWRVFDAKGQVLGRLASQIAVVLQGKDKPTYAPHVENGDMCVVLNA 73
Query: 61 KDIAVTGRKLTDKVYYWHTG------KEL*RIRWPKTLQMLFAKLFCA*FQKTTY--VMD 112
KDI+VTGRK+TDK+YYWHTG + + + K + K + D
Sbjct: 74 KDISVTGRKMTDKIYYWHTGYIGHLKERRLKDQMEKDPTEVIRKAVMRMLPRNRLRDDRD 133
Query: 113 RKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAE 172
RKLRIF GSEHPF DRPLEP+ MPPR VREMRP+ARRA+IRAQKK + + + + E
Sbjct: 134 RKLRIFSGSEHPFHDRPLEPFAMPPRQVREMRPQARRALIRAQKKEQDRAAASTKDDKDE 193
Query: 173 EMKNSKK 179
E KK
Sbjct: 194 EEDKGKK 200
>UniRef100_Q60E11 Putative ribosomal protein I [Oryza sativa]
Length = 191
Score = 207 bits (527), Expect = 1e-52
Identities = 108/186 (58%), Positives = 135/186 (72%), Gaps = 9/186 (4%)
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
+A AG++RI LDGLRWRVFDAKGQ+LGRLASQIA V+ GKDKPTY P+ ++GDMC+VLNA
Sbjct: 2 KALAGLRRINLDGLRWRVFDAKGQVLGRLASQIAVVLQGKDKPTYAPHVENGDMCVVLNA 61
Query: 61 KDIAVTGRKLTDKVYYWHTG--------KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVMD 112
KDI+VTGRK+TDK+YYWHTG + ++ T + A + + D
Sbjct: 62 KDISVTGRKMTDKIYYWHTGYIGHLKERRLKDQMEKDPTEAIRKAVMRMLPRNRLRDDRD 121
Query: 113 RKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAE 172
RKLRIF G+EHPF DRPLEP++MP R VREMRPRARRA++RAQKK EQ + A ++ E
Sbjct: 122 RKLRIFSGNEHPFHDRPLEPFMMPRRQVREMRPRARRALLRAQKK-EQDRAAAASTKDEE 180
Query: 173 EMKNSK 178
KN+K
Sbjct: 181 NAKNAK 186
>UniRef100_Q9SGJ2 Putative ribosomal protein L13 [Arabidopsis thaliana]
Length = 137
Score = 139 bits (351), Expect = 3e-32
Identities = 64/80 (80%), Positives = 71/80 (88%)
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
+A AGIKRI LDGLRWRVFDA+GQ+LGRLASQI+TV+ KDKPTY PNRDDGD+CIVLNA
Sbjct: 17 KAVAGIKRINLDGLRWRVFDARGQVLGRLASQISTVLQAKDKPTYCPNRDDGDICIVLNA 76
Query: 61 KDIAVTGRKLTDKVYYWHTG 80
K+I TGRKLTDK Y WHTG
Sbjct: 77 KEIGFTGRKLTDKFYRWHTG 96
>UniRef100_Q9SEK5 Ribosomal protein I [Ceratopteris richardii]
Length = 141
Score = 124 bits (311), Expect = 1e-27
Identities = 67/133 (50%), Positives = 84/133 (62%), Gaps = 8/133 (6%)
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
RA AG+ +++GLRWRVFDAKGQILGRLASQI+ V+ GKDKPTY P+ + GD+C+V+NA
Sbjct: 7 RAMAGLTPERIEGLRWRVFDAKGQILGRLASQISVVLQGKDKPTYAPHLEKGDICVVINA 66
Query: 61 KDIAVTGRKLTDKVYYWHTG------KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVMD-- 112
K + TGRKLTDK Y WHTG + + + K + K K D
Sbjct: 67 KHVCFTGRKLTDKFYRWHTGYIGHLKERSLKEQMXKDPTEVIRKAVVRMLPKNRLRDDRA 126
Query: 113 RKLRIFPGSEHPF 125
RKLRIF +HPF
Sbjct: 127 RKLRIFVXDQHPF 139
>UniRef100_Q67JX6 50S ribosomal protein L13 [Symbiobacterium thermophilum]
Length = 143
Score = 92.8 bits (229), Expect = 4e-18
Identities = 50/128 (39%), Positives = 77/128 (60%), Gaps = 12/128 (9%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA+G+ LGRLAS++A ++ GK KPT+TP+ D GD I++NA+ I VTG+K+TDKV
Sbjct: 15 KWYVVDAEGKTLGRLASKVAAILRGKHKPTFTPHVDCGDFVIIVNAEKIQVTGKKVTDKV 74
Query: 75 YYWHTG----------KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHP 124
YY H+G +++ R + L++ + + M RKL+++ G EHP
Sbjct: 75 YYRHSGYPGGQKATTFEQMIARRPERVLELAIKGMLP--HNRLGRQMYRKLKVYAGPEHP 132
Query: 125 FGDRPLEP 132
+ EP
Sbjct: 133 HAAQKPEP 140
>UniRef100_UPI000033130D UPI000033130D UniRef100 entry
Length = 138
Score = 90.5 bits (223), Expect = 2e-17
Identities = 40/65 (61%), Positives = 52/65 (79%)
Query: 16 WRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVY 75
W + DA+ +LGRLA++IATV+ GK KPTY P +D+GD +V+NAK+IAVTG KL DK+Y
Sbjct: 11 WFIIDAQDAVLGRLATKIATVLCGKHKPTYLPYKDNGDYIVVINAKNIAVTGNKLNDKIY 70
Query: 76 YWHTG 80
Y HTG
Sbjct: 71 YKHTG 75
>UniRef100_Q9X1G5 50S ribosomal protein L13 [Thermotoga maritima]
Length = 149
Score = 90.5 bits (223), Expect = 2e-17
Identities = 51/127 (40%), Positives = 75/127 (58%), Gaps = 10/127 (7%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA G++LGRLA++IA ++MGK KP YTP+ D GD IV+NA + +TG+KL KV
Sbjct: 21 KWYVVDASGKVLGRLATRIAKILMGKHKPNYTPHVDTGDYVIVVNADKVVLTGKKLDQKV 80
Query: 75 YYWHTG-----KEL*R----IRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHPF 125
YYWH+G K L + P+ L L K +K ++ ++L+++ EHP
Sbjct: 81 YYWHSGYPGGLKSLTARQMLEKHPERLIWLAVKRMLPKNRKGRKML-KRLKVYASPEHPH 139
Query: 126 GDRPLEP 132
+ EP
Sbjct: 140 QAQKPEP 146
>UniRef100_Q60E13 Hypothetical protein OSJNBa0004B23.10 [Oryza sativa]
Length = 160
Score = 89.4 bits (220), Expect = 4e-17
Identities = 41/58 (70%), Positives = 49/58 (83%)
Query: 2 AAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLN 59
A A ++RI LDGLRW VFDAKGQ+LGRLASQIA V+ GKDKPTY P+ ++ DMC+VLN
Sbjct: 102 ALADLRRINLDGLRWCVFDAKGQVLGRLASQIAVVLQGKDKPTYAPHVENRDMCVVLN 159
>UniRef100_Q982W8 50S ribosomal protein L13 [Rhizobium loti]
Length = 154
Score = 88.2 bits (217), Expect = 9e-17
Identities = 48/120 (40%), Positives = 70/120 (58%), Gaps = 11/120 (9%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W + DA+G ++GRLA+ IA + GK KPT+TP+ DDGD IV+NA + TG+K TDKV
Sbjct: 14 KWILIDAEGLVVGRLATVIANHLRGKHKPTFTPHVDDGDNVIVINADKVVFTGKKFTDKV 73
Query: 75 YYWHTG----------KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHP 124
YYWHTG ++L R+P+ + + M + LR++ G+EHP
Sbjct: 74 YYWHTGHPGGIKERTARQLLEGRFPERVVEKAVERMVPRGPLGRRQM-KNLRVYAGAEHP 132
>UniRef100_Q8R7Y8 Ribosomal protein L13 [Thermoanaerobacter tengcongensis]
Length = 143
Score = 88.2 bits (217), Expect = 9e-17
Identities = 42/81 (51%), Positives = 60/81 (73%), Gaps = 2/81 (2%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA+G++LGRLASQIA ++MGK KPTYTP+ D GD +V+NA + +TG+KL DK
Sbjct: 14 KWFVIDAEGKVLGRLASQIAKILMGKHKPTYTPHVDTGDFVVVINADKVVLTGKKLEDKY 73
Query: 75 YYWHTGKE--L*RIRWPKTLQ 93
Y ++TG L +I++ K +Q
Sbjct: 74 YKYYTGYPGGLKQIQYKKLMQ 94
>UniRef100_Q6HPM6 Ribosomal protein L13 [Bacillus thuringiensis]
Length = 145
Score = 88.2 bits (217), Expect = 9e-17
Identities = 51/129 (39%), Positives = 75/129 (57%), Gaps = 12/129 (9%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA+GQ LGRLAS++A+++ GK+KPT+TP+ D GD I++NA+ I +TG KL DK+
Sbjct: 15 KWYVVDAEGQTLGRLASEVASILRGKNKPTFTPHVDTGDHVIIINAEKIHLTGNKLNDKI 74
Query: 75 YYWHTGK----------EL*RIRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHP 124
YY HT E+ R +P + L K + V +KL ++ G+EHP
Sbjct: 75 YYRHTNHPGGLKQRTALEM-RTNYPVQMLELAIKGMLPKGRLGRQV-SKKLNVYAGAEHP 132
Query: 125 FGDRPLEPY 133
+ E Y
Sbjct: 133 HQAQKPEVY 141
>UniRef100_Q65P73 50S ribosomal protein L13 [Bacillus licheniformis]
Length = 145
Score = 88.2 bits (217), Expect = 9e-17
Identities = 51/129 (39%), Positives = 75/129 (57%), Gaps = 12/129 (9%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA G+ LGRL++++A+++ GK KPTYTP+ D GD I++NA+ I +TG+KLTDK+
Sbjct: 15 KWLVVDAAGKTLGRLSTEVASILRGKHKPTYTPHVDTGDHVIIINAEKIELTGKKLTDKI 74
Query: 75 YYWHT----------GKEL*RIRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHP 124
YY HT E+ R +P+ + L K M +KL ++ GSEHP
Sbjct: 75 YYRHTQHPGGLKSRTALEM-RTNYPEKMLELAIKGMLP-KGSLGRQMFKKLNVYRGSEHP 132
Query: 125 FGDRPLEPY 133
+ E Y
Sbjct: 133 HQAQKPEVY 141
>UniRef100_Q63H58 Ribosomal protein L13 [Bacillus cereus]
Length = 145
Score = 88.2 bits (217), Expect = 9e-17
Identities = 50/128 (39%), Positives = 74/128 (57%), Gaps = 10/128 (7%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA+GQ LGRLAS++A+++ GK+KPT+TP+ D GD I++NA+ I +TG KL DK+
Sbjct: 15 KWYVVDAEGQTLGRLASEVASILRGKNKPTFTPHVDTGDHVIIINAEKIHLTGNKLNDKM 74
Query: 75 YYWHTGKE---------L*RIRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHPF 125
YY HT + R +P + L K + V +KL ++ G+EHP
Sbjct: 75 YYRHTNHPGGLKQRTALVLRTNYPVQMLELAIKGMLPKGRLGRQV-SKKLNVYAGAEHPH 133
Query: 126 GDRPLEPY 133
+ E Y
Sbjct: 134 QAQKPEVY 141
>UniRef100_Q8RGG9 LSU ribosomal protein L13P [Fusobacterium nucleatum]
Length = 144
Score = 87.4 bits (215), Expect = 2e-16
Identities = 53/117 (45%), Positives = 71/117 (60%), Gaps = 10/117 (8%)
Query: 16 WRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVY 75
W +DA+GQILGRLA +IA +MGK+K T+TP+ D GD +V N + I VTG+KLTDKVY
Sbjct: 17 WHHYDAEGQILGRLAVEIAKKLMGKEKLTFTPHIDGGDYVVVTNVEKIVVTGKKLTDKVY 76
Query: 76 YWHTG-------KEL*RI--RWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEH 123
Y H+G ++L I + P+ L ML K K +LR+F G+EH
Sbjct: 77 YNHSGFPGGIRARKLGEILAKKPEELLMLAVKRMLP-KNKLGRQQLTRLRVFAGAEH 132
>UniRef100_Q65T20 50S ribosomal protein L13 [Mannheimia succiniciproducens]
Length = 147
Score = 87.4 bits (215), Expect = 2e-16
Identities = 50/118 (42%), Positives = 68/118 (57%), Gaps = 12/118 (10%)
Query: 16 WRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVY 75
W V DA G+ LGRLA+++A+ + GK K YTP+ D GD IV+NA +AVTGRK TDKVY
Sbjct: 20 WYVVDATGKTLGRLATELASRLRGKHKAEYTPHVDTGDYIIVINADKVAVTGRKETDKVY 79
Query: 76 YWHTG----------KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEH 123
YWHTG KE+ R +++ + M RKL+++ GS+H
Sbjct: 80 YWHTGYVGGIKQATFKEMIARRPEAVIEIAVKGMLPK--GPLGRAMFRKLKVYAGSQH 135
>UniRef100_Q9KGD5 50S ribosomal protein L13 [Bacillus halodurans]
Length = 145
Score = 87.0 bits (214), Expect = 2e-16
Identities = 38/66 (57%), Positives = 52/66 (78%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA+GQ LGRLAS++A+++ GK KPTYTP+ D GD I++NA+ I +TG KL DK+
Sbjct: 15 KWYVVDAEGQTLGRLASEVASILRGKHKPTYTPHVDTGDHVIIINAEKIHLTGNKLQDKI 74
Query: 75 YYWHTG 80
YY H+G
Sbjct: 75 YYRHSG 80
>UniRef100_O67722 50S ribosomal protein L13 [Aquifex aeolicus]
Length = 144
Score = 86.3 bits (212), Expect = 3e-16
Identities = 50/127 (39%), Positives = 73/127 (57%), Gaps = 10/127 (7%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA G+ LGRLAS+IA ++ GK KP Y P+ D GD IV+NA+ I VTG+KL K
Sbjct: 14 KWWVVDATGKTLGRLASEIAKILRGKHKPYYQPDVDCGDFVIVINAEKIRVTGKKLEQKK 73
Query: 75 YYWHT----GKEL*RIRW-----PKTLQMLFAKLFCA*FQKTTYVMDRKLRIFPGSEHPF 125
YYWH+ G + ++W P+ + L K + + M +KL+++ G EHP
Sbjct: 74 YYWHSRYPGGLKERTLKWMLENKPEEVIRLAVKRMLP-KNRLGHRMLKKLKVYRGPEHPH 132
Query: 126 GDRPLEP 132
+ +P
Sbjct: 133 QAQKPQP 139
>UniRef100_UPI000027A8AF UPI000027A8AF UniRef100 entry
Length = 157
Score = 85.9 bits (211), Expect = 4e-16
Identities = 38/66 (57%), Positives = 49/66 (73%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W + DA G +LGRLAS IA + GK KPTYTP+ D GD IV+NA+ + +TG+K DK+
Sbjct: 17 KWLLIDADGVVLGRLASIIAKYLRGKHKPTYTPHMDMGDNVIVVNAEKVELTGKKRDDKI 76
Query: 75 YYWHTG 80
YYWHTG
Sbjct: 77 YYWHTG 82
>UniRef100_Q5WLM9 50S ribosomal protein L13 [Bacillus clausii]
Length = 145
Score = 85.9 bits (211), Expect = 4e-16
Identities = 39/65 (60%), Positives = 50/65 (76%)
Query: 15 RWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKV 74
+W V DA+GQ LGRLAS++A+++ GK KPT+TP+ D GD IVLNA I +TG KL DK+
Sbjct: 15 KWYVVDAEGQTLGRLASEVASILRGKHKPTFTPHVDTGDHVIVLNASKIQLTGNKLQDKI 74
Query: 75 YYWHT 79
YY HT
Sbjct: 75 YYRHT 79
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.327 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 280,968,280
Number of Sequences: 2790947
Number of extensions: 10836850
Number of successful extensions: 70331
Number of sequences better than 10.0: 601
Number of HSP's better than 10.0 without gapping: 574
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 69377
Number of HSP's gapped (non-prelim): 821
length of query: 181
length of database: 848,049,833
effective HSP length: 119
effective length of query: 62
effective length of database: 515,927,140
effective search space: 31987482680
effective search space used: 31987482680
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC140670.5


