

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.16 + phase: 0
(235 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
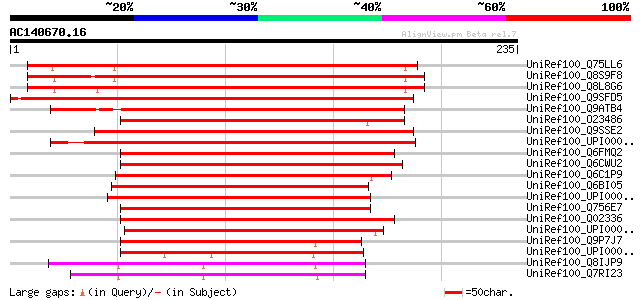
UniRef100_Q75LL6 Putative transcriptional adaptor [Oryza sativa] 262 5e-69
UniRef100_Q8S9F8 Transcriptional adaptor [Zea mays] 257 2e-67
UniRef100_Q8L8G6 Histone acetyltransferase complex component [Ze... 256 3e-67
UniRef100_Q9SFD5 F17A17.8 protein [Arabidopsis thaliana] 236 5e-61
UniRef100_Q9ATB4 Transcriptional adaptor ADA2b [Arabidopsis thal... 235 8e-61
UniRef100_O23486 SIMILARITY to probable transcriptional adaptor ... 217 2e-55
UniRef100_Q9SSE2 MLP3.19 protein [Arabidopsis thaliana] 216 5e-55
UniRef100_UPI000028E952 UPI000028E952 UniRef100 entry 174 2e-42
UniRef100_Q6FMQ2 Candida glabrata strain CBS138 chromosome K com... 139 8e-32
UniRef100_Q6CWU2 Kluyveromyces lactis strain NRRL Y-1140 chromos... 137 3e-31
UniRef100_Q6C1P9 Yarrowia lipolytica chromosome F of strain CLIB... 134 2e-30
UniRef100_Q6BI05 Similar to CA3340|CaADA2 Candida albicans CaADA... 133 4e-30
UniRef100_UPI000042C941 UPI000042C941 UniRef100 entry 131 1e-29
UniRef100_Q756E7 AER318Cp [Ashbya gossypii] 131 2e-29
UniRef100_Q02336 Transcriptional adapter 2 [Saccharomyces cerevi... 131 2e-29
UniRef100_UPI0000498CF8 UPI0000498CF8 UniRef100 entry 130 3e-29
UniRef100_Q9P7J7 SPCC24B10.08c protein [Schizosaccharomyces pombe] 130 4e-29
UniRef100_UPI0000219BE7 UPI0000219BE7 UniRef100 entry 128 1e-28
UniRef100_Q8IJP9 ADA2-like protein [Plasmodium falciparum] 127 2e-28
UniRef100_Q7RI23 ADA2-like protein [Plasmodium yoelii yoelii] 126 5e-28
>UniRef100_Q75LL6 Putative transcriptional adaptor [Oryza sativa]
Length = 570
Score = 262 bits (670), Expect = 5e-69
Identities = 122/186 (65%), Positives = 153/186 (81%), Gaps = 5/186 (2%)
Query: 9 SRSSPIAADD---NRSKRKKVALNADNLETSSAAGMGITTDG-KVSLYHCNYCNKDISGK 64
SR P + DD +RSKR++VA + D ++ SAA G G K +LYHCNYCNKDISGK
Sbjct: 4 SRGVPNSGDDETNHRSKRRRVASSGDAPDSLSAACGGAGEGGGKKALYHCNYCNKDISGK 63
Query: 65 IRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEAL 124
IRIKC+ C DFDLC+ECFSVG E+TPH+SNHPYRVMDNLSFPLICPDW+A+EE+LLLE +
Sbjct: 64 IRIKCSKCPDFDLCVECFSVGAEVTPHRSNHPYRVMDNLSFPLICPDWNADEEILLLEGI 123
Query: 125 DMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAK-G 183
+MYG GNW +VA+++GTK+K+QCIDHY T Y+NSPC+P+PD+SH +GKN++ELLAMAK
Sbjct: 124 EMYGLGNWAEVAEHVGTKTKAQCIDHYTTAYMNSPCYPLPDMSHVNGKNRKELLAMAKVQ 183
Query: 184 NQVKKG 189
+ KKG
Sbjct: 184 GESKKG 189
>UniRef100_Q8S9F8 Transcriptional adaptor [Zea mays]
Length = 565
Score = 257 bits (657), Expect = 2e-67
Identities = 122/190 (64%), Positives = 153/190 (80%), Gaps = 7/190 (3%)
Query: 9 SRSSPIAADDN---RSKRKKVALNADNLETSSAAGMGITTDG--KVSLYHCNYCNKDISG 63
SR + DD+ RSKR++VA D ++ SA G+G +G K +LYHCNYCNKDISG
Sbjct: 4 SRGVQNSGDDDTVHRSKRRRVASGGDATDSVSA-GIGGAGEGGGKKALYHCNYCNKDISG 62
Query: 64 KIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEA 123
KIRIKC+ C DFDLC+ECFSVG E+TPH+SNHPY+VMDNLSFPLICPDW+A+EE+LLLE
Sbjct: 63 KIRIKCSKCPDFDLCVECFSVGAEVTPHRSNHPYKVMDNLSFPLICPDWNADEEILLLEG 122
Query: 124 LDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAK- 182
++MYG GNW +VA+++GTKSK QCIDHY T Y+NSPC+P+PD+SH +GKN++ELLAMAK
Sbjct: 123 IEMYGLGNWLEVAEHVGTKSKLQCIDHYTTAYMNSPCYPLPDMSHVNGKNRKELLAMAKV 182
Query: 183 GNQVKKGLLL 192
+ KKG L
Sbjct: 183 QGESKKGTSL 192
>UniRef100_Q8L8G6 Histone acetyltransferase complex component [Zea mays]
Length = 565
Score = 256 bits (655), Expect = 3e-67
Identities = 120/189 (63%), Positives = 152/189 (79%), Gaps = 5/189 (2%)
Query: 9 SRSSPIAADDN---RSKRKKVALNADNLETSSAA-GMGITTDGKVSLYHCNYCNKDISGK 64
SR + DD+ RSKR++V+ D ++ SA+ G GK +LYHCNYCNKDISGK
Sbjct: 4 SRGVLSSGDDDTGHRSKRRRVSSGGDATDSISASIGGAGEGGGKKALYHCNYCNKDISGK 63
Query: 65 IRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEAL 124
IRIKC+ C DFDLC+ECFSVG E+TPH+SNHPY+VMDNLSFPLICPDW+A+EE+LLLE +
Sbjct: 64 IRIKCSKCPDFDLCVECFSVGAEVTPHRSNHPYKVMDNLSFPLICPDWNADEEILLLEGI 123
Query: 125 DMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAK-G 183
+MYG GNW +VA+++GTKSK QCIDHY + Y+NSPC+P+PD+SH +GKN++ELLAMAK
Sbjct: 124 EMYGLGNWLEVAEHVGTKSKLQCIDHYTSAYMNSPCYPLPDMSHVNGKNRKELLAMAKVQ 183
Query: 184 NQVKKGLLL 192
+ KKG LL
Sbjct: 184 GESKKGTLL 192
>UniRef100_Q9SFD5 F17A17.8 protein [Arabidopsis thaliana]
Length = 548
Score = 236 bits (601), Expect = 5e-61
Identities = 107/187 (57%), Positives = 143/187 (76%), Gaps = 1/187 (0%)
Query: 1 MGRCCRAASRSSPIAADDNRSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKD 60
MGR + ASR + + +SKRKK++L +N S + G+ + K LY CNYC+KD
Sbjct: 1 MGRS-KLASRPAEEDLNPGKSKRKKISLGPENAAASISTGIEAGNERKPGLYCCNYCDKD 59
Query: 61 ISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLL 120
+SG +R KCAVC DFDLC+ECFSVGVEL HK++HPYRVMDNLSF L+ DW+A+EE+LL
Sbjct: 60 LSGLVRFKCAVCMDFDLCVECFSVGVELNRHKNSHPYRVMDNLSFSLVTSDWNADEEILL 119
Query: 121 LEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAM 180
LEA+ YGFGNW +VAD++G+K+ ++CI H+N+ Y+ SPCFP+PDLSH GK+K+ELLAM
Sbjct: 120 LEAIATYGFGNWKEVADHVGSKTTTECIKHFNSAYMQSPCFPLPDLSHTIGKSKDELLAM 179
Query: 181 AKGNQVK 187
+K + VK
Sbjct: 180 SKDSAVK 186
>UniRef100_Q9ATB4 Transcriptional adaptor ADA2b [Arabidopsis thaliana]
Length = 487
Score = 235 bits (599), Expect = 8e-61
Identities = 105/164 (64%), Positives = 134/164 (81%), Gaps = 4/164 (2%)
Query: 20 RSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCI 79
R+++KK A N +N E++S G GK Y+C+YC KDI+GKIRIKCAVC DFDLCI
Sbjct: 17 RTRKKKNAANVENFESTSLVP-GAEGGGK---YNCDYCQKDITGKIRIKCAVCPDFDLCI 72
Query: 80 ECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNI 139
EC SVG E+TPHK +HPYRVM NL+FPLICPDWSA++EMLLLE L++YG GNW +VA+++
Sbjct: 73 ECMSVGAEITPHKCDHPYRVMGNLTFPLICPDWSADDEMLLLEGLEIYGLGNWAEVAEHV 132
Query: 140 GTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKG 183
GTKSK QC++HY +Y+NSP FP+PD+SH +GKN++EL AMAKG
Sbjct: 133 GTKSKEQCLEHYRNIYLNSPFFPLPDMSHVAGKNRKELQAMAKG 176
>UniRef100_O23486 SIMILARITY to probable transcriptional adaptor ADA2 [Arabidopsis
thaliana]
Length = 480
Score = 217 bits (553), Expect = 2e-55
Identities = 94/139 (67%), Positives = 117/139 (83%), Gaps = 7/139 (5%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
Y+C+YC KDI+GKIRIKCAVC DFDLCIEC SVG E+TPHK +HPYRVM NL+FPLICPD
Sbjct: 31 YNCDYCQKDITGKIRIKCAVCPDFDLCIECMSVGAEITPHKCDHPYRVMGNLTFPLICPD 90
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVP------- 164
WSA++EMLLLE L++YG GNW +VA+++GTKSK QC++HY +Y+NSP FP+P
Sbjct: 91 WSADDEMLLLEGLEIYGLGNWAEVAEHVGTKSKEQCLEHYRNIYLNSPFFPLPEKRLFSQ 150
Query: 165 DLSHFSGKNKEELLAMAKG 183
D+SH +GKN++EL AMAKG
Sbjct: 151 DMSHVAGKNRKELQAMAKG 169
>UniRef100_Q9SSE2 MLP3.19 protein [Arabidopsis thaliana]
Length = 527
Score = 216 bits (549), Expect = 5e-55
Identities = 93/148 (62%), Positives = 121/148 (80%)
Query: 40 GMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRV 99
G+ + K LY CNYC+KD+SG +R KCAVC DFDLC+ECFSVGVEL HK++HPYRV
Sbjct: 18 GIEAGNERKPGLYCCNYCDKDLSGLVRFKCAVCMDFDLCVECFSVGVELNRHKNSHPYRV 77
Query: 100 MDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSP 159
MDNLSF L+ DW+A+EE+LLLEA+ YGFGNW +VAD++G+K+ ++CI H+N+ Y+ SP
Sbjct: 78 MDNLSFSLVTSDWNADEEILLLEAIATYGFGNWKEVADHVGSKTTTECIKHFNSAYMQSP 137
Query: 160 CFPVPDLSHFSGKNKEELLAMAKGNQVK 187
CFP+PDLSH GK+K+ELLAM+K + VK
Sbjct: 138 CFPLPDLSHTIGKSKDELLAMSKDSAVK 165
>UniRef100_UPI000028E952 UPI000028E952 UniRef100 entry
Length = 170
Score = 174 bits (440), Expect = 2e-42
Identities = 81/169 (47%), Positives = 113/169 (65%), Gaps = 7/169 (4%)
Query: 20 RSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCI 79
+ KR++VA T +A ++ K +L+ C YC KDISG +RIKCA C + LC+
Sbjct: 2 KPKRRRVA-------TENAMTKANPSEPKRALFSCVYCQKDISGVVRIKCAECAEMHLCV 54
Query: 80 ECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNI 139
+CFSVGVE PH++ H Y V+D+LSFPL PDW A+EE+LLLEA++MYG GNW +V++++
Sbjct: 55 DCFSVGVEPHPHRACHAYHVVDDLSFPLFTPDWGADEELLLLEAIEMYGLGNWTEVSEHV 114
Query: 140 GTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKGNQVKK 188
GTKSK +C HY VYVNSP P+PD++ GK +G + K+
Sbjct: 115 GTKSKQRCHAHYFDVYVNSPTTPLPDMTKVLGKGHAREEPKEEGGEKKR 163
>UniRef100_Q6FMQ2 Candida glabrata strain CBS138 chromosome K complete sequence
[Candida glabrata]
Length = 445
Score = 139 bits (349), Expect = 8e-32
Identities = 53/127 (41%), Positives = 87/127 (67%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++RI CAVC ++DLC+ CFS G+ H+ H YR+++ S+P++CPD
Sbjct: 5 FHCDVCSSDCTHRVRISCAVCPEYDLCVPCFSQGLYNGNHRPFHDYRIIETNSYPILCPD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+ L++ G GNW+D+A++IG++ K + DHY Y+NSP +P+PD++
Sbjct: 65 WGADEELALIKGAQSLGLGNWHDIAEHIGSREKDEVRDHYIEYYINSPFYPIPDITKDIQ 124
Query: 172 KNKEELL 178
+EE L
Sbjct: 125 VPQEEFL 131
>UniRef100_Q6CWU2 Kluyveromyces lactis strain NRRL Y-1140 chromosome B of strain NRRL
Y- 1140 of Kluyveromyces lactis [Kluyveromyces lactis]
Length = 435
Score = 137 bits (344), Expect = 3e-31
Identities = 52/131 (39%), Positives = 88/131 (66%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++RI CAVC ++DLC+ CF+ G+ H+ +H Y+V++ S+P++C D
Sbjct: 5 FHCDICSADCTNRVRITCAVCPEYDLCVPCFAKGLYNGKHRPSHDYKVIETNSYPILCED 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+LL++ G GNW D+AD+IG++ K + DHY Y+NS +P+PD++
Sbjct: 65 WGADEELLLIKGAQTLGLGNWQDIADHIGSRDKEEVYDHYLKFYLNSEHYPIPDITKDIN 124
Query: 172 KNKEELLAMAK 182
+ +E+ L K
Sbjct: 125 EPQEQFLEKRK 135
>UniRef100_Q6C1P9 Yarrowia lipolytica chromosome F of strain CLIB99 of Yarrowia
lipolytica [Yarrowia lipolytica]
Length = 463
Score = 134 bits (337), Expect = 2e-30
Identities = 56/136 (41%), Positives = 87/136 (63%), Gaps = 8/136 (5%)
Query: 50 SLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLIC 109
S +HC+ C+ D + +RI+CA CQD+DLC++CFS G HK H Y +++ ++P+
Sbjct: 5 SRFHCDVCSCDCTNLVRIRCAECQDYDLCVQCFSQGASSGVHKPYHNYMIIEQHAYPIFT 64
Query: 110 PDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDL--- 166
DW A+EE+LL+E +M G GNW D+AD+IG +SK + +HY VY++SP +P+P +
Sbjct: 65 EDWGADEELLLIEGAEMQGLGNWQDIADHIGGRSKEEVGEHYKEVYLDSPDYPLPPMKRK 124
Query: 167 -----SHFSGKNKEEL 177
+ F+ KE L
Sbjct: 125 FEITAAEFAANKKERL 140
>UniRef100_Q6BI05 Similar to CA3340|CaADA2 Candida albicans CaADA2 [Debaryomyces
hansenii]
Length = 447
Score = 133 bits (334), Expect = 4e-30
Identities = 51/119 (42%), Positives = 83/119 (68%)
Query: 48 KVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPL 107
K LYHC+ C+ D + +IRI+CA+C D+DLC+ CF+ G HK H Y+V++ ++P+
Sbjct: 4 KTRLYHCDVCSSDCTNRIRIQCAICADYDLCVPCFASGSATGDHKPWHDYQVIEQNTYPI 63
Query: 108 ICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDL 166
DW A+EE+LL++ + +G GNW D+AD+IG +SK + +HY +Y+NS +P+P++
Sbjct: 64 FDKDWGADEELLLVQGCETFGLGNWQDIADHIGNRSKDEVKNHYFDIYLNSKEYPLPEM 122
>UniRef100_UPI000042C941 UPI000042C941 UniRef100 entry
Length = 445
Score = 131 bits (330), Expect = 1e-29
Identities = 48/122 (39%), Positives = 84/122 (68%)
Query: 46 DGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSF 105
D + L+HC+ C+ D + +IRI+CA+C D+DLC+ CF+ G+ HK H Y++++ ++
Sbjct: 2 DSRTKLFHCDVCSSDCTNRIRIQCAICTDYDLCVPCFAAGLTTGDHKPWHDYQIIEQNTY 61
Query: 106 PLICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPD 165
P+ DW A+EE+LL++ + G GNW D+AD+IG +SK + +HY +Y+ S +P+P+
Sbjct: 62 PIFDRDWGADEELLLIQGCETSGLGNWADIADHIGNRSKEEVAEHYFKIYLESKDYPLPE 121
Query: 166 LS 167
++
Sbjct: 122 MN 123
>UniRef100_Q756E7 AER318Cp [Ashbya gossypii]
Length = 435
Score = 131 bits (329), Expect = 2e-29
Identities = 48/116 (41%), Positives = 78/116 (66%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++RI CAVC ++DLC+ CFS G+ H+ H YR+++ S+P++C D
Sbjct: 5 FHCDICSSDCTNRVRISCAVCPEYDLCVPCFSQGLYNGNHRPYHDYRIIETNSYPILCDD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLS 167
W A+EE L++ G GNW D+ADNIG++ K + +HY Y+ S +P+PD++
Sbjct: 65 WGADEEQALIKGAQTLGLGNWQDIADNIGSREKEEVYEHYVKYYLKSDFYPIPDIT 120
>UniRef100_Q02336 Transcriptional adapter 2 [Saccharomyces cerevisiae]
Length = 434
Score = 131 bits (329), Expect = 2e-29
Identities = 48/127 (37%), Positives = 83/127 (64%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
+HC+ C+ D + ++R+ CA+C ++DLC+ CFS G H+ H YR+++ S+P++CPD
Sbjct: 5 FHCDVCSADCTNRVRVSCAICPEYDLCVPCFSQGSYTGKHRPYHDYRIIETNSYPILCPD 64
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W A+EE+ L++ G GNW D+AD+IG++ K + +HY Y+ S +P+PD++
Sbjct: 65 WGADEELQLIKGAQTLGLGNWQDIADHIGSRGKEEVKEHYLKYYLESKYYPIPDITQNIH 124
Query: 172 KNKEELL 178
++E L
Sbjct: 125 VPQDEFL 131
>UniRef100_UPI0000498CF8 UPI0000498CF8 UniRef100 entry
Length = 293
Score = 130 bits (327), Expect = 3e-29
Identities = 59/121 (48%), Positives = 79/121 (64%), Gaps = 1/121 (0%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWS 113
CN CNK I+ RI C C +FDLC+ECFS G E+ HK+NH YRV+ +L FPL+ DW
Sbjct: 12 CNSCNKVITTMTRITCVECDNFDLCLECFSQGKEIGKHKNNHSYRVIPSLHFPLLSSDWG 71
Query: 114 AEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSH-FSGK 172
A+EE++LLEA++ G NW +V + + TK+ +C HY Y+NS P+PDL F K
Sbjct: 72 ADEELMLLEAIEQKGLDNWPEVENFVKTKTAKECRSHYYDYYLNSKTHPLPDLEESFLKK 131
Query: 173 N 173
N
Sbjct: 132 N 132
>UniRef100_Q9P7J7 SPCC24B10.08c protein [Schizosaccharomyces pombe]
Length = 437
Score = 130 bits (326), Expect = 4e-29
Identities = 52/113 (46%), Positives = 76/113 (67%), Gaps = 1/113 (0%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPD 111
YHCN C +DI+ I I+C C DFDLCI CF+ G L H +HPYR+++ S+P+ +
Sbjct: 6 YHCNVCAQDITRSIHIRCVECVDFDLCIPCFTSGASLGTHHPSHPYRIIETNSYPIFDEN 65
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIG-TKSKSQCIDHYNTVYVNSPCFPV 163
W A+EE+LL++A + G GNW D+AD +G ++K +C DHY Y+ S C+P+
Sbjct: 66 WGADEELLLIDACETLGLGNWADIADYVGNARTKEECRDHYLKTYIESDCYPL 118
>UniRef100_UPI0000219BE7 UPI0000219BE7 UniRef100 entry
Length = 546
Score = 128 bits (321), Expect = 1e-28
Identities = 57/117 (48%), Positives = 81/117 (68%), Gaps = 4/117 (3%)
Query: 52 YHCNYCNKDISGKIRIKCA--VCQDFDLCIECFSVGVELTPHK-SNHPYRVMDNLSFPLI 108
YHC+ C+ DI+ +RI+CA C D+DLC+ CF+ G + H+ + HPYRV++ SFP+
Sbjct: 19 YHCDVCSVDITSTVRIRCAHSACNDYDLCVNCFAQGSSSSNHQPATHPYRVIEQNSFPIF 78
Query: 109 CPDWSAEEEMLLLEALDMYGFGNWNDVADNI-GTKSKSQCIDHYNTVYVNSPCFPVP 164
DW A+EE+LLLE ++YG G+W D+AD+I G + K + DHY YVNSP FP+P
Sbjct: 79 EEDWGADEELLLLEGAEIYGLGSWADIADHIGGYRHKDEVRDHYINAYVNSPRFPLP 135
>UniRef100_Q8IJP9 ADA2-like protein [Plasmodium falciparum]
Length = 2578
Score = 127 bits (319), Expect = 2e-28
Identities = 65/158 (41%), Positives = 88/158 (55%), Gaps = 11/158 (6%)
Query: 19 NRSKRKKVALNADNLETSSAAGMGITTDGKV--SLYHCNYCNKDISGKIRIKCAVCQDFD 76
N +R A N+ +S + T+ + S YHC+ CNKDI+ IRI+CA C DFD
Sbjct: 1492 NEMQRTNDAFKGGNIIEASNVSLEAITEEDIFDSNYHCDICNKDITHTIRIRCAECVDFD 1551
Query: 77 LCIECFSVGVEL----TPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNW 132
LC+ CFS G E+ H + H Y + FPL +WSAEEE+LLL+ + +GFGNW
Sbjct: 1552 LCVNCFSTGKEMKSDKCEHYNYHNYIPIPKYDFPLYKLNWSAEEELLLLDGISKFGFGNW 1611
Query: 133 NDVADNIG-----TKSKSQCIDHYNTVYVNSPCFPVPD 165
VAD + TK+ +C HY Y+ S C P+PD
Sbjct: 1612 EQVADLVNSVANITKTDRECESHYYNFYLKSNCAPLPD 1649
>UniRef100_Q7RI23 ADA2-like protein [Plasmodium yoelii yoelii]
Length = 2228
Score = 126 bits (316), Expect = 5e-28
Identities = 65/147 (44%), Positives = 85/147 (57%), Gaps = 10/147 (6%)
Query: 29 NADNLETSSAAGMGITTDGKV-SLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVE 87
N +N ++ G+T D S YHC+ CNKDI+ IRI+CA C DFDLCI CFS G E
Sbjct: 1251 NNNNNNNNNTTIEGVTEDDIFDSNYHCDICNKDITHTIRIRCADCVDFDLCINCFSSGKE 1310
Query: 88 L----TPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGT-- 141
+ H + H Y + FPL +WSAEEE+LLL+ + YGFGNW VAD + +
Sbjct: 1311 IKSEKCEHYNYHNYIPIPKYDFPLYKLNWSAEEELLLLDGISKYGFGNWEQVADLVNSVA 1370
Query: 142 ---KSKSQCIDHYNTVYVNSPCFPVPD 165
K+ +C HY Y+ S C P+PD
Sbjct: 1371 KIPKTNKECETHYYNYYLKSSCAPLPD 1397
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.135 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 404,567,288
Number of Sequences: 2790947
Number of extensions: 16719681
Number of successful extensions: 41467
Number of sequences better than 10.0: 584
Number of HSP's better than 10.0 without gapping: 323
Number of HSP's successfully gapped in prelim test: 261
Number of HSP's that attempted gapping in prelim test: 40863
Number of HSP's gapped (non-prelim): 713
length of query: 235
length of database: 848,049,833
effective HSP length: 123
effective length of query: 112
effective length of database: 504,763,352
effective search space: 56533495424
effective search space used: 56533495424
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC140670.16


