

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140550.12 + phase: 0
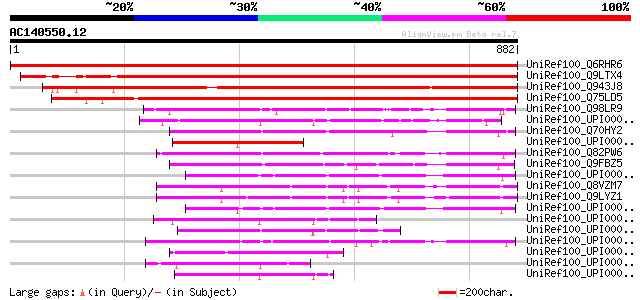
(882 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6RHR6 DMI1 protein [Medicago truncatula] 1736 0.0
UniRef100_Q9LTX4 Emb|CAB86048.1 [Arabidopsis thaliana] 1095 0.0
UniRef100_Q943J8 P0039A07.8 protein [Oryza sativa] 1071 0.0
UniRef100_Q75LD5 Expressed protein [Oryza sativa] 1002 0.0
UniRef100_Q98LR9 Probable secreted protein [Rhizobium loti] 256 3e-66
UniRef100_UPI00002BCA85 UPI00002BCA85 UniRef100 entry 218 5e-55
UniRef100_Q70HY2 Hypothetical protein [Streptomyces parvulus] 210 2e-52
UniRef100_UPI0000314DF4 UPI0000314DF4 UniRef100 entry 203 2e-50
UniRef100_Q82PW6 Hypothetical protein [Streptomyces avermitilis] 203 2e-50
UniRef100_Q9FBZ5 Putative lipoprotein [Streptomyces coelicolor] 187 1e-45
UniRef100_UPI00002D6154 UPI00002D6154 UniRef100 entry 182 3e-44
UniRef100_Q8VZM7 Hypothetical protein At5g02940 [Arabidopsis tha... 182 3e-44
UniRef100_Q9LYZ1 Hypothetical protein F9G14_250 [Arabidopsis tha... 176 2e-42
UniRef100_UPI00002F7A23 UPI00002F7A23 UniRef100 entry 172 4e-41
UniRef100_UPI000029AC2D UPI000029AC2D UniRef100 entry 169 3e-40
UniRef100_UPI0000296BB6 UPI0000296BB6 UniRef100 entry 167 1e-39
UniRef100_UPI0000335672 UPI0000335672 UniRef100 entry 165 5e-39
UniRef100_UPI00002AD0BD UPI00002AD0BD UniRef100 entry 137 2e-30
UniRef100_UPI000026A417 UPI000026A417 UniRef100 entry 130 2e-28
UniRef100_UPI000030E759 UPI000030E759 UniRef100 entry 129 3e-28
>UniRef100_Q6RHR6 DMI1 protein [Medicago truncatula]
Length = 882
Score = 1736 bits (4495), Expect = 0.0
Identities = 882/882 (100%), Positives = 882/882 (100%)
Query: 1 MAKSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQW 60
MAKSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQW
Sbjct: 1 MAKSNEESSNLNVMNKPPLKKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQW 60
Query: 61 NYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITK 120
NYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITK
Sbjct: 61 NYPSFLGIGSTSRKRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITK 120
Query: 121 QQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQE 180
QQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQE
Sbjct: 121 QQQQHSTSSPIFYLLVICCIILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQE 180
Query: 181 EVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAY 240
EVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAY
Sbjct: 181 EVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAY 240
Query: 241 MVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAET 300
MVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAET
Sbjct: 241 MVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAET 300
Query: 301 EGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKL 360
EGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKL
Sbjct: 301 EGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKL 360
Query: 361 GSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLK 420
GSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLK
Sbjct: 361 GSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLK 420
Query: 421 KVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLV 480
KVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLV
Sbjct: 421 KVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLV 480
Query: 481 GGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILI 540
GGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILI
Sbjct: 481 GGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILI 540
Query: 541 SFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPR 600
SFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPR
Sbjct: 541 SFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPR 600
Query: 601 IRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVF 660
IRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVF
Sbjct: 601 IRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVF 660
Query: 661 GLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDI 720
GLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDI
Sbjct: 661 GLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDI 720
Query: 721 QSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYV 780
QSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYV
Sbjct: 721 QSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYV 780
Query: 781 LSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRT 840
LSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRT
Sbjct: 781 LSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRT 840
Query: 841 RKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
RKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE
Sbjct: 841 RKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
>UniRef100_Q9LTX4 Emb|CAB86048.1 [Arabidopsis thaliana]
Length = 824
Score = 1095 bits (2832), Expect = 0.0
Identities = 575/863 (66%), Positives = 677/863 (77%), Gaps = 51/863 (5%)
Query: 20 KKTKTLPSLNLRVSVTPPNPNDNNGIGGTSTTKTDFSEQQWNYPSFLGIGSTSRKRRQPP 79
+++KTLP+ + R +P P G T + D S P+ +P
Sbjct: 13 QRSKTLPTNSYRRFTSPVIPTFRYDDGDTVSYDGDDSSNLPTVPN---------PEEKPV 63
Query: 80 PPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPIFYLLVICC 139
P PS+ P S IT+ Q S + + L C
Sbjct: 64 PVPSQSP----------------------------SQRITRLWTQFSLT----HCLKFIC 91
Query: 140 IILVPYSAYLQYKLAKLKDMKLQLCGQIDFCSRNGKTSIQEEVDDDDNADSRTIALYIVL 199
Y +L+ K+++L+ + L + + S N + + +SR + + V+
Sbjct: 92 SCSFTYVMFLRSKVSRLEAENIILLTRCNSSSDNNEM---------EETNSRAVVFFSVI 142
Query: 200 FTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLC 259
T +LPF+LY YLD L + N LRRT KEDVPLKKR+AY +DV FS+YPYAKLLALL
Sbjct: 143 ITFVLPFLLYMYLDDLSHVKNLLRRTNQKKEDVPLKKRLAYSLDVCFSVYPYAKLLALLL 202
Query: 260 ATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLI 319
AT+ LI +GGLALYAV+ + EALW SWT+VAD+G+HA+ G G RIVSV+ISAGGMLI
Sbjct: 203 ATVVLIVYGGLALYAVSDCGVDEALWLSWTFVADSGSHADRVGVGARIVSVAISAGGMLI 262
Query: 320 FAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVI 379
FA MLGL+SDAIS+ VDSLRKGKSEV+E NH+LILGWSDKLGSLLKQLAIANKS+GGGV+
Sbjct: 263 FATMLGLISDAISKMVDSLRKGKSEVLESNHILILGWSDKLGSLLKQLAIANKSIGGGVV 322
Query: 380 VVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENA 439
VVLAE++KEEME DIAK EFD MGTSVICRSGSPLILADLKKVSVS ARAIIVL +DENA
Sbjct: 323 VVLAERDKEEMETDIAKFEFDLMGTSVICRSGSPLILADLKKVSVSNARAIIVLGSDENA 382
Query: 440 DQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMI 499
DQSDARALRVVLSL GVKEG +GHVVVEM DLDNEPLVKLVGGE IETVVAHDVIGRLMI
Sbjct: 383 DQSDARALRVVLSLTGVKEGWKGHVVVEMCDLDNEPLVKLVGGERIETVVAHDVIGRLMI 442
Query: 500 QCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKI 559
QCALQPGLAQIWEDILGFENAEFYIK+WP+LD F+D+LISFP+AIPCGVKVAAD GKI
Sbjct: 443 QCALQPGLAQIWEDILGFENAEFYIKKWPQLDGYCFEDVLISFPNAIPCGVKVAAD-GKI 501
Query: 560 VINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRD 619
V+NP D+YVL++GDE+LVIAEDDDTYAPG LPEVR +FP+++DPPKYPEKILFCGWRRD
Sbjct: 502 VLNPSDDYVLKEGDEILVIAEDDDTYAPGSLPEVRMCHFPKMQDPPKYPEKILFCGWRRD 561
Query: 620 IDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRH 679
IDDMI VLEA LAPGSELWMFNEVP++ERE+KL L++ L NIKLVHR+GNAVIRRH
Sbjct: 562 IDDMIKVLEALLAPGSELWMFNEVPDQEREKKLTDAGLNISKLVNIKLVHRQGNAVIRRH 621
Query: 680 LESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSG 739
LESLPLETFDSILILA++S+E+S+ HSDSRSLATLLLIRDIQS+RLPY+D KS++LR+SG
Sbjct: 622 LESLPLETFDSILILAEQSLENSIVHSDSRSLATLLLIRDIQSKRLPYKDAKSSALRISG 681
Query: 740 FSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQI 799
F + WIR+MQQASDKSI+ISEILDSRT+NLVSVSRISDYVLSNELVSMALAMVAEDKQI
Sbjct: 682 FPNCCWIRKMQQASDKSIVISEILDSRTKNLVSVSRISDYVLSNELVSMALAMVAEDKQI 741
Query: 800 NRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIIN 859
NRVL+ELFAE+GNE+CI+PAEFY++DQEE+CFYDIM R R R+EI+IGYRLA E+A+IN
Sbjct: 742 NRVLKELFAEKGNELCIRPAEFYIYDQEEVCFYDIMRRARQRQEIIIGYRLAGMEQAVIN 801
Query: 860 PSEKSVPRKWSLDDVFVVLASGE 882
P++KS KWSLDDVFVV+AS +
Sbjct: 802 PTDKSKLTKWSLDDVFVVIASSQ 824
>UniRef100_Q943J8 P0039A07.8 protein [Oryza sativa]
Length = 927
Score = 1072 bits (2771), Expect = 0.0
Identities = 573/847 (67%), Positives = 660/847 (77%), Gaps = 38/847 (4%)
Query: 57 EQQWNYPSFLGIGSTS----RKRRQPPPPP------SKPPVNLIPPHPRPLSVNDHNKTT 106
++ W YPSFLG ++ R ++Q P S+ P PP +S + K+
Sbjct: 98 DRDWCYPSFLGPHASRPRPPRSQQQTPTTTAAAAADSRSPTPAAPPQTASVSQREEEKSL 157
Query: 107 SSLLPQPS------SSSITKQQQQHSTSSPIFYLLVICCIILVPYS-AYLQYKLAKLKDM 159
+S++ +P S S QQ+ YL+++ + ++ +S A Q+ A +
Sbjct: 158 ASVVKRPMLLDERRSLSPPPPQQRAPRFDLSPYLVLMLVVTVISFSLAIWQWMKATVLQE 217
Query: 160 KLQLCGQIDFCSRNGKTSI----QEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYL 215
K++ C + T + D N+ +A + +P L KY+D L
Sbjct: 218 KIRSCCSVSTVDCKTTTEAFKINGQHGSDFINSADWNLASCSRMLVFAIPVFLVKYIDQL 277
Query: 216 PQIINFLRRTESNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAV 275
+ R S +E+VPLKKR+AY VDVFFS +PYAKLLALL AT+ LIA GG+ALY V
Sbjct: 278 RRRNTDSIRLRSTEEEVPLKKRIAYKVDVFFSGHPYAKLLALLLATIILIASGGIALYVV 337
Query: 276 TGGSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKV 335
+G EALW SWT+VAD+GNHA+ G G RIVSVSIS+GGML+FA MLGLVSDAISEKV
Sbjct: 338 SGSGFLEALWLSWTFVADSGNHADQVGLGPRIVSVSISSGGMLVFATMLGLVSDAISEKV 397
Query: 336 DSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIA 395
DS RKGKSE GSLLKQLAIANKS+GGGV+VVLAE++KEEMEMDI
Sbjct: 398 DSWRKGKSE----------------GSLLKQLAIANKSIGGGVVVVLAERDKEEMEMDIG 441
Query: 396 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAG 455
KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA+DENADQSDARALRVVLSL G
Sbjct: 442 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLASDENADQSDARALRVVLSLTG 501
Query: 456 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 515
VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL
Sbjct: 502 VKEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 561
Query: 516 GFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEV 575
GFENAEFYIKRWPELD + F D+LISFPDA+PCGVK+A+ GKI++NPD++YVL++GDEV
Sbjct: 562 GFENAEFYIKRWPELDGMRFGDVLISFPDAVPCGVKIASKAGKILMNPDNDYVLQEGDEV 621
Query: 576 LVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGS 635
LVIAEDDDTY P LP+VRKG+ P I PPKYPEKILFCGWRRDI DMIMVLEAFLAPGS
Sbjct: 622 LVIAEDDDTYVPASLPQVRKGFLPNIPTPPKYPEKILFCGWRRDIHDMIMVLEAFLAPGS 681
Query: 636 ELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILA 695
ELWMFNEVPEKERERKL G +D++GL NIKLVH+EGNAVIRRHLESLPLETFDSILILA
Sbjct: 682 ELWMFNEVPEKERERKLTDGGMDIYGLTNIKLVHKEGNAVIRRHLESLPLETFDSILILA 741
Query: 696 DESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDK 755
DESVEDS+ HSDSRSLATLLLIRDIQS+RLP ++ KS LR +GF H+SWIREMQ ASDK
Sbjct: 742 DESVEDSIVHSDSRSLATLLLIRDIQSKRLPSKELKS-PLRYNGFCHSSWIREMQHASDK 800
Query: 756 SIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 815
SIIISEILDSRTRNLVSVS+ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC
Sbjct: 801 SIIISEILDSRTRNLVSVSKISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMC 860
Query: 816 IKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVF 875
I+ AEFYL++QEEL F+DIM+R R R E+VIGYRLAN ++AIINP +KS RKWSLDDVF
Sbjct: 861 IRSAEFYLYEQEELSFFDIMVRARERDEVVIGYRLANDDQAIINPEQKSEIRKWSLDDVF 920
Query: 876 VVLASGE 882
VV++ G+
Sbjct: 921 VVISKGD 927
>UniRef100_Q75LD5 Expressed protein [Oryza sativa]
Length = 893
Score = 1002 bits (2591), Expect = 0.0
Identities = 524/827 (63%), Positives = 630/827 (75%), Gaps = 22/827 (2%)
Query: 74 KRRQPPPPPSKPPVNLIPPHPRPLSVNDHNKTTSSLLPQPSSSSITKQQQQHSTSSPI-- 131
+++Q PPP+ PP PR V + T + +++++ Q H + S
Sbjct: 71 QQKQQQPPPTTPPPAPRRRDPRYAGVRRGDVRTLTAEKAAAAAAVPTAAQVHGSKSAASA 130
Query: 132 ----------FYLLVICCIILVPYSAYLQYKLAKLKDM------KLQLCGQIDFCSRNGK 175
+V+C LV ++ L ++ LK KLQ C +
Sbjct: 131 TTLRWSGMVSVAAIVLCFSSLVRSNSSLHDQVHHLKAQLAEATTKLQSCITESSMDMSSI 190
Query: 176 TSIQEEVDDDDNADSRTIALYIVLFTLILPFVLYKYLDYLPQIINFLRRTESNKEDVPLK 235
S Q N + +L + L TL P ++ KY+D + LR ++ ++E+VP+
Sbjct: 191 LSYQSNNSTSQNRGLKNFSLLLSLSTLYAPLLILKYMDLFLK----LRSSQDSEEEVPIN 246
Query: 236 KRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADAG 295
KR+AY VD+F S+ PYAK L LL ATL LI GGLALY V S+ + LW SWT+VAD+G
Sbjct: 247 KRLAYRVDIFLSLQPYAKPLVLLVATLLLIGLGGLALYGVNDDSLLDCLWLSWTFVADSG 306
Query: 296 NHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILG 355
NHA EG G ++VSVSIS GGML+FAMMLGLV+D+ISEK DSLRKG+SEVIE++H L+LG
Sbjct: 307 NHANAEGFGPKLVSVSISIGGMLVFAMMLGLVTDSISEKFDSLRKGRSEVIEQSHTLVLG 366
Query: 356 WSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLI 415
WSDKLGSLL Q+AIAN+S+GGG IVV+AEK+KEEME DIAK+EFD GT++ICRSGSPLI
Sbjct: 367 WSDKLGSLLNQIAIANESLGGGTIVVMAEKDKEEMEADIAKMEFDLKGTAIICRSGSPLI 426
Query: 416 LADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEP 475
LADLKKVSVSKARAI+VLA + NADQSDARALR VLSL GVKEGLRGH+VVE+SDLDNE
Sbjct: 427 LADLKKVSVSKARAIVVLAEEGNADQSDARALRTVLSLTGVKEGLRGHIVVELSDLDNEV 486
Query: 476 LVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLF 535
LVKLVGG+L+ETVVAHDVIGRLMIQCA QPGLAQIWEDILGFEN EFYIKRWP+LD + F
Sbjct: 487 LVKLVGGDLVETVVAHDVIGRLMIQCARQPGLAQIWEDILGFENCEFYIKRWPQLDGMQF 546
Query: 536 KDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPEVRK 595
+D+LISFPDAIPCG+KVA+ GGKI++NPDD YVL++GDEVLVIAEDDDTYAP PLP+V +
Sbjct: 547 EDVLISFPDAIPCGIKVASYGGKIILNPDDFYVLQEGDEVLVIAEDDDTYAPAPLPKVMR 606
Query: 596 GYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNEVPEKERERKLAAG 655
GY P+ PK PE+ILFCGWRRD++DMIMVL+AFLAPGSELWMFN+VPE +RERKL G
Sbjct: 607 GYLPKDFVVPKSPERILFCGWRRDMEDMIMVLDAFLAPGSELWMFNDVPEMDRERKLIDG 666
Query: 656 ELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLL 715
LD LENI LVHREGNAVIRRHLESLPLE+FDSILILADESVEDS +DSRSLATLL
Sbjct: 667 GLDFSRLENITLVHREGNAVIRRHLESLPLESFDSILILADESVEDSAIQADSRSLATLL 726
Query: 716 LIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSR 775
LIRDIQ++RLP+R+ + + F SWI EMQQASDKS+IISEILD RT+NL+SVS+
Sbjct: 727 LIRDIQAKRLPFREAMVSHVTRGSFCEGSWIGEMQQASDKSVIISEILDPRTKNLLSVSK 786
Query: 776 ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIM 835
ISDYVLSNELVSMALAMVAED+QIN VLEELFAE+GNEM I+PA+ YL + EEL F+++M
Sbjct: 787 ISDYVLSNELVSMALAMVAEDRQINDVLEELFAEQGNEMQIRPADLYLREDEELNFFEVM 846
Query: 836 IRGRTRKEIVIGYRLANQERAIINPSEKSVPRKWSLDDVFVVLASGE 882
+RGR RKEIVIGYRL + ERAIINP +K R+WS DVFVV+ E
Sbjct: 847 LRGRQRKEIVIGYRLVDAERAIINPPDKVSRRRWSAKDVFVVITEKE 893
>UniRef100_Q98LR9 Probable secreted protein [Rhizobium loti]
Length = 633
Score = 256 bits (653), Expect = 3e-66
Identities = 202/669 (30%), Positives = 327/669 (48%), Gaps = 67/669 (10%)
Query: 234 LKKRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVT-----GG---SMAEALW 285
L R+ Y D + P A L+ L LI A AVT GG + EA W
Sbjct: 7 LGTRLRYGFDKSMAAGPIA-LIGWLAVVSLLIIIAAAAFLAVTRIAPEGGEPLNFFEAFW 65
Query: 286 HSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEV 345
S D+G G R+V + ++ G+ + + ++G++S + K+D LRKG+S V
Sbjct: 66 ESLMRTLDSGTMGGDTGWAFRLVMLVVTLAGIFVVSALIGVLSAGVDGKLDELRKGRSRV 125
Query: 346 IERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTS 405
+E +H +IL WS + ++ +L IAN S IVV+A +K ME +IA T
Sbjct: 126 LESDHTIILNWSPSIFDVISELVIANASRRRPRIVVMANMDKVAMEDEIAAKVGKLGNTR 185
Query: 406 VICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRG--- 462
+ICRSG P L DL V+ +R++IVL+ D D D++ ++ VL+L V + R
Sbjct: 186 IICRSGDPTDLYDLAIVNPQTSRSVIVLSPD--GDDPDSQVIKTVLAL--VNDPSRRTDP 241
Query: 463 -HVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAE 521
++ E+ D N + ++VGG ++ V+A +I R+++ + Q GL+ ++ ++L F+ E
Sbjct: 242 YNIAAEIRDGKNAEVARVVGGAEVQLVLADQLISRIVVHSSRQSGLSGVYSELLDFDGCE 301
Query: 522 FYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAED 581
Y PEL F + ++++ G + GG++ +NP V+ ++IAED
Sbjct: 302 IYTTTQPELAGKTFGEAVMAYEHCALIG--LCDQGGRVNLNPPSELVIGKDMRAIIIAED 359
Query: 582 DDTYAPGPLPEVRKGYFPRIRDP---PKYPEKILFCGWRRDIDDMIMVLEAFLAPGSELW 638
D PG K IRDP PE+ L GW R + L ++APGS L
Sbjct: 360 DAAIRPGSAG--IKIDTAAIRDPRPVEAKPERTLILGWNRRGPIITYELSRYVAPGSILT 417
Query: 639 MFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADES 698
+ + P E+E AG L V G +N+ + R + L SL + ++D +L+L S
Sbjct: 418 IAADTPGLEQE---VAG-LPVAG-DNLSVSCRITDTSSSTALSSLDVPSYDHVLVLG-YS 471
Query: 699 VEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSII 758
+ +D+R+L TLL +R I L +S I
Sbjct: 472 ETMAAQPADTRTLVTLLHLRKI---------ADDAGLHIS-------------------I 503
Query: 759 ISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKP 818
+SE++D R R L +V++ D+V+SN LVS+ LA +E++ + + ++L E+G+E+ ++P
Sbjct: 504 VSEMIDVRNRELAAVTKADDFVVSNRLVSLMLAQASENQYLAAIFDDLLDEQGSEIYMRP 563
Query: 819 AEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLAN----QERAI----INPSEKSVPRKWS 870
Y+ + L F+ I R R EI IGYR +RA+ +NP KS +
Sbjct: 564 VADYVAIDQPLTFWTIAESARLRGEIAIGYRRIRAGDADQRALGGVTVNPL-KSEALTYL 622
Query: 871 LDDVFVVLA 879
+D +VLA
Sbjct: 623 PEDRIIVLA 631
>UniRef100_UPI00002BCA85 UPI00002BCA85 UniRef100 entry
Length = 661
Score = 218 bits (556), Expect = 5e-55
Identities = 181/660 (27%), Positives = 327/660 (49%), Gaps = 65/660 (9%)
Query: 227 SNKEDVPLKKRVAYMVDVFFSIYPYAKLLALLCATLF-------LIAFGGLALYAVTGGS 279
S + PL R+ Y +D F S + LALLC LF + + L T S
Sbjct: 2 STNKKYPLNLRLRYAIDNFMSKGSSSIFLALLCLFLFGFLIMVLIRSIANTFLPDETLSS 61
Query: 280 MAEALWHSWTYVADAGNHAETEGTGQRIVSVSISAG---GMLIFAMMLGLVSDAISEKVD 336
AE W + V + G+ AET+G ++ G G+++F+ M+ ++ K+D
Sbjct: 62 WAEIPWRVYVAVME-GSAAETDGDSNWAAKLTSIIGVMVGLILFSSMVAFITSVFEAKLD 120
Query: 337 SLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAK 396
LR+G+S V+E++H LILG+ D++ ++++L AN+S IV+LAE +KEEM+ I
Sbjct: 121 ELRRGRSLVLEKDHTLILGFGDRILEVIRELIEANESEPDAAIVILAENDKEEMDNIIRD 180
Query: 397 LEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAA------DENADQSDARALRVV 450
DF+ T +I RSG + +L+KV A+++I++ + +E + +DA L+ +
Sbjct: 181 NISDFVTTRIITRSGIVTNINNLRKVMAETAKSVIIMNSAASWRPEEEKNLADALVLKSI 240
Query: 451 LSLAGVKEG-LRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCAL-QPGLA 508
+S+ V +G +V E+ + L + + ++ + V+ R++ Q AL + GL+
Sbjct: 241 MSIIAVCDGNEHPPIVCEIHSDRDRNLAENISNGTVKALNEVSVLSRMIAQLALSRNGLS 300
Query: 509 QIWEDILGFENAEFYIKR----WPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPD 564
++ D++GF+ EFY + W L F + L F + P G+ A G+IV+NP
Sbjct: 301 IVYSDMVGFDGNEFYFYKPDDGWG--GKLNFGESLNRFNSSTPMGIHNA--NGEIVLNPP 356
Query: 565 DNYVLRDGDEVLVIAEDDDT--YAPGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDD 622
+ + D DE++V AEDD T Y + P P +I W
Sbjct: 357 KDTPINDDDELIVFAEDDSTIFYFNESVFAPSLNTIPSSEISPS-SHRIALLNWTTKTGI 415
Query: 623 MIMVLEAFLAPGSELWMF--NEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHL 680
++ L ++L SEL+ + +++PE + L + + + NI + + N + + L
Sbjct: 416 ILEKLCSYLPANSELFTYVSDKLPEMD---GLKSSLSESYPGINIMINEVDLNDLDK--L 470
Query: 681 ESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGF 740
+ + + FDSILIL+ ++ D+ ++ L+ IR I + D S
Sbjct: 471 DDIEPQNFDSILILSPGGT--TIEEMDAYVISLLIRIRQILISKSNKNDKNS-------- 520
Query: 741 SHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQIN 800
N W + +I+E++DS +++ S + D+++SN+ VS +A V+E+
Sbjct: 521 --NKWPK----------LITEVMDSDNIDIILNSGVEDFMVSNQFVSQIMAQVSEEPMAL 568
Query: 801 RVLEELFAEEGNEMCIKPAEFYLFDQEE-----LCFYDIMIRGRTRKEIVIGYRLANQER 855
V ++LF EG+E+ IKPA FY FD E + + + + + R E+++G ++ +++R
Sbjct: 569 DVYDDLFQAEGSELYIKPASFY-FDFSEKDSITIPYGECVQAAQLRNEVILGLQIHSEQR 627
>UniRef100_Q70HY2 Hypothetical protein [Streptomyces parvulus]
Length = 638
Score = 210 bits (534), Expect = 2e-52
Identities = 167/620 (26%), Positives = 290/620 (45%), Gaps = 56/620 (9%)
Query: 279 SMAEALWHSWTYVADAGNHAETEGTGQR-IVSVSISAGGMLIFAMMLGLVSDAISEKVDS 337
S+AE L W + GT R ++SV ++ +L + ++GL++ A++E++ S
Sbjct: 56 SLAERLAEVWRLTGETLRLGGATGTPLRAMLSVLLALVTLLYVSTLVGLITTALTERLTS 115
Query: 338 LRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKL 397
LR+G+S V+E+ H ++LGWS+++ +++ +L AN + G +VVLA+++K ME +
Sbjct: 116 LRRGRSTVLEQGHAVVLGWSEQVFTVVSELVAANVNQRGAAVVVLADRDKTVMEESLGTK 175
Query: 398 EFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVK 457
GT +ICRSG A L S + A ++VL DE +DA ++ +L+L
Sbjct: 176 VGSCGGTRLICRSGPTTDPAVLPLTSPATAGVVLVLPPDE--PHADAEVVKTLLALRAAL 233
Query: 458 EGL--RGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDIL 515
G R VV + D L G + + V RL++Q A +PG++ + ++L
Sbjct: 234 AGAKPRPPVVAAVRDDRYRLAACLAAGPDGVVLESDTVTARLIVQAARRPGISLVHRELL 293
Query: 516 GFENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEV 575
F EFY+ P L F ++L+S+ G+ G ++NP + D +
Sbjct: 294 DFAGDEFYLISEPALTGRPFGEVLLSYSTTSVVGLM---RGCTPLLNPPPTTPVAPDDLL 350
Query: 576 LVIAEDDDTYAPGPLPEVRKGYFPRIRDPPKYP-EKILFCGWRRDIDDMIMVLEAFLAPG 634
+VI DDDT E + R P P E+IL GW R ++ L PG
Sbjct: 351 VVITGDDDTARLDDCAESVEKAAVASRPPTPAPAERILLLGWNRRAPLVVDQLHRRARPG 410
Query: 635 SELWMFNEVPEKERERKLAAGELDVFGLEN----IKLVHREGNAVIRRHLESLPLETFDS 690
S + + E P + R+++ E D EN + L G+ L L + ++DS
Sbjct: 411 SAVDVVAE-PGEATIREISEAEADSGNGENGGNGLSLALHHGDITRPETLRRLDVHSYDS 469
Query: 691 ILILA-DESVEDSVAHSDSRSLATLLLIRDIQS---RRLPYRDTKSTSLRLSGFSHNSWI 746
+++L D + D+R+L TLLL+R ++ R LP
Sbjct: 470 VIVLGRDPAPGQPPDDPDNRTLVTLLLLRQLEEATGRELP-------------------- 509
Query: 747 REMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEEL 806
+++E++D R R L + +D ++S +L+ + ++ +++++ + V EEL
Sbjct: 510 -----------VVTELIDDRNRALAPIGPGADVIISGKLIGLLMSQISQNRHLAAVFEEL 558
Query: 807 FAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYR------LANQERAIINP 860
F+ EG + ++PA YL F ++ R R E IGYR INP
Sbjct: 559 FSAEGAGVRLRPATDYLLPGSTTSFATVVAAARRRGECAIGYRDHADASTRPHYGVRINP 618
Query: 861 SEKSVPRKWSLDDVFVVLAS 880
++ R+W+ +D VV+ +
Sbjct: 619 PKRE-RRRWTAEDEVVVIGT 637
>UniRef100_UPI0000314DF4 UPI0000314DF4 UniRef100 entry
Length = 236
Score = 203 bits (517), Expect = 2e-50
Identities = 108/236 (45%), Positives = 163/236 (68%), Gaps = 8/236 (3%)
Query: 284 LWHSWTYVADAGNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKS 343
+W ++ + D G+H E E QR + +I+ G++IF+ ++G++ DA++EK++ L+KGKS
Sbjct: 1 MWEVYSMIMDPGSHVELESAPQRAMGSAITWIGVIIFSTIVGIILDAVTEKMEDLKKGKS 60
Query: 344 EVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDI------AKL 397
V+E +H LI+GW+DK L +++A AN+S GGGVIVVL +KEE+EM A +
Sbjct: 61 TVVEADHTLIIGWTDKTAGLCREIADANESEGGGVIVVLDPLDKEELEMIYRNQVRPADM 120
Query: 398 EFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA-ADENADQSDARALRVVLSLAGV 456
D T V+ RSGS L DL +VS + AR+I++L+ ++ D +DA LRVVL+L +
Sbjct: 121 APDGNVTKVVFRSGSALRATDLARVSATTARSIVLLSDYGQDPDLADADILRVVLTLRTL 180
Query: 457 KEG-LRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIW 511
G L GH+V E+ D+DNEPLV LVGG+ +ETVV+HD+IGRLM+ A QPG+A+++
Sbjct: 181 NGGELSGHIVCEVRDVDNEPLVSLVGGKHVETVVSHDIIGRLMLMAARQPGIARVF 236
>UniRef100_Q82PW6 Hypothetical protein [Streptomyces avermitilis]
Length = 656
Score = 203 bits (517), Expect = 2e-50
Identities = 180/642 (28%), Positives = 298/642 (46%), Gaps = 61/642 (9%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG--SMAEALWHSWTYVADAGNHAETEGTGQR-IVSVS 311
L ++C + ++ L ++ G S++ L W A+ GT R ++S
Sbjct: 35 LVIICLAV-VVPVSALLVWTDPGSPRSLSGRLAAVWRSSAETLRLGTVTGTPLRMLLSAL 93
Query: 312 ISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIAN 371
+ +L + ++G+++ ++E++ L +G+S V+E+ H ++LGWSD++ +++ +L A
Sbjct: 94 LGLVALLCVSTLVGVITTGLAERLAELSRGRSTVLEQGHAVVLGWSDQVSTVVGELVAAQ 153
Query: 372 KSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSVSKARAII 431
S +VVLAE++K EME +A T ++CRSG L VS A ++
Sbjct: 154 SSYRPRAVVVLAERDKTEMERALAAHVGPAGRTRLVCRSGPASDPGVLALVSPQTASTVL 213
Query: 432 VLAADENADQSDARALRVVLSLAGV-KEGLRGHVVVE--MSDLDNEPLVKLVGGELIETV 488
VL + E +DA LRV+L+L V EG G V+ + D P +L G +
Sbjct: 214 VLPSGE--PTADAEVLRVLLALRAVLGEGTGGPPVLAAVLDDRYRAP-ARLAAGPRGTVL 270
Query: 489 VAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPC 548
V RL+ QC +PGL+ + D+L F EF++ F L+ A C
Sbjct: 271 ETDTVTARLIAQCVGRPGLSLVLRDLLDFAGDEFHLAEATAFHGGPFGAALLG--HATSC 328
Query: 549 GVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPLPE-VRKGYFPRIRDPPKY 607
V + G+ ++NP ++ G ++V+ DD + P V + PP+
Sbjct: 329 VVGLLTAEGRTLLNPPAATLVAPGSRLVVLTRDDGSARPEDCRHLVEPSAIAMAQPPPED 388
Query: 608 PEKILFCGWRRDIDDMIMVLEAFLAPGSELWMFNE--VPEKERERKLAAGELDVFGLENI 665
+L GW R ++ L PGS L + + VP RE + A L V ++
Sbjct: 389 AAHLLLLGWNRRAPLVVNQLRRTARPGSVLDVVTDRAVPGPTREPESGAARLAV-RFQSA 447
Query: 666 KLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRDIQSRRL 725
L E L L L +D +++L E D D R+L TLL +R ++ R
Sbjct: 448 DLSRPE-------TLLGLDLAPYDGLIVLGPEP-GDGPDRPDDRTLVTLLALRLLEER-- 497
Query: 726 PYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNEL 785
+G REM+ +I+E+ D R R L V+ SD ++S EL
Sbjct: 498 ------------TG-------REMR-------VITELTDDRNRPLAPVNPGSDVIVSGEL 531
Query: 786 VSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIV 845
+ LA +A+++ + V +ELF+ +G+ +C++ A Y+ E F ++ R R E
Sbjct: 532 TGLLLAQIAQNRHLAPVFDELFSADGSTICLRQAGHYVRTGCETTFATVVAAARDRGECA 591
Query: 846 IGYRLANQERAI-------INPSEKSVPRKWSLDDVFVVLAS 880
IGYR + RAI +NP +KS R+WS DD VV+A+
Sbjct: 592 IGYR-RHDRRAIPPDHGIRLNP-DKSERREWSADDQVVVVAT 631
>UniRef100_Q9FBZ5 Putative lipoprotein [Streptomyces coelicolor]
Length = 672
Score = 187 bits (475), Expect = 1e-45
Identities = 156/614 (25%), Positives = 270/614 (43%), Gaps = 57/614 (9%)
Query: 279 SMAEALWHSWTYVADAGNHAETEGTGQR-IVSVSISAGGMLIFAMMLGLVSDAISEKVDS 337
S+ E L W A+ G R ++SV + +L + ++G+++ + ++++
Sbjct: 57 SLTERLVAVWRTSAETLRLGGVTGAPLRMLLSVFLGLIALLCVSTLVGVITTGLGDRLEE 116
Query: 338 LRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKL 397
LR+G+S V+E+ H ++LGWSD++ +++ ++ I+ G + VLA+++ M D+
Sbjct: 117 LRRGRSRVLEKGHAVVLGWSDQVFTVVGEMVISQVGRVRGAVAVLADRDSAVMASDLNAA 176
Query: 398 EFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSL-AGV 456
G V+CR+G+P+ A L ++ + A ++VL D++AD DA +RV+L+L A +
Sbjct: 177 LGVTRGVRVVCRTGAPIDPAALALLTPAAAHCVLVLPGDDDAD--DAEVVRVLLALRALL 234
Query: 457 KEGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILG 516
G VV + D +L G + RL++Q A PGL + D+L
Sbjct: 235 GAGAGPPVVAAVRDERFLTAARLAAGPRGFVLDVESTAARLLVQAARHPGLVRALRDLLD 294
Query: 517 FENAEFYIKRWPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVL 576
AEF++ P+ L F +I + +A C V A G+ ++ P GD ++
Sbjct: 295 LTGAEFHVVHAPDALGLTFAEISSRYEEA--CAVGYLAADGRALLTPASGARCGPGDRLI 352
Query: 577 VIAEDDDTYAPGPLPEVRKGYFPRIR----DPPKYPEKILFCGWRRDIDDMIMVLEAFLA 632
V+A DD P P + D + K L GW R ++ L
Sbjct: 353 VVARDD---RPPVAKREGTAVDPTVMADRPDRQRSFSKTLLLGWNRRAPLVMESLSRTAQ 409
Query: 633 PGSELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSIL 692
PGS L V + + AG + E + + + G L +L L +DS++
Sbjct: 410 PGSHL----HVVSGADDGPVTAGAVGPTDDERVAVTYHVGEPTRPDTLRALDLFGYDSVI 465
Query: 693 ILADESVEDSVAHSDSRSLATLLLIRDIQ---SRRLPYRDTKSTSLRLSGFSHNSWIREM 749
LA ++ A D + L TLL +R ++ R LP
Sbjct: 466 ALAPDAGPHR-ARPDDQLLLTLLNLRAVEEETGRALP----------------------- 501
Query: 750 QQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAE 809
+++E+ D R+ L + D V+ EL S+ + +A++ + V EELFA
Sbjct: 502 --------VVAELTDHRSLALAPLGPEGDAVVRGELTSLVMTRIAQNTGMAAVFEELFAA 553
Query: 810 EGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGY-----RLANQERAIINPSEKS 864
G + ++PA Y+ F ++ TR E VIGY R +E + K+
Sbjct: 554 RGGALALRPASHYVLPGRVASFATVVASALTRGECVIGYRAHDARTPRREADVRLAPGKA 613
Query: 865 VPRKWSLDDVFVVL 878
R WS D +V+
Sbjct: 614 ERRVWSSSDEILVV 627
>UniRef100_UPI00002D6154 UPI00002D6154 UniRef100 entry
Length = 590
Score = 182 bits (463), Expect = 3e-44
Identities = 139/587 (23%), Positives = 283/587 (47%), Gaps = 55/587 (9%)
Query: 307 IVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLGSLLKQ 366
+++ ++ G+L+ ++++G++ AI+++++S+R G + E+NH L+LGWS + + K+
Sbjct: 43 LLNFFVAMTGLLVSSIVIGIIVQAITDRIESIRNGTGFIDEKNHQLVLGWSSGIDLVFKE 102
Query: 367 LAIANKSVGGGVIVVLAE-KEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSVS 425
+AN++ IV + E + I+K +I + G L+ V+V
Sbjct: 103 FLLANENQKNSNIVFMDNINPVEANQRIISKFPDKKDYQRIIPKQGKTSDRKYLEIVNVD 162
Query: 426 KARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGELI 485
+AR+I + + + D +A+ +++ KEG + ++V +M+ +N L K++GG+
Sbjct: 163 QARSIFI---NHDDDMQSIKAIAAIVNNPNRKEG-KYNIVSKMNVKENYELAKIIGGDET 218
Query: 486 ETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPDA 545
+ + D++GR+ Q LQ GLA ++ +++ F+ E Y + L + D ++S+ +
Sbjct: 219 KVLFFGDMLGRVGAQSCLQSGLAHVYLELVNFDGDEIYFHKEASLVGKSYADSVLSYDTS 278
Query: 546 IPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTY---APGPLPEVRKGYFPRIR 602
G++ G ++INP + D D ++ ++ DDDT LP+ K I+
Sbjct: 279 SVIGIE---RNGNVIINPSGKEKIMDDDLIIAVSMDDDTVIKDGKNILPDESK-MVKEIK 334
Query: 603 DPPKYPEKILFCGWRRDIDDMIMV---LEAFLAPGSELWMFNEVPEKERERKLAAGELDV 659
K +F D+ + ++ L +L GS + + NE + ++
Sbjct: 335 IQRKKDNIFIFGHNEGDLSRLKVMCDHLSKYLDDGSSICLVNE----NKNSLDIVDQVSS 390
Query: 660 FGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLLLIRD 719
+ IK+ EG+ R LESL +E ILIL+ + + + D+++L TL+ +RD
Sbjct: 391 SSEKEIKISFIEGDYRSREFLESLQIENGTKILILSPYNGLNDIDSCDAKTLFTLINLRD 450
Query: 720 IQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDY 779
I+ R + I+ +E+++S + S ++ DY
Sbjct: 451 IEKRE----------------------------NKNFILTTELINSNNAEIFSSNKDDDY 482
Query: 780 VLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGR 839
+ S ++V L ++E+ +++ ++L + +G+E+ KP Y+ E + FY I
Sbjct: 483 LYSEDIVKSMLVQISENPALDKFFDDLASPQGSEIYFKPLINYVDLSEPIDFYTICKSAL 542
Query: 840 TRKEIVIGYRLANQER-------AIINPSEKSVPRKWSLDDVFVVLA 879
+ + IGY++A+ E +INP KS R ++ +D+ +V A
Sbjct: 543 DKNQTAIGYQIADNEHLKELDCGIVINP-VKSEKRLFNKEDMLIVFA 588
>UniRef100_Q8VZM7 Hypothetical protein At5g02940 [Arabidopsis thaliana]
Length = 813
Score = 182 bits (463), Expect = 3e-44
Identities = 156/685 (22%), Positives = 317/685 (45%), Gaps = 98/685 (14%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG-SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSIS 313
+ LL + GGL + S+ + LW +W + +A H E + +R++ ++
Sbjct: 163 VVLLITCFSFVIIGGLFFFKFRKDTSLEDCLWEAWACLVNADTHLEQKTRFERLIGFVLA 222
Query: 314 AGGMLIFAMMLGLVSDAISEKVDSLRKGKS-EVIERNHVLILGWSDKLGSLLKQL----- 367
G++ ++ +L +++ + +R+G +V+E +H++I G + L +LKQL
Sbjct: 223 IWGIVFYSRLLSTMTEQFRYHMKKVREGAHMQVLESDHIIICGINSHLPFILKQLNSYQQ 282
Query: 368 ---AIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSV 424
+ + ++++++ ++EM+ DF ++ +S S + ++ +
Sbjct: 283 HAVRLGTTTARKQTLLLMSDTPRKEMDKLAEAYAKDFDQLDILTKSCSLNMTKSFERAAA 342
Query: 425 SKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGEL 484
ARAII+L + + D A VL+L +++ +VE+S + L+K + G
Sbjct: 343 CMARAIIILPTKGDRYEVDTDAFLSVLALEPIQKMESIPTIVEVSSSNMYDLLKSISGLK 402
Query: 485 IETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPD 544
+E V + +L +QC+ Q L +I+ +L + F + +P L + ++ + + F +
Sbjct: 403 VEPV--ENSTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLTGMKYRQLRLGFQE 460
Query: 545 AIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA-------------------EDDDTY 585
+ CG+ GK+ +P+D+ L + D++L IA E DDT
Sbjct: 461 VVVCGI---LRDGKVNFHPNDDEELMETDKLLFIAPLKKDFLYTDMKTENMTVDETDDTR 517
Query: 586 APGPLPEVRKGYFPRIRDPP-----------KYP-EKILFCGWRRDIDDMIMVLEAFLAP 633
+ E +K +I P K P E IL GWR D+ +MI +++L P
Sbjct: 518 KQ--VYEEKKSRLEKIITRPSKSLSKGSDSFKGPKESILLLGWRGDVVNMIKEFDSYLGP 575
Query: 634 GSELWMFNEVPEKER---ERKLAAGELDVFGLENIKLVHREGNAV-----------IRRH 679
GS L + ++VP ++R ++ +A G+ ++NI++ H GN + ++
Sbjct: 576 GSSLEILSDVPLEDRRGVDQSIATGK-----IKNIQVSHSVGNHMDYDTLKESIMHMQNK 630
Query: 680 LESLPLETFDSILILAD-ESVEDSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLS 738
E + +I++++D + + + +D +S TLLL I + +L
Sbjct: 631 YEKGEEDIRLTIVVISDRDLLLGDPSRADKQSAYTLLLAETICN-------------KLG 677
Query: 739 GFSHNSWIREMQQASDKSIIISEILDSRT-RNLVSVSRISDYVLSNELVSMALAMVAEDK 797
HN + SEI+D++ + + + ++ + E++S+ A VAE+
Sbjct: 678 VKVHN--------------LASEIVDTKLGKQITRLKPSLTFIAAEEVMSLVTAQVAENS 723
Query: 798 QINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAI 857
++N V +++ EG+E+ +K E Y+ + E F ++ R R+E+ IGY ++ I
Sbjct: 724 ELNEVWKDILDAEGDEIYVKDIELYMKEGENPSFTELSERAWLRREVAIGYIKGGKK--I 781
Query: 858 INPSEKSVPRKWSLDDVFVVLASGE 882
INP K+ P ++D +V++ E
Sbjct: 782 INPVPKTEPVSLEMEDSLIVISELE 806
>UniRef100_Q9LYZ1 Hypothetical protein F9G14_250 [Arabidopsis thaliana]
Length = 776
Score = 176 bits (447), Expect = 2e-42
Identities = 153/684 (22%), Positives = 314/684 (45%), Gaps = 88/684 (12%)
Query: 255 LALLCATLFLIAFGGLALYAVTGG-SMAEALWHSWTYVADAGNHAETEGTGQRIVSVSIS 313
+ LL + GGL + S+ + LW +W + +A H E + +R++ ++
Sbjct: 118 VVLLITCFSFVIIGGLFFFKFRKDTSLEDCLWEAWACLVNADTHLEQKTRFERLIGFVLA 177
Query: 314 AGGMLIFAMMLGLVSDAISEKVDSLRKGKS-EVIERNHVLILGWSDKLGSLLKQL----- 367
G++ ++ +L +++ + +R+G +V+E +H++I G + L +LKQL
Sbjct: 178 IWGIVFYSRLLSTMTEQFRYHMKKVREGAHMQVLESDHIIICGINSHLPFILKQLNSYQQ 237
Query: 368 ---AIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKKVSV 424
+ + ++++++ ++EM+ DF ++ +S + + ++ +
Sbjct: 238 HAVRLGTTTARKQTLLLMSDTPRKEMDKLAEAYAKDFDQLDILTKSLN--MTKSFERAAA 295
Query: 425 SKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVGGEL 484
ARAII+L + + D A VL+L +++ +VE+S + L+K + G
Sbjct: 296 CMARAIIILPTKGDRYEVDTDAFLSVLALEPIQKMESIPTIVEVSSSNMYDLLKSISGLK 355
Query: 485 IETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILISFPD 544
+E V + +L +QC+ Q L +I+ +L + F + +P L + ++ + + F +
Sbjct: 356 VEPV--ENSTSKLFVQCSRQKDLIKIYRHLLNYSKNVFNLCSFPNLTGMKYRQLRLGFQE 413
Query: 545 AIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIA-------------------EDDDTY 585
+ CG+ GK+ +P+D+ L + D++L IA E DDT
Sbjct: 414 VVVCGI---LRDGKVNFHPNDDEELMETDKLLFIAPLKKDFLYTDMKTENMTVDETDDTR 470
Query: 586 APGPLPEVRKGYFPRIRDPP-----------KYP-EKILFCGWRRDIDDMIMVLEAFLAP 633
+ E +K +I P K P E IL GWR D+ +MI +++L P
Sbjct: 471 KQ--VYEEKKSRLEKIITRPSKSLSKGSDSFKGPKESILLLGWRGDVVNMIKEFDSYLGP 528
Query: 634 GSELWMFNEVPEKER---ERKLAAGELDVFGLENIKLVHREGNAV-----------IRRH 679
GS L + ++VP ++R ++ +A G++ +NI++ H GN + ++
Sbjct: 529 GSSLEILSDVPLEDRRGVDQSIATGKI-----KNIQVSHSVGNHMDYDTLKESIMHMQNK 583
Query: 680 LESLPLETFDSILILADESVE-DSVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLS 738
E + +I++++D + + +D +S TLLL I ++ L
Sbjct: 584 YEKGEEDIRLTIVVISDRDLLLGDPSRADKQSAYTLLLAETICNK-------------LG 630
Query: 739 GFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQ 798
HN + K ++ D++ + + ++ + E++S+ A VAE+ +
Sbjct: 631 VKVHNLASEIVDTKLGKQLLKH---DTKPYQITRLKPSLTFIAAEEVMSLVTAQVAENSE 687
Query: 799 INRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIMIRGRTRKEIVIGYRLANQERAII 858
+N V +++ EG+E+ +K E Y+ + E F ++ R R+E+ IGY ++ II
Sbjct: 688 LNEVWKDILDAEGDEIYVKDIELYMKEGENPSFTELSERAWLRREVAIGYIKGGKK--II 745
Query: 859 NPSEKSVPRKWSLDDVFVVLASGE 882
NP K+ P ++D +V++ E
Sbjct: 746 NPVPKTEPVSLEMEDSLIVISELE 769
>UniRef100_UPI00002F7A23 UPI00002F7A23 UniRef100 entry
Length = 624
Score = 172 bits (436), Expect = 4e-41
Identities = 132/591 (22%), Positives = 289/591 (48%), Gaps = 65/591 (10%)
Query: 307 IVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLGSLLKQ 366
++++ ++ G+ + ++++G++ AI+++++S+R+G + E NH LILGWS + + ++
Sbjct: 79 LLNLLVTLTGLFVSSIVIGIIVTAITDRIESVRQGTGYIDESNHQLILGWSSGITLVFRE 138
Query: 367 LAIANKSVGGGVIVVLAEKEKEEMEMDIA-----KLEFDFMGTSVICRSGSPLILADLKK 421
+AN++ IV + E I K ++ +I + G+ L+
Sbjct: 139 FLLANENQKDTNIVFMDNINPVEANQRIITQFPNKKDYQ----RIIPKQGNISDRKHLEI 194
Query: 422 VSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSDLDNEPLVKLVG 481
V+V KAR+I + D D +A+ +++ ++ + ++V +M+ +N L K++G
Sbjct: 195 VNVDKARSIFINHED---DMESIKAIAAIVNNPNRRKE-KYNIVSKMNIKENYELAKIIG 250
Query: 482 GELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPELDDLLFKDILIS 541
G+ + + D++ R+ Q LQ GLA ++ +++ F+ E Y + P L + D ++S
Sbjct: 251 GDETKVLFFGDMLARVGAQSCLQSGLANVYLELVNFDGDEIYFHKEPTLVGKNYGDAVLS 310
Query: 542 FPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTY---APGPLPEVRKGYF 598
+ + G++ G +++NP+ N + + D ++ I+ DDDT +P K
Sbjct: 311 YDTSSVIGIE---RDGNVIVNPNANEEIMNNDSIIAISMDDDTVIKDGKDIIPAANK-IA 366
Query: 599 PRIRDPPKYPEKILFCGWRRDIDDMIMV---LEAFLAPGSELWMFNEVPEKERERKLAAG 655
++ + K +F D+ + ++ L ++ GS + + NE + L+
Sbjct: 367 NKVSNNNKTENIFIFGHSEGDLSRLKVICNHLIKYIDDGSSICLVNEDSD-----TLSLI 421
Query: 656 ELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVEDSVAHSDSRSLATLL 715
E D+ +NIK+ +GN R LE++ +++ D +LIL+ + + + SD+ +L TL+
Sbjct: 422 E-DIRSDKNIKVSFIQGNFRSREFLENIQIKSGDKVLILSPFNGTNDIDSSDAETLFTLI 480
Query: 716 LIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISEILDSRTRNLVSVSR 775
+RDI+ R+ + ++ +E+++S+ + + +
Sbjct: 481 NLRDIEKRK----------------------------NKNFVLTTELINSKNAEIFASNM 512
Query: 776 ISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEFYLFDQEELCFYDIM 835
DY+ S ++V L ++E+ +++ +L + EG+E+ KP Y+ E + FY +
Sbjct: 513 NDDYLYSEDIVQSMLVQISENPTMDKFFNDLASPEGSEIYFKPITDYIDISEPIDFYTVC 572
Query: 836 IRGRTRKEIVIGYRLANQE-------RAIINPSEKSVPRKWSLDDVFVVLA 879
+ + IGY++A+ E ++NP KS + ++ +D +V A
Sbjct: 573 KSALNKGQTAIGYQIADNEYLKELDSGIVLNP-VKSEEKLFNSEDKLIVFA 622
>UniRef100_UPI000029AC2D UPI000029AC2D UniRef100 entry
Length = 460
Score = 169 bits (429), Expect = 3e-40
Identities = 120/411 (29%), Positives = 217/411 (52%), Gaps = 33/411 (8%)
Query: 251 YAKLLALLCATLFLIAFGGLAL--YAVTGGSMAE----ALWHSWTYVADAGNHAETEGTG 304
Y L LL T+ I GGL + Y V E +WH+++ + D+G AE +
Sbjct: 27 YVFLALLLMFTIAFIVMGGLRVIVYLVFHDPNIEDPLQLIWHAFSQITDSGALAELDTDS 86
Query: 305 Q---RIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLILGWSDKLG 361
++V ++ + G+++F+ M+ ++ +++ LRKGKS+VIE +H+LI+G+S+++
Sbjct: 87 NISGKLVGIATISIGIVLFSSMVAFITQEFDKRLAMLRKGKSKVIEEDHILIVGFSERVI 146
Query: 362 SLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTSVICRSGSPLILADLKK 421
+++++ IAN+S GVI +LA+ +KEEM+ + D T VI RSGS L +L+
Sbjct: 147 DIIREIVIANES-DEGVIAILADMDKEEMDDYLRDRIMDLQSTRVITRSGSTTSLLNLRN 205
Query: 422 VSVSKARAIIVL----AADEN--ADQSDARALRVVLSLAGVK-EGLRGHVVVEMSDLDNE 474
+ A +++VL A+D N + +DA+ ++ ++++ K E VV E+
Sbjct: 206 AGIDLAHSVVVLNTAKASDSNDVKEAADAKVIKTIMAVVAAKGEDSFPPVVAEVHIDRYR 265
Query: 475 PLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKR----WPEL 530
L + + I T+ D++ R+++Q + GL+ ++ D++GFE EFY R W
Sbjct: 266 KLAETIVEGKITTLNEADLLARMLVQTSRCEGLSMVYMDLVGFEGNEFYFFRPDQGWGTQ 325
Query: 531 DDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDT--YAPG 588
+ F ++ F + +P GV+ + G I +NP +Y L+D D+V+V+AEDD T Y+
Sbjct: 326 N---FGELQFHFMETVPLGVRNSK--GMITMNPPLDYALKDDDDVIVLAEDDSTINYSTQ 380
Query: 589 PL--PEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVLEAFLAPGSEL 637
+ P+ R R P EK + GW ++ +++ GS +
Sbjct: 381 QVIEPKTRSYSTDRANIP---VEKHMIVGWNNKAPIVLREYASYMIKGSSV 428
>UniRef100_UPI0000296BB6 UPI0000296BB6 UniRef100 entry
Length = 411
Score = 167 bits (424), Expect = 1e-39
Identities = 115/413 (27%), Positives = 213/413 (50%), Gaps = 39/413 (9%)
Query: 293 DAGNHAETEGTGQRIVSVS-ISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHV 351
D G EG+ ++S+ I+ G+ I ++++G+++ I + SLRKG+S+V+E+NH
Sbjct: 5 DPGTMGGDEGSWGFLLSLLFITIIGIFILSVLIGIITTGIENAIISLRKGRSKVLEKNHT 64
Query: 352 LILGWSDKLGSLLKQLAIANKSVGGGV-IVVLAEKEKEEMEMDIAKLEFDFMGTSVICRS 410
L+LGW++++ S++ +L I+N G IV+LA K+K EME I + + T +ICRS
Sbjct: 65 LLLGWNNRIMSVISELIISNNQYNGKFSIVILAPKDKVEMEDSIREKIGNSGNTKIICRS 124
Query: 411 GSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEG--LRGHVVVEM 468
G + DL+ +++ A++II+ D + +D + +++L++ + R H+V E
Sbjct: 125 GDASVKIDLELTNINHAKSIIIFPPD--SAYADIQIFKIILAIVNRYDSNEKRLHMVTEA 182
Query: 469 SDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIK--- 525
+DN L+K+VG +E + +++I R++ Q A QPGLA ++ ++L F N +FY K
Sbjct: 183 RYIDNIDLIKMVGDNQVEVIYPNEIIARIISQTARQPGLAVLYSELLSF-NGKFYYKDIR 241
Query: 526 -RW--------------PELDDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLR 570
W EL L+ D ++S+ G V K ++NPD Y ++
Sbjct: 242 SAWYDEASGDEIYYHPQKELIGQLYSDAVLSYETCSVIG--VFNSDNKNILNPDMEYEIQ 299
Query: 571 DGDEVLVIAEDDDTY---APGPLPEVRKGYFPRIRDPPKYPEKILFCGWRRDIDDMIMVL 627
D++++IAED + + + + I P K + IL W R +I +
Sbjct: 300 MNDKMILIAEDKEKIVYTSNSDIKIIENKIVNTISIPDK--DNILIINWNRLTPVIITEI 357
Query: 628 EAFLAPGSELWMFNEVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHL 680
+++ PGS + + + E K +DV +EN + + G+ +++
Sbjct: 358 DSYAVPGSSITILS-------ENKNIDESVDVVNIENCIIYTKNGDTTSPKYI 403
>UniRef100_UPI0000335672 UPI0000335672 UniRef100 entry
Length = 627
Score = 165 bits (418), Expect = 5e-39
Identities = 144/664 (21%), Positives = 302/664 (44%), Gaps = 63/664 (9%)
Query: 236 KRVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALYAVTGGSMAEALWHSWTYVADA- 294
K++ Y +D S ++ + L+ + ++ F L Y A + YV A
Sbjct: 5 KKLRYKIDKSLSRSIWSVISLLILLVIAIVLFISLIDYFFISHDKASFHNTFFDYVIAAI 64
Query: 295 -GNHAETEGTGQRIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSEVIERNHVLI 353
GN E +++ + + + ++++GL+ + + E+++S+R G+S V+E NH ++
Sbjct: 65 KGNTIENSSFVNILLNTVMIGTSLFVTSVVIGLIVNNLRERIESIRNGRSFVLESNHQIV 124
Query: 354 LGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKL---EFDFMGTSVICRS 410
LGWS + ++++ +AN++ +V+L E M + K+ D+ +I +
Sbjct: 125 LGWSQGIHLIIREFLLANENNKNTKLVILDEMNPVLMNEKLRKMFSKRKDY--ERIIPKQ 182
Query: 411 GSPLILADLKKVSVSKARAIIVLAADENADQSDARALRVVLSLAGVKEGLRGHVVVEMSD 470
GS +L+ ++++++R+I++ D D + + + +++ + ++ +++
Sbjct: 183 GSTFDRDNLQTININESRSILINHLD---DMTTIKTMAAIINNPD-RRSEPYNISTKINS 238
Query: 471 LDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGFENAEFYIKRWPEL 530
N L +++GGE + D++ R+ Q LQ GLA ++ +++ F+ E Y L
Sbjct: 239 RKNFELARIIGGEESTVLYFGDMLSRIDAQTCLQSGLANVFLELVNFDGDEIYFTNESSL 298
Query: 531 DDLLFKDILISFPDAIPCGVKVAADGGKIVINPDDNYVLRDGDEVLVIAEDDDTYAPGPL 590
++ F++ ++S+ + G++ A++ I+++P + +++ GD+++ I+EDDD +
Sbjct: 299 ENRTFREAVLSYDTSSIIGIQRASE---IILSPSFDEIIKAGDKLISISEDDDKVLKDGI 355
Query: 591 PEVRKGYFPRIR-----DPPKYPEKILFCGWRRDIDDMIMVLE----AFLAPGSELWMFN 641
G + + K E++ G D I VL +L GSE+++ N
Sbjct: 356 NLPDSGLYNEKNIIKGIEKNKKIERVFVFGCSEDDLSKIRVLSKNLMKYLDSGSEIFLVN 415
Query: 642 EVPEKERERKLAAGELDVFGLENIKLVHREGNAVIRRHLESLPLETFDSILILADESVED 701
+ + K E++ ++ H G+ R L +L L D ILIL+ + +
Sbjct: 416 D----RNDTKALIEEIE--NKNHLVFNHISGDVRSRDFLNTLDLRNGDKILILSPFNGLN 469
Query: 702 SVAHSDSRSLATLLLIRDIQSRRLPYRDTKSTSLRLSGFSHNSWIREMQQASDKSIIISE 761
+ D+ L TL+ +RDIQ + T S + +E
Sbjct: 470 EIESCDADVLFTLINLRDIQVK---------TGKNFS-------------------VTTE 501
Query: 762 ILDSRTRNLVSVSRISDYVLSNELVSMALAMVAEDKQINRVLEELFAEEGNEMCIKPAEF 821
+++S+ + R DY+ S +V L +AE+ + + L + +G E+ +
Sbjct: 502 LINSKNAEIFQSERDDDYLYSEVIVQSMLVQIAENPSLGDFFDYLVSPKGAEIYFRSISD 561
Query: 822 YLFDQEELCFYDIMIRGRTRKEIVIGY-RLANQERAIINPSE-----KSVPRKWSLDDVF 875
Y+ + + FY + + E IGY R +NQE +N KS R +S +D
Sbjct: 562 YVDISKPIDFYSVCESAGLKGETAIGYQRTSNQELKSLNAGVNINPLKSKKRTFSTNDKV 621
Query: 876 VVLA 879
+V A
Sbjct: 622 IVFA 625
>UniRef100_UPI00002AD0BD UPI00002AD0BD UniRef100 entry
Length = 314
Score = 137 bits (344), Expect = 2e-30
Identities = 89/314 (28%), Positives = 171/314 (54%), Gaps = 15/314 (4%)
Query: 278 GSMAEALWHSWTYVADAGNHAETEGTGQRIVSVSI--SAGGMLIFAMMLGLVSDAISEKV 335
G + LW S ++ D G +E I + + + G++I +++G V + I +
Sbjct: 2 GFLDTLLW-SLKHILDPGAWSEDYSAPLMITLIGLVNTITGLIIVGILIGFVVNFIQNAM 60
Query: 336 DSLRKGKSEVIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIA 395
D L++G +V E H LILGW+ K+ S+L L NK +V+L+ + + + ++
Sbjct: 61 DELKRGSIDVHESGHFLILGWNRKVLSILTFLEELNKKQS---VVLLSNADIDLVSEEVR 117
Query: 396 KLEFDFMGTSVICRSGSPLILADLKKVSVSKARAIIVLA--ADENADQS-DARALRVVLS 452
+E +F +VI + GSP + A+L++V++ ++ ++++LA DE++DQS D ++ ++
Sbjct: 118 HVERNFRNVTVIPQHGSPTLAAELERVAIRESSSVVLLADEVDESSDQSNDIPTIKSLMQ 177
Query: 453 LAGVK-EGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIW 511
L V G R ++VVE+++ +N +V+L G + V + D + + ++QCA G A+I+
Sbjct: 178 LDNVSWYGKRPNIVVEVANKENLDMVELASGASVPVVSSADFVSKTLVQCARYQGYAEIY 237
Query: 512 EDILGFENAEFYIKRWPELDDLLFKDILISFPDAIPCGV-----KVAADGGKIVINPDDN 566
I FE+ E I++ ++ LF ++ AI GV K + V+NP+ +
Sbjct: 238 STIFAFEDNEIQIRKMEGIEGKLFGELAEQLESAILLGVSWTEIKNGVERRVAVLNPEPD 297
Query: 567 YVLRDGDEVLVIAE 580
Y L + +E++V+ E
Sbjct: 298 YDLVEDEELIVLRE 311
>UniRef100_UPI000026A417 UPI000026A417 UniRef100 entry
Length = 317
Score = 130 bits (327), Expect = 2e-28
Identities = 95/307 (30%), Positives = 168/307 (53%), Gaps = 23/307 (7%)
Query: 237 RVAYMVDVFFSIYPYAKLLALLCATLFLIAFGGLALY-AVTGGSMAEALWHSW------T 289
R+ Y +D F S + LALLC LFL F + L ++ M + SW
Sbjct: 1 RLRYAIDNFMSKGSSSIFLALLC--LFLFGFLVMVLIRSIANTFMPDETLSSWGEIPWRV 58
Query: 290 YVADA-GNHAETEGTGQRIVSVSISAG---GMLIFAMMLGLVSDAISEKVDSLRKGKSEV 345
YVA G+ AET+G ++ G G+++F+ M+ ++ + K+D LR+G+S V
Sbjct: 59 YVAVMEGSAAETDGDSNWAAKITSILGVLVGLVLFSSMVAFITSVFTAKLDELRRGRSLV 118
Query: 346 IERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGTS 405
+E +H LILG+ D++ ++++L AN+S IV+LA+ +KEEM+ I DF+ T
Sbjct: 119 LENDHTLILGFGDRILEVIRELIEANESEPDAAIVILADNDKEEMDNMIRDNISDFVTTR 178
Query: 406 VICRSGSPLILADLKKVSVSKARAIIVLAA------DENADQSDARALRVVLSLAGVKEG 459
VI RSG + +L+KV + A+++I++ + ++ + +DA L+ ++S+ V +G
Sbjct: 179 VITRSGVVTNINNLRKVMAANAKSVIIMNSAASWRPEDEKNLADALVLKSIMSIIAVCDG 238
Query: 460 -LRGHVVVEM-SDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCAL-QPGLAQIWEDILG 516
+V E+ SD D + G+ ++ + V+ R++ Q AL + GL+ ++ D++G
Sbjct: 239 DEHPPIVCEIHSDRDRNLAENISNGD-VKALNEVSVLSRMIAQLALSRNGLSVVYSDMVG 297
Query: 517 FENAEFY 523
F+ EFY
Sbjct: 298 FDGNEFY 304
>UniRef100_UPI000030E759 UPI000030E759 UniRef100 entry
Length = 286
Score = 129 bits (325), Expect = 3e-28
Identities = 84/290 (28%), Positives = 162/290 (54%), Gaps = 20/290 (6%)
Query: 288 WTYVADAGNHAETEGTGQ---RIVSVSISAGGMLIFAMMLGLVSDAISEKVDSLRKGKSE 344
++ + D+G AE + ++V ++ + G+++F+ M+ ++ +++ LRKGKS
Sbjct: 1 FSQITDSGALAELDTDSNIAGKLVGIATISIGIVLFSSMVAFITQEFDKRLAMLRKGKSM 60
Query: 345 VIERNHVLILGWSDKLGSLLKQLAIANKSVGGGVIVVLAEKEKEEMEMDIAKLEFDFMGT 404
VIE++H+LI+G+S+++ +++++ IAN+S GVI +LA+ +KEEM+ + D T
Sbjct: 61 VIEKDHILIVGFSERVIDIIREIVIANES-DEGVIAILADMDKEEMDDYLRDRIMDLQST 119
Query: 405 SVICRSGSPLILADLKKVSVSKARAIIVL----AADEN--ADQSDARALRVVLSLAGVK- 457
VI RSGS L +L+ + A +++VL A+D N + +DA+ ++ ++++ K
Sbjct: 120 RVITRSGSTTSLLNLRNAGIDLAHSVVVLNTAKASDSNDVKEAADAKVIKTIMAVVAAKG 179
Query: 458 EGLRGHVVVEMSDLDNEPLVKLVGGELIETVVAHDVIGRLMIQCALQPGLAQIWEDILGF 517
E VV E+ L + + I T+ D++ R+++Q + GL+ ++ D++GF
Sbjct: 180 EDSFPPVVAEVHIDRYRKLAETIVEGKITTLNEADLLARMLVQTSRCEGLSMVYMDLVGF 239
Query: 518 ENAEFYIKR----WPELDDLLFKDILISFPDAIPCGVKVAADGGKIVINP 563
E EFY R W + F ++ F + +P GV+ + G I +NP
Sbjct: 240 EGNEFYFFRPDQGWGTQN---FGELQFHFMETVPLGVRNSK--GMITMNP 284
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,479,761,602
Number of Sequences: 2790947
Number of extensions: 65695916
Number of successful extensions: 283095
Number of sequences better than 10.0: 396
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 319
Number of HSP's that attempted gapping in prelim test: 280424
Number of HSP's gapped (non-prelim): 1563
length of query: 882
length of database: 848,049,833
effective HSP length: 136
effective length of query: 746
effective length of database: 468,481,041
effective search space: 349486856586
effective search space used: 349486856586
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC140550.12


