

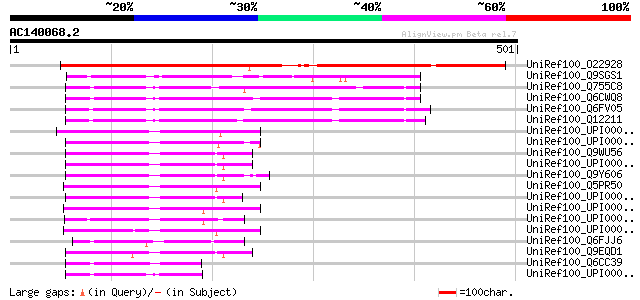
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140068.2 + phase: 0
(501 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O22928 Putative pseudouridylate synthase [Arabidopsis ... 490 e-137
UniRef100_Q9SGS1 T23E18.5 [Arabidopsis thaliana] 117 1e-24
UniRef100_Q755C8 AFL105Cp [Ashbya gossypii] 112 2e-23
UniRef100_Q6CWQ8 Similar to sp|Q12211 Saccharomyces cerevisiae Y... 111 6e-23
UniRef100_Q6FV05 Similar to sp|Q12211 Saccharomyces cerevisiae Y... 107 8e-22
UniRef100_Q12211 Pseudouridylate synthase 1 [Saccharomyces cerev... 107 8e-22
UniRef100_UPI00002B27F4 UPI00002B27F4 UniRef100 entry 101 4e-20
UniRef100_UPI00003AAFB4 UPI00003AAFB4 UniRef100 entry 100 7e-20
UniRef100_Q9WU56 tRNA pseudouridine synthase A [Mus musculus] 100 2e-19
UniRef100_UPI000017E82B UPI000017E82B UniRef100 entry 99 2e-19
UniRef100_Q9Y606 tRNA pseudouridine synthase A [Homo sapiens] 99 3e-19
UniRef100_Q5PR50 Hypothetical protein [Brachydanio rerio] 98 5e-19
UniRef100_UPI0000368C42 UPI0000368C42 UniRef100 entry 98 6e-19
UniRef100_UPI0000363FE8 UPI0000363FE8 UniRef100 entry 94 7e-18
UniRef100_UPI00004993EF UPI00004993EF UniRef100 entry 94 7e-18
UniRef100_UPI000019EB4C UPI000019EB4C UniRef100 entry 92 3e-17
UniRef100_Q6FJJ6 Similar to sp|P53167 Saccharomyces cerevisiae Y... 90 1e-16
UniRef100_Q9EQD1 Pseudouridine synthase 1 [Mus musculus] 89 4e-16
UniRef100_Q6CC39 Similar to sp|Q12211 Saccharomyces cerevisiae Y... 86 2e-15
UniRef100_UPI0000234E9A UPI0000234E9A UniRef100 entry 85 6e-15
>UniRef100_O22928 Putative pseudouridylate synthase [Arabidopsis thaliana]
Length = 510
Score = 490 bits (1262), Expect = e-137
Identities = 251/452 (55%), Positives = 320/452 (70%), Gaps = 39/452 (8%)
Query: 51 WQPFRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKI 110
W+ +RKKKVV+RI Y GT+YRGLQ+QRD S+ T+E ELE AI+KAGGIR+SN GDL KI
Sbjct: 57 WESYRKKKVVIRIGYVGTDYRGLQIQRDDPSIKTIEGELEVAIYKAGGIRDSNYGDLHKI 116
Query: 111 GWARSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQ 170
GWARSSRTDKGVHSLAT ++ KMEIPE +W +DP G LA ++ +LP +I+V S+LP+
Sbjct: 117 GWARSSRTDKGVHSLATSISLKMEIPETAWKDDPQGTVLAKCISKHLPENIRVFSVLPSN 176
Query: 171 RSFDPRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSR 230
R FDPR+EC LRKYSYLLP D++GI++ ++DEIDYH+++FN+ILKEFEG +PFHNYT R
Sbjct: 177 RRFDPRRECTLRKYSYLLPVDVLGIKNSFTSDEIDYHITDFNEILKEFEGEYPFHNYTQR 236
Query: 231 SRYRR-----------HIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQ 279
SRYRR PR P K E + E +EEE +VD
Sbjct: 237 SRYRRKSEQKIKQRNGRPPREPKSIKASESEFREENHIEIGEEEEEKEVD---------- 286
Query: 280 NQKSSGTSDSCEPVIETGNKGNLHDQSSSSVV-RAKWLYEPDEADRLNASHFRRVFRCSS 338
G SD E V+ D +S V RAKWLYEPDE D+++ +HFR+VFRC S
Sbjct: 287 -----GESD--EHVVTP-------DSDNSQVYSRAKWLYEPDETDKISGAHFRKVFRCRS 332
Query: 339 GKLEKSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPL 398
GKLE S+G ++EI I GESFMLHQIRKM+ TAVAVKR+LLPRDII LSL+KF+RI+LPL
Sbjct: 333 GKLENSLGFGFVEISIWGESFMLHQIRKMIGTAVAVKRELLPRDIIRLSLNKFTRIVLPL 392
Query: 399 APSEVLLLRGNSFSMRTKPGNVTRPEMQTIVESEEINKVVDDFYRSVMLPQVSKFLDPSR 458
APSEVL+LRGNSF +R P RP M+ ESEE+ K +++FYR+VM+PQVS FLD +
Sbjct: 393 APSEVLILRGNSFEVRRLP---ERPGMEATGESEEVEKEIEEFYRAVMVPQVSIFLDSEK 449
Query: 459 VPWVEWIEKFDAHTSIPNDQLDEVRKARNLWK 490
PW EW++ D + + +++L++VRK WK
Sbjct: 450 SPWKEWVDHLDRNDGLIDEELEDVRKGWEEWK 481
>UniRef100_Q9SGS1 T23E18.5 [Arabidopsis thaliana]
Length = 463
Score = 117 bits (292), Expect = 1e-24
Identities = 105/377 (27%), Positives = 167/377 (43%), Gaps = 53/377 (14%)
Query: 57 KKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSS 116
+KV + A+ G Y+G+Q T+E ELE A+F AG + ES G + +ARS+
Sbjct: 22 QKVAIIFAFCGVGYQGMQKNP---GAKTIEGELEEALFHAGAVPESIRGKPKLYDFARSA 78
Query: 117 RTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDPR 176
RTDKGV ++ +V+ + + DP GF N +NS LP I++ SF +
Sbjct: 79 RTDKGVSAVGQVVSGRFIV-------DPLGFV--NRLNSNLPNQIRIFGYKHVTPSFSSK 129
Query: 177 KECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSRSRYRRH 236
K C R+Y YLLP + SH + + + + +K FE S R
Sbjct: 130 KFCDRRRYVYLLPVFALDPISHRDRETVMASLGPGEEYVKCFEC----------SERGRK 179
Query: 237 IPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCEPVIET 296
IP G + + ++ D + + AL +I+ + SS + C +E
Sbjct: 180 IPPGLVGKWKG---TNFGTKSLDFQSDISSNNSSALRSDIKIE-ALSSNLAGLCSVDVEV 235
Query: 297 G----NKGNLHDQSSSSVVRAKWLYEPDEADRL-----------NASHF----------- 330
G + L+ SS + V++K+ Y E +R N +F
Sbjct: 236 GRIQEDSCKLNTNSSETKVKSKFCYGEKEKERFSRILSCYVGSYNFHNFTTRTKADDPTA 295
Query: 331 -RRVFRCSSGKLEKSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLS 389
R++ ++ + G +I+ + G+SFMLHQIRKM+ AVA+ R +I + S
Sbjct: 296 NRQIISFTANTVINLDGIDFIKCEVLGKSFMLHQIRKMMGLAVAIMRNCASESLIQSAFS 355
Query: 390 KFSRIILPLAPSEVLLL 406
K I +P+AP L L
Sbjct: 356 KDVNITVPMAPEVGLYL 372
>UniRef100_Q755C8 AFL105Cp [Ashbya gossypii]
Length = 528
Score = 112 bits (280), Expect = 2e-23
Identities = 101/355 (28%), Positives = 158/355 (44%), Gaps = 30/355 (8%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+KV + + Y GT Y G+Q T+E EL A KAG I +N DL+K G+ R+
Sbjct: 66 KRKVAVMVGYCGTGYHGMQYNPPNR---TIEAELFEAFVKAGAISRANSTDLKKNGFMRA 122
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGVH+ +++ K+ I EDP A+ + +N +LP I+V I ++FD
Sbjct: 123 ARTDKGVHAGGNVISLKLII------EDP---AIKDKINEHLPPGIRVWGISRVNKAFDC 173
Query: 176 RKECVLRKYSYLLPA-DIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSR---S 231
RK C R Y YLLP IG + ++ +++ + E P + S
Sbjct: 174 RKLCGSRWYEYLLPTYSFIGPKPNT-------YLARTIEQCGEAASEKPDRDQESLDFWE 226
Query: 232 RYRRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCE 291
+R+ + + + + I++ +ED DE +E +++S S +
Sbjct: 227 AFRKAVDEKFTQEEQDAIVNYVAPSKEDFDENSGLYQKVKQYKQMENAHRRSYRVSSAKL 286
Query: 292 PVIETGNKGNLHDQSSSSVVRAKWLYEPDEADRLNASHFRRVFRCSSGKLEKSMGHSYIE 351
K L + + K +P + R F K E ++
Sbjct: 287 ARFREAMKQYLGPHNFHNYTLGKDFKDPSTVRFMKDITVSRPFVIGEMKTE------WVS 340
Query: 352 IYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLL 406
I I G+SFMLHQIRKM++ A V R + I S +I +P AP+ LLL
Sbjct: 341 IKIHGQSFMLHQIRKMISMATLVARCNCSPERIAQSYGP-QKINIPKAPALGLLL 394
>UniRef100_Q6CWQ8 Similar to sp|Q12211 Saccharomyces cerevisiae YPL212c PUS1
pseudouridine synthase 1 [Kluyveromyces lactis]
Length = 553
Score = 111 bits (277), Expect = 6e-23
Identities = 98/353 (27%), Positives = 161/353 (44%), Gaps = 27/353 (7%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+KV + I Y GT Y G+Q + T+EKEL A +AG I ++N DL+K G+ R+
Sbjct: 98 KRKVAVMIGYCGTGYHGMQYNPPND---TIEKELFEAFVRAGAISKANSTDLKKNGFQRA 154
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGVH+ +++ K+ I EDP + +N+ LP I+V I ++FD
Sbjct: 155 ARTDKGVHAGGNVISLKLII------EDP---EVKEKINNELPDQIRVWDISRVNKAFDC 205
Query: 176 RKECVLRKYSYLLPA-DIIGIQ-SHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSRSRY 233
RK C R Y YLLP +IG + S +EI+ E +++++ F S
Sbjct: 206 RKMCSSRWYEYLLPTYSLIGPKPSTYLYNEIEASKKEIPNVIQDDVESAQFWEAFSTEAE 265
Query: 234 RRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCEPV 293
+ E ++A+ ++ DE +E +++++ S
Sbjct: 266 SKFTKEEL------EEIAAFVMPKDSFDENNETYQKSKAFKKLEAEHKRAYRISKERLDR 319
Query: 294 IETGNKGNLHDQSSSSVVRAKWLYEPDEADRLNASHFRRVFRCSSGKLEKSMGHSYIEIY 353
+ K + + K +P +A F + S + ++ I
Sbjct: 320 FRSALKQYEGTFNFHNFTLGKDFNDP------SAKRFMKQITVSEPFVIGEAKTEWVSIK 373
Query: 354 IEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLL 406
I G+SFMLHQIRKM++ A + R P + I + + +I +P AP+ LLL
Sbjct: 374 IHGQSFMLHQIRKMISMATLITRCFSPLERIRQAYGQ-EKINIPKAPALGLLL 425
>UniRef100_Q6FV05 Similar to sp|Q12211 Saccharomyces cerevisiae YPL212c PUS1
pseudouridine synthase 1 [Candida glabrata]
Length = 549
Score = 107 bits (267), Expect = 8e-22
Identities = 98/364 (26%), Positives = 164/364 (44%), Gaps = 28/364 (7%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+KV + I Y GT Y G+Q T+E L A AG I ++N DL+K + R+
Sbjct: 86 KRKVAVMIGYCGTGYHGMQYNPPN---PTIEAALFKAFVDAGAISKANSNDLKKNNFMRA 142
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGVH+ +++ KM I EDP + + +N+ LP I++ I ++FD
Sbjct: 143 ARTDKGVHAGGNLISLKMII------EDP---EIMDKINANLPEGIRIWDIERVNKAFDC 193
Query: 176 RKECVLRKYSYLLPA-DIIGIQSHS-SNDEIDYHMSEFNDILKE-FEGGHPFHNYTSRSR 232
RK C R Y YLLP +IG + + N++I+ +E +L E E + N+ + ++
Sbjct: 194 RKMCSSRWYEYLLPTYSLIGPKPGTILNNDIESSKTELPGVLDEDLESKEFWENFEAAAK 253
Query: 233 YRRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCEP 292
+ + + I +E+ D++E +E +++ S+
Sbjct: 254 ------KEFTPEEIAAIKDYSPPPREEFDDQEELYQKVKQWKLLENTHRRQYRISEMKLN 307
Query: 293 VIETGNKGNLHDQSSSSVVRAKWLYEPDEADRLNASHFRRVFRCSSGKLEKSMGHSYIEI 352
L + + K +P +A F + + S + ++ I
Sbjct: 308 KFRAAMNQYLGAHNFHNFTLGKEFKDP------SAIRFMKEIKVSDPFVIGDAKAEWVSI 361
Query: 353 YIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLLRGNSFS 412
I G+SFMLHQIRKM++ A + R P + I+ + +I +P AP+ LLL F
Sbjct: 362 KIHGQSFMLHQIRKMISMATLIARCGTPVERISQAFGP-QKINIPKAPALGLLLEAPVFE 420
Query: 413 MRTK 416
K
Sbjct: 421 SYNK 424
>UniRef100_Q12211 Pseudouridylate synthase 1 [Saccharomyces cerevisiae]
Length = 544
Score = 107 bits (267), Expect = 8e-22
Identities = 101/358 (28%), Positives = 158/358 (43%), Gaps = 26/358 (7%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+KV + + Y GT Y G+Q T+E L A +AG I + N DL+K G+ R+
Sbjct: 74 KRKVAVMVGYCGTGYHGMQYNPPN---PTIESALFKAFVEAGAISKDNSNDLKKNGFMRA 130
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGVH+ +++ KM I EDP + +N LP I+V I ++FD
Sbjct: 131 ARTDKGVHAGGNLISLKMII------EDP---DIKQKINEKLPEGIRVWDIERVNKAFDC 181
Query: 176 RKECVLRKYSYLLPA-DIIGIQSHS-SNDEIDYHMSEFNDILKEFEGGHPFHNYTSRSRY 233
RK C R Y YLLP +IG + S +I+ +E +L E F +
Sbjct: 182 RKMCSSRWYEYLLPTYSLIGPKPGSILYRDIEESKTELPGVLDEDLESKEFW-----EEF 236
Query: 234 RRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCEPV 293
++ + S + IL+ +++ D E +E +++ S +
Sbjct: 237 KKDANEKFSTEEIEAILAYVPPARDEFDINEELYQKVKKYKQLENAHRRRYRISAAKLAK 296
Query: 294 IETGNKGNLHDQSSSSVVRAKWLYEPDEADRLNASHFRRVFRCSSGKLEKSMGHSYIEIY 353
L + + K EP +A F + + S + +I I
Sbjct: 297 FRASTSQYLGAHNFHNFTLGKDFKEP------SAIRFMKDIKVSDPFVIGDAQTEWISIK 350
Query: 354 IEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLLRGNSF 411
I G+SFMLHQIRKMV+ A + R P + I+ + + +I +P AP+ LLL F
Sbjct: 351 IHGQSFMLHQIRKMVSMATLITRCGCPVERISQAYGQ-QKINIPKAPALGLLLEAPVF 407
>UniRef100_UPI00002B27F4 UPI00002B27F4 UniRef100 entry
Length = 375
Score = 101 bits (252), Expect = 4e-20
Identities = 61/209 (29%), Positives = 109/209 (51%), Gaps = 17/209 (8%)
Query: 47 QSYSWQPFRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGD 106
QS + + KKKV + +AY+G Y G+Q + T+E +L +A+ K+G I E++ D
Sbjct: 65 QSQEDRRYPKKKVALLLAYSGKGYYGMQRNPGSSEFRTIEDDLVAALVKSGCIPENHAED 124
Query: 107 LEKIGWARSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISI 166
++K+ + R +RTDKGV + +V+ K+ + ++ LA +N +LP I+V+ +
Sbjct: 125 MKKMSFQRCARTDKGVSAAGQVVSLKLWLIDD----------LAEKINQHLPPQIRVLGV 174
Query: 167 LPAQRSFDPRKECVLRKYSYLLPADIIGIQ-SHSSNDEIDYH-----MSEFNDILKEFEG 220
+ F+ + C R Y+Y+LP + S +++D + + N + ++G
Sbjct: 175 KRVTQGFNSKNNCDARTYAYMLPTVAFAPKDSDAASDAASFRLQPETLQRVNRLFALYKG 234
Query: 221 GHPFHNYTSRSRYRRHIPRR-PSHSKCGE 248
H FHN+TS+ RR +H CGE
Sbjct: 235 THNFHNFTSQKAPNDPSARRYVTHVCCGE 263
Score = 34.3 bits (77), Expect = 8.6
Identities = 14/23 (60%), Positives = 18/23 (77%)
Query: 352 IYIEGESFMLHQIRKMVATAVAV 374
I + G+SFMLHQIRKM+ +AV
Sbjct: 275 ITVRGQSFMLHQIRKMIGLVIAV 297
>UniRef100_UPI00003AAFB4 UPI00003AAFB4 UniRef100 entry
Length = 362
Score = 100 bits (250), Expect = 7e-20
Identities = 56/203 (27%), Positives = 106/203 (51%), Gaps = 25/203 (12%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+K+V+ +AY+G Y G+Q + T+E +L SA+ ++G I E++ D++K+ + R
Sbjct: 45 KRKIVLLMAYSGKGYHGMQRNLGSSQFKTIEDDLVSALVRSGCIPENHGEDMKKMSFQRC 104
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGV + +V+ K+ + ++ + +N++LP I+++ + F+
Sbjct: 105 ARTDKGVSAAGQIVSLKVRLIDD----------ILEKINNHLPSHIRILGLKRVTGGFNS 154
Query: 176 RKECVLRKYSYLLPADIIGIQSHSSNDEID----YHMSEFNDILKEFEGGHPFHNYTSRS 231
+ +C R Y+Y+LP + H +E + + N +L ++G H FHN+TS+
Sbjct: 155 KNKCDARTYAYMLPTFAFAHKDHDVQEEAHRLSRETLEKVNSLLARYKGTHNFHNFTSQK 214
Query: 232 RYRRHIPRRPSHSK------CGE 248
P+ PS + CGE
Sbjct: 215 G-----PKDPSARRYIMEMYCGE 232
Score = 42.7 bits (99), Expect = 0.024
Identities = 22/61 (36%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G + I ++G+SFM+HQIRKM+ +A+ + P II S + ++ +P AP L+
Sbjct: 238 GIEFAVIKVKGQSFMMHQIRKMIGLVIAIVKGYAPESIIERSWGE-EKVDVPKAPGLGLV 296
Query: 406 L 406
L
Sbjct: 297 L 297
>UniRef100_Q9WU56 tRNA pseudouridine synthase A [Mus musculus]
Length = 393
Score = 99.8 bits (247), Expect = 2e-19
Identities = 57/190 (30%), Positives = 101/190 (53%), Gaps = 16/190 (8%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+K+V+ +AY+G Y G+Q + T+E +L SA+ +AG I E++ D+ K+ + R
Sbjct: 49 KRKIVLLMAYSGKGYHGMQRNLGSSQFRTIEDDLVSALVQAGCIPENHGTDMRKMSFQRC 108
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGV + +V+ K+ + ++ + + +NS+LP I+++ + F+
Sbjct: 109 ARTDKGVSAAGQVVSLKVWLIDD----------ILDKINSHLPSHIRILGLKRVTGGFNS 158
Query: 176 RKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMS-----EFNDILKEFEGGHPFHNYTSR 230
+ +C R Y Y+LP + DE Y +S + N +L ++G H FHN+TS+
Sbjct: 159 KNKCDARTYCYMLPTFAFAHKDRDVQDE-SYRLSAETLQQVNRLLACYKGTHNFHNFTSQ 217
Query: 231 SRYRRHIPRR 240
R RR
Sbjct: 218 KGPREPSARR 227
Score = 42.4 bits (98), Expect = 0.031
Identities = 22/61 (36%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G + I ++G+SFM+HQIRKMV VA+ + P ++ S + ++ +P AP L+
Sbjct: 242 GLEFAVIKVKGQSFMMHQIRKMVGLVVAIVKGYAPESVLERSWGE-EKVDVPKAPGLGLV 300
Query: 406 L 406
L
Sbjct: 301 L 301
>UniRef100_UPI000017E82B UPI000017E82B UniRef100 entry
Length = 393
Score = 99.4 bits (246), Expect = 2e-19
Identities = 57/190 (30%), Positives = 101/190 (53%), Gaps = 16/190 (8%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+K+V+ +AY+G Y G+Q + T+E +L SA+ +AG I E++ D+ K+ + R
Sbjct: 49 KRKIVLLMAYSGKGYHGMQRNLGSSQFRTIEDDLVSALVQAGCIPENHGTDMRKMSFQRC 108
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGV + +V+ K+ + ++ + + +NS+LP I+++ + F+
Sbjct: 109 ARTDKGVSAAGQVVSLKVWLIDD----------ILDKINSHLPSHIRILGLKRVTGGFNS 158
Query: 176 RKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMS-----EFNDILKEFEGGHPFHNYTSR 230
+ +C R Y Y+LP + DE Y +S + N +L ++G H FHN+TS+
Sbjct: 159 KNKCDARTYCYMLPTFAFAHKDRDVQDE-SYRLSAETLQQVNRLLGCYKGTHNFHNFTSQ 217
Query: 231 SRYRRHIPRR 240
R RR
Sbjct: 218 KGPREPSARR 227
Score = 42.4 bits (98), Expect = 0.031
Identities = 22/61 (36%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G + I ++G+SFM+HQIRKMV VA+ + P ++ S + ++ +P AP L+
Sbjct: 242 GLEFAVIKVKGQSFMMHQIRKMVGLVVAIVKGYAPESVLERSWGE-EKVDVPKAPGLGLV 300
Query: 406 L 406
L
Sbjct: 301 L 301
>UniRef100_Q9Y606 tRNA pseudouridine synthase A [Homo sapiens]
Length = 399
Score = 99.0 bits (245), Expect = 3e-19
Identities = 62/206 (30%), Positives = 105/206 (50%), Gaps = 23/206 (11%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+K+V+ +AY+G Y G+Q + T+E +L SA+ ++G I E++ D+ K+ + R
Sbjct: 55 KRKIVLLMAYSGKGYHGMQRNVGSSQFKTIEDDLVSALVRSGCIPENHGEDMRKMSFQRC 114
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGV + +V+ K+ + ++ + +NS+LP I+++ + F+
Sbjct: 115 ARTDKGVSAAGQVVSLKVWLIDD----------ILEKINSHLPSHIRILGLKRVTGGFNS 164
Query: 176 RKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMS-----EFNDILKEFEGGHPFHNYTSR 230
+ C R Y YLLP + DE Y +S + N +L ++G H FHN+TS+
Sbjct: 165 KNRCDARTYCYLLPTFAFAHKDRDVQDE-TYRLSAETLQQVNRLLACYKGTHNFHNFTSQ 223
Query: 231 SRYRRHIPRRPSHSKCGEILSAYNSE 256
P+ P S C IL Y E
Sbjct: 224 KG-----PQDP--SACRYILEMYCEE 242
Score = 41.6 bits (96), Expect = 0.054
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G + I ++G+SFM+HQIRKMV VA+ + P ++ S ++ +P AP L+
Sbjct: 248 GLEFAVIRVKGQSFMMHQIRKMVGLVVAIVKGYAPESVLERSWGT-EKVDVPKAPGLGLV 306
Query: 406 L 406
L
Sbjct: 307 L 307
>UniRef100_Q5PR50 Hypothetical protein [Brachydanio rerio]
Length = 376
Score = 98.2 bits (243), Expect = 5e-19
Identities = 59/200 (29%), Positives = 101/200 (50%), Gaps = 15/200 (7%)
Query: 54 FRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWA 113
+ KKKV + +AY+G Y G+Q + T+E EL +A+ A I E + +++K+ +
Sbjct: 37 YPKKKVALLMAYSGKGYYGMQRNARNSQFKTIEDELVTALVGARCIPEGHAEEMKKMSFQ 96
Query: 114 RSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSF 173
RS+RTDKGV ++ +V+ K+ + EN + +N++LP I+V+ F
Sbjct: 97 RSARTDKGVSAVGRVVSLKVWMIEN----------ILEKINAHLPPRIRVLGCKRVTGGF 146
Query: 174 DPRKECVLRKYSYLLPADIIGIQSHSSNDE----IDYHMSEFNDILKEFEGGHPFHNYTS 229
+ + C R YSY+LP + ++ D + N + ++G H FHN+TS
Sbjct: 147 NSKNNCYARTYSYMLPTVAFSPKDYNQEDTSFRLTSDTLQTVNRLFCLYKGTHNFHNFTS 206
Query: 230 RSRYR-RHIPRRPSHSKCGE 248
+ +R R +H CGE
Sbjct: 207 QKGHRDPSAMRYITHMSCGE 226
Score = 35.0 bits (79), Expect = 5.0
Identities = 28/113 (24%), Positives = 52/113 (45%), Gaps = 5/113 (4%)
Query: 352 IYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLLRGNSF 411
I + G+SFM+HQIRKM+ +AV + + ++ S + ++ + AP L+L F
Sbjct: 238 ITVRGQSFMMHQIRKMIGLVIAVVKGYVDEGVMEKSWGE-EKVGVSKAPGLGLVLERVHF 296
Query: 412 SMRTKPGNVTRPEMQTIVESEEINKVVDDFYRSVMLPQV--SKFLDPSRVPWV 462
K + +E E + + DF + P + ++ + S V W+
Sbjct: 297 DGYNK--RFGGDGIHETLEWTEQEEAIADFKEKHIYPTIVDTELNERSMVNWM 347
>UniRef100_UPI0000368C42 UPI0000368C42 UniRef100 entry
Length = 399
Score = 97.8 bits (242), Expect = 6e-19
Identities = 55/180 (30%), Positives = 96/180 (52%), Gaps = 16/180 (8%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+K+V+ +AY+G Y G+Q + T+E +L SA+ ++G I E++ D+ K+ + R
Sbjct: 55 KRKIVLLMAYSGKGYHGMQRNVGSSQFKTIEDDLVSALVRSGCIPENHGEDMRKMSFQRC 114
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGV + +V+ K+ + E+ + +NS+LP I+++ + F+
Sbjct: 115 ARTDKGVSAAGQVVSLKVWLIED----------ILEKINSHLPSHIRILGLKRVTGGFNS 164
Query: 176 RKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMS-----EFNDILKEFEGGHPFHNYTSR 230
+ C R Y YLLP + DE Y +S + N +L ++G H FHN+TS+
Sbjct: 165 KNRCDARTYCYLLPTFAFAHKDRDVQDE-TYRLSAETLQQVNRLLACYKGTHNFHNFTSQ 223
Score = 41.6 bits (96), Expect = 0.054
Identities = 22/61 (36%), Positives = 35/61 (57%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G + I ++G+SFM+HQIRKMV VA+ + P ++ S ++ +P AP L+
Sbjct: 248 GLEFAVIRVKGQSFMMHQIRKMVGLVVAIVKGYAPESVLERSWGT-EKVDVPKAPGLGLV 306
Query: 406 L 406
L
Sbjct: 307 L 307
>UniRef100_UPI0000363FE8 UPI0000363FE8 UniRef100 entry
Length = 345
Score = 94.4 bits (233), Expect = 7e-18
Identities = 56/202 (27%), Positives = 101/202 (49%), Gaps = 17/202 (8%)
Query: 54 FRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWA 113
+ KKKV + +AY+G Y G+Q + T+E +L +A+ K+G I +++ D++K+ +
Sbjct: 3 YPKKKVALLMAYSGKGYYGMQRNPGSSEFRTIEDDLVAALVKSGCIPQNHGDDMKKMSFQ 62
Query: 114 RSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSF 173
R +RTDKGV + +V+ K+ + ++ L +N +LP I+++ + + F
Sbjct: 63 RCARTDKGVSAAGQVVSLKLLLIDD----------LTEKINEHLPPQIRILGVKRVTQGF 112
Query: 174 DPRKECVLRKYSYLLPA------DIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNY 227
+ + C R Y+Y+LP D + S + N + ++G H FHN+
Sbjct: 113 NSKNNCDARTYAYMLPTVAFSPKDFDAAGTAPSFRLQPETLQRVNHLFAHYKGTHNFHNF 172
Query: 228 TSRSRYRRHIPRR-PSHSKCGE 248
TS+ RR +H CGE
Sbjct: 173 TSQKAPNDPSARRYITHMLCGE 194
Score = 41.2 bits (95), Expect = 0.070
Identities = 33/124 (26%), Positives = 56/124 (44%), Gaps = 5/124 (4%)
Query: 352 IYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLLRGNSF 411
I + G+SFMLHQIRKM+ +AV + +++ S ++ +P AP L+L F
Sbjct: 206 ITVRGQSFMLHQIRKMIGLVIAVIKGYAGEEVMERSWG-LQKVDVPKAPGLGLVLEKVHF 264
Query: 412 SMRTKPGNVTRPEMQTIVESEEINKVVDDFYRSVMLPQV--SKFLDPSRVPWVEWIEKFD 469
K + +E EE + V F + + P + ++ + S V W+ + D
Sbjct: 265 DRYNK--RFGGDGLHERLEWEEQEEAVQAFKEAFIYPTIMETERQEGSMVSWMNTLPIHD 322
Query: 470 AHTS 473
S
Sbjct: 323 FEAS 326
>UniRef100_UPI00004993EF UPI00004993EF UniRef100 entry
Length = 401
Score = 94.4 bits (233), Expect = 7e-18
Identities = 56/188 (29%), Positives = 94/188 (49%), Gaps = 26/188 (13%)
Query: 55 RKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWAR 114
+K KV + +Y G Y GL N+ T+E E+E+A++KA I E N DL+K+GW+R
Sbjct: 39 KKFKVAVIFSYVGRGYFGLMYLN--NNPKTIEGEIENAMYKAKCISEDNKHDLKKVGWSR 96
Query: 115 SSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFD 174
+RTDKGV + +++ KM + N +NS+LP I++ ++ SF+
Sbjct: 97 CARTDKGVSAANNLISLKMGLDGN----------YIERINSFLPETIRIQDVVKVSHSFN 146
Query: 175 PRKECVLRKYSYLLPA----------DIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPF 224
+ R Y Y+ P+ I ++ + ++IDY FN ++ + H F
Sbjct: 147 AKTRVDSRTYEYIFPSYAIQKIDSEHPEINVEYNPQQEDIDY----FNSVISNYVKTHNF 202
Query: 225 HNYTSRSR 232
NYT ++
Sbjct: 203 QNYTFNTK 210
Score = 41.6 bits (96), Expect = 0.054
Identities = 37/143 (25%), Positives = 68/143 (46%), Gaps = 7/143 (4%)
Query: 338 SGKLEKSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLP----RDIITLSLSKFS- 392
S +L + G + I + I G+SFML QIR+M+ A+ + LP +I+ S + +
Sbjct: 227 SAQLVQRNGLNVIAVSIHGQSFMLWQIRRMIGFAIVLTNFRLPIEQMNEIMKTSFTLINW 286
Query: 393 RIILPLAPSEVLLLRGNSF-SMRTKPGNVTRPEMQTIVESEEINKVVDDFYRSVMLPQVS 451
R ++P AP+ L L F + + T +T + SEE K +F + +
Sbjct: 287 RSVIPTAPAHGLFLDQCHFGNHNERLQRDTSNNSKTNINSEETLKKCMEFKEKYIWNDII 346
Query: 452 KFLDPSRVPWVEWIEKFDAHTSI 474
F + P+ +++++ +T I
Sbjct: 347 NF-EKEEQPFAKYMKEQVNYTKI 368
>UniRef100_UPI000019EB4C UPI000019EB4C UniRef100 entry
Length = 353
Score = 92.0 bits (227), Expect = 3e-17
Identities = 58/200 (29%), Positives = 99/200 (49%), Gaps = 16/200 (8%)
Query: 54 FRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWA 113
+ KKKV + +AY+G Y G+Q + T+E EL +A+ A I E + +++K+ +
Sbjct: 18 YPKKKVALLMAYSGKGYYGMQRNARNSQFKTIEDELVTALVGARCIPEGHAEEMKKMSFQ 77
Query: 114 RSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSF 173
+S+RTD GV S +V+ K+ + EN + +N++LP I+V+ F
Sbjct: 78 KSARTDXGV-SAGQVVSLKVWMIEN----------ILEKINAHLPPQIRVLGCKRVTGGF 126
Query: 174 DPRKECVLRKYSYLLPADIIGIQSHSSNDE----IDYHMSEFNDILKEFEGGHPFHNYTS 229
+ + C R YSY+LP + ++ D + N + ++G H FHN+TS
Sbjct: 127 NSKNNCYARTYSYMLPTVAFSPKDYNQEDTSFRLTSDTLQTVNRLFGLYKGTHNFHNFTS 186
Query: 230 RSRYR-RHIPRRPSHSKCGE 248
+ +R R +H CGE
Sbjct: 187 QKGHRDPSAMRYITHMSCGE 206
Score = 39.7 bits (91), Expect = 0.20
Identities = 30/113 (26%), Positives = 53/113 (46%), Gaps = 5/113 (4%)
Query: 352 IYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLLLRGNSF 411
I + G+SFM+HQIRKM+ +AV + + +I S + ++ +P AP L+L F
Sbjct: 218 ITVRGQSFMMHQIRKMIGLVIAVVKGYVDEGVIEKSWGE-EKVGVPKAPGLGLVLERVHF 276
Query: 412 SMRTKPGNVTRPEMQTIVESEEINKVVDDFYRSVMLPQV--SKFLDPSRVPWV 462
K + +E E + + DF + P + ++ + S V W+
Sbjct: 277 DGYNK--RFGGDGIHETLEWTEQEEAIADFKEKHIYPTIVDTELNEKSMVNWM 327
>UniRef100_Q6FJJ6 Similar to sp|P53167 Saccharomyces cerevisiae YGL063w PUS2
pseudouridine synthase 2 [Candida glabrata]
Length = 317
Score = 90.1 bits (222), Expect = 1e-16
Identities = 54/173 (31%), Positives = 89/173 (51%), Gaps = 18/173 (10%)
Query: 63 IAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSSRTDKGV 122
+ Y GT Y+G+Q + T++ EL A+ I +SN D K ++R++RTDKGV
Sbjct: 5 LGYLGTGYQGIQYNPPAH---TIDNELFKALVATNCISQSNSDDFSKSSFSRATRTDKGV 61
Query: 123 HSLATMVAFKM---EIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDPRKEC 179
H+L +VA KM E+P+ E +N LP DI++ ++P ++ F+ R
Sbjct: 62 HALTNLVALKMQRYEVPQQVKAE----------LNEILPDDIRIWDVVPVKKKFNARVMA 111
Query: 180 VLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSRSR 232
R Y Y++P ++ HS D ++ F +I F G H ++N+T R +
Sbjct: 112 GTRHYRYVIPTFLLQCGPHS--DLSTSQLARFKNIWNMFLGTHNYYNFTPRRK 162
>UniRef100_Q9EQD1 Pseudouridine synthase 1 [Mus musculus]
Length = 411
Score = 88.6 bits (218), Expect = 4e-16
Identities = 57/208 (27%), Positives = 101/208 (48%), Gaps = 34/208 (16%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+K+V+ +AY+G Y G+Q + T+E +L SA+ +AG I E++ D+ K+ + R
Sbjct: 49 KRKIVLLMAYSGKGYHGMQRNLGSSQFRTIEDDLVSALVQAGCIPENHGTDMRKMSFQRC 108
Query: 116 SRTDK------------------GVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYL 157
+RTDK GV + +V+ K+ + ++ + + +NS+L
Sbjct: 109 ARTDKPLRTSACFCLYTGFLFHWGVSAAGQVVSLKVWLIDD----------ILDKINSHL 158
Query: 158 PCDIKVISILPAQRSFDPRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMS-----EFN 212
P I+++ + F+ + +C R Y Y+LP + DE Y +S + N
Sbjct: 159 PSHIRILGLKRVTGGFNSKNKCDARTYCYMLPTFAFAHKDRDVQDE-SYRLSAETLQQVN 217
Query: 213 DILKEFEGGHPFHNYTSRSRYRRHIPRR 240
+L ++G H FHN+TS+ R RR
Sbjct: 218 RLLACYKGTHNFHNFTSQKGPREPSARR 245
Score = 42.4 bits (98), Expect = 0.031
Identities = 22/61 (36%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G + I ++G+SFM+HQIRKMV VA+ + P ++ S + ++ +P AP L+
Sbjct: 260 GLEFAVIKVKGQSFMMHQIRKMVGLVVAIVKGYAPESVLERSWGE-EKVDVPKAPGLGLV 318
Query: 406 L 406
L
Sbjct: 319 L 319
>UniRef100_Q6CC39 Similar to sp|Q12211 Saccharomyces cerevisiae YPL212c PUS1
pseudouridine synthase 1 [Yarrowia lipolytica]
Length = 528
Score = 86.3 bits (212), Expect = 2e-15
Identities = 46/134 (34%), Positives = 74/134 (54%), Gaps = 12/134 (8%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+KV I Y GT Y G+Q+ Q T+E ++ A KAG I ++N D +K + R+
Sbjct: 84 KRKVACMIGYCGTGYHGMQLNPPQK---TIEGDIFQAFVKAGAISQNNADDPKKSAFMRA 140
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGVH+ +++ KM I + + + +NS+LP ++V + ++F+
Sbjct: 141 ARTDKGVHAAGNVISLKMIIEDEN---------IVEKINSHLPEQLRVWGVSRTNKAFEC 191
Query: 176 RKECVLRKYSYLLP 189
RK C R Y YL+P
Sbjct: 192 RKLCSSRVYEYLMP 205
Score = 45.4 bits (106), Expect = 0.004
Identities = 27/62 (43%), Positives = 41/62 (65%), Gaps = 2/62 (3%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVA-TAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVL 404
G ++ I I G+SFMLHQIRKM++ A++V+ P+ +I + K +RI +P AP+ L
Sbjct: 355 GTEWVSIKIHGQSFMLHQIRKMISMVALSVRCNADPQKLIPQTFEK-ARINIPKAPALGL 413
Query: 405 LL 406
LL
Sbjct: 414 LL 415
>UniRef100_UPI0000234E9A UPI0000234E9A UniRef100 entry
Length = 574
Score = 84.7 bits (208), Expect = 6e-15
Identities = 50/135 (37%), Positives = 75/135 (55%), Gaps = 12/135 (8%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
KKKV + I Y+GT Y+G+Q+ + T+E EL +A AG I ++N D +K R
Sbjct: 78 KKKVAVLIGYSGTGYKGMQLSTTEK---TIEGELFTAFVAAGAISKANAADPKKSSLVRC 134
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
+RTDKGVH+ +V+ K+ + EDP + +N +L I+V IL A +SF
Sbjct: 135 ARTDKGVHAAGNIVSLKLIV------EDP---DIVQKINDHLSPQIRVWGILVANKSFSS 185
Query: 176 RKECVLRKYSYLLPA 190
+ C R Y YL+P+
Sbjct: 186 YQMCDSRIYEYLIPS 200
Score = 40.4 bits (93), Expect = 0.12
Identities = 25/61 (40%), Positives = 35/61 (56%), Gaps = 1/61 (1%)
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPLAPSEVLL 405
G ++ + + G+SFM+HQIRKMVA A V R I S +F ++ +P AP LL
Sbjct: 392 GTEWLSLKVHGQSFMMHQIRKMVAMATMVVRCGCDPARINDSYGEF-KMAIPKAPGLGLL 450
Query: 406 L 406
L
Sbjct: 451 L 451
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 859,902,715
Number of Sequences: 2790947
Number of extensions: 36762428
Number of successful extensions: 95362
Number of sequences better than 10.0: 504
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 440
Number of HSP's that attempted gapping in prelim test: 94586
Number of HSP's gapped (non-prelim): 719
length of query: 501
length of database: 848,049,833
effective HSP length: 132
effective length of query: 369
effective length of database: 479,644,829
effective search space: 176988941901
effective search space used: 176988941901
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 77 (34.3 bits)
Medicago: description of AC140068.2


