

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.9 - phase: 0 /pseudo
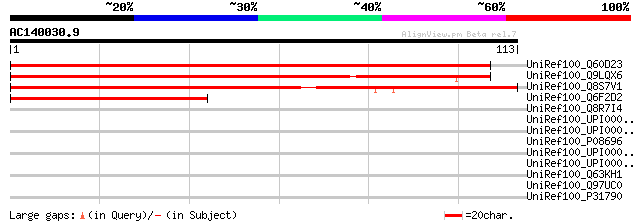
(113 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q60D23 Putative TPR domain containing protein [Solanum... 139 2e-32
UniRef100_Q9LQX6 T24P13.14 [Arabidopsis thaliana] 132 2e-30
UniRef100_Q8S7V1 Hypothetical protein OSJNBa0091P11.26 [Oryza sa... 96 3e-19
UniRef100_Q6F2D2 Putative TPR domain containing protein [Solanum... 67 7e-11
UniRef100_Q8R7I4 Hypothetical protein [Thermoanaerobacter tengco... 32 2.6
UniRef100_UPI0000345B43 UPI0000345B43 UniRef100 entry 31 5.9
UniRef100_UPI00002E81AA UPI00002E81AA UniRef100 entry 31 5.9
UniRef100_P08696 Bacteriocin BCN5 [Clostridium perfringens] 31 5.9
UniRef100_UPI0000312612 UPI0000312612 UniRef100 entry 31 7.7
UniRef100_UPI00002926A5 UPI00002926A5 UniRef100 entry 31 7.7
UniRef100_Q63KH1 Hypothetical protein [Burkholderia pseudomallei] 31 7.7
UniRef100_Q97UC0 Hypothetical protein [Sulfolobus solfataricus] 31 7.7
UniRef100_P31790 Retrovirus-related Gag polyprotein [Mouse intra... 31 7.7
>UniRef100_Q60D23 Putative TPR domain containing protein [Solanum demissum]
Length = 536
Score = 139 bits (349), Expect = 2e-32
Identities = 61/107 (57%), Positives = 81/107 (75%)
Query: 1 EQMKRWKVRGKEKGYLRASFWSLYSEAYGSERCMKRWGRRIPALDAVVDSITDVVGGDHR 60
E M+RW VRGK KGYLR+SFW +YSE Y SER ++RWG ++P +D V+DS+ + VG D R
Sbjct: 425 ENMRRWMVRGKGKGYLRSSFWRVYSEVYESERLLRRWGSKVPLMDNVLDSVVNAVGSDER 484
Query: 61 VLKILMEELKRKGGGSGGGNLEMEKVFKLAREVYGKVVKKQAMRTLL 107
++K+LM LKRK G G G +EMEK KL R +YGK++KKQ +RT+L
Sbjct: 485 IVKLLMRGLKRKNGHKGNGIVEMEKAMKLGRGLYGKIMKKQTLRTIL 531
>UniRef100_Q9LQX6 T24P13.14 [Arabidopsis thaliana]
Length = 969
Score = 132 bits (332), Expect = 2e-30
Identities = 64/108 (59%), Positives = 84/108 (77%), Gaps = 2/108 (1%)
Query: 1 EQMKRWKVRGKEKGYLRASFWSLYSEAYGSERCMKRWGRRIPALDAVVDSITDVVGGDHR 60
E MKRWKV+GK+KG LRAS+W +Y E Y SER MKRWGR+IP ++ VVDS++DVVG D R
Sbjct: 860 EGMKRWKVKGKDKGLLRASYWGVYDEIYNSERLMKRWGRKIPTMEVVVDSVSDVVGSDER 919
Query: 61 VLKILMEELKRKGGGSGGGNLEMEKVFKLAREVYGKVV-KKQAMRTLL 107
++K+ +E + +K GG +EMEK+ KL + VYGKVV KK+AM+TLL
Sbjct: 920 LMKMAVEGMMKKHGGF-SNIVEMEKIMKLGKGVYGKVVSKKKAMKTLL 966
>UniRef100_Q8S7V1 Hypothetical protein OSJNBa0091P11.26 [Oryza sativa]
Length = 536
Score = 95.5 bits (236), Expect = 3e-19
Identities = 49/115 (42%), Positives = 76/115 (65%), Gaps = 5/115 (4%)
Query: 1 EQMKRWKVRGKEKGYLRASFWSLYSEAYGSERCMKRWGRRIPALDAVVDSITDVVGGDHR 60
E+M++ V+ + K +LRASFWS YS + S++ +++WGRR+P AV +S+ +GG+
Sbjct: 419 ERMRKSMVKERRKAFLRASFWSAYSALFDSDKLVRKWGRRVPGEAAVAESVAGAIGGNES 478
Query: 61 VLKILMEELKRKGGGSGGGN-LEME-KVFKLAREVYGKVVKKQAMRTLLELCIAA 113
VL+ + L+ G+G GN LE+E KV ++ R YG+VVK+QAMR L L + A
Sbjct: 479 VLRAM---LRGADNGNGCGNRLEVEDKVVRIGRATYGRVVKRQAMRALFRLTLDA 530
>UniRef100_Q6F2D2 Putative TPR domain containing protein [Solanum demissum]
Length = 438
Score = 67.4 bits (163), Expect = 7e-11
Identities = 26/44 (59%), Positives = 35/44 (79%)
Query: 1 EQMKRWKVRGKEKGYLRASFWSLYSEAYGSERCMKRWGRRIPAL 44
E M+RW VRGK KGYLR+SFW +YSE Y SER +++WG ++P +
Sbjct: 388 ENMRRWMVRGKGKGYLRSSFWRVYSEVYESERSLRKWGSKVPLM 431
>UniRef100_Q8R7I4 Hypothetical protein [Thermoanaerobacter tengcongensis]
Length = 112
Score = 32.3 bits (72), Expect = 2.6
Identities = 21/67 (31%), Positives = 34/67 (50%), Gaps = 13/67 (19%)
Query: 58 DHRVLKILMEELKR-------------KGGGSGGGNLEMEKVFKLAREVYGKVVKKQAMR 104
D+ KI ME+LKR + GSG G ++ E+VFKL ++V G + + +
Sbjct: 26 DYLSTKIGMEKLKRYYEIAKAFVQAVEQQVGSGNGPIKKEQVFKLLKKVIGNKLTDEEID 85
Query: 105 TLLELCI 111
L+E +
Sbjct: 86 KLIEAAV 92
>UniRef100_UPI0000345B43 UPI0000345B43 UniRef100 entry
Length = 177
Score = 31.2 bits (69), Expect = 5.9
Identities = 17/38 (44%), Positives = 24/38 (62%), Gaps = 2/38 (5%)
Query: 23 LYSEAYGS--ERCMKRWGRRIPALDAVVDSITDVVGGD 58
L EAYG+ + C+KR G IPA + +D ++VV GD
Sbjct: 39 LEREAYGAALDACIKRLGLDIPAYERYLDWASNVVRGD 76
>UniRef100_UPI00002E81AA UPI00002E81AA UniRef100 entry
Length = 197
Score = 31.2 bits (69), Expect = 5.9
Identities = 17/38 (44%), Positives = 24/38 (62%), Gaps = 2/38 (5%)
Query: 23 LYSEAYGS--ERCMKRWGRRIPALDAVVDSITDVVGGD 58
L EAYG+ + C+KR G IPA + +D ++VV GD
Sbjct: 39 LEREAYGAALDACIKRLGLDIPAYERYLDWASNVVRGD 76
>UniRef100_P08696 Bacteriocin BCN5 [Clostridium perfringens]
Length = 890
Score = 31.2 bits (69), Expect = 5.9
Identities = 28/101 (27%), Positives = 44/101 (42%), Gaps = 9/101 (8%)
Query: 15 YLRASFWSLYSEAYGSER-CMKRWGRRIPALDAVVD------SITDVVGGDHRVLKILME 67
Y R F YG R M W + I A A+++ S +DVV G R L+ ++
Sbjct: 490 YFRNQFGFGQRSGYGDNRGFMIGWAKSIGAKAALIELPGSTKSHSDVVNG--RYLQKIIN 547
Query: 68 ELKRKGGGSGGGNLEMEKVFKLAREVYGKVVKKQAMRTLLE 108
+ GGSGG + ++ E G+V+ Q+ + E
Sbjct: 548 AVTNLIGGSGGSSSGGSSFSDVSYEATGEVINVQSFLNVRE 588
>UniRef100_UPI0000312612 UPI0000312612 UniRef100 entry
Length = 318
Score = 30.8 bits (68), Expect = 7.7
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query: 23 LYSEAYGS--ERCMKRWGRRIPALDAVVDSITDVVGGD 58
L EAYG+ + C+KR G +PA + +D ++VV GD
Sbjct: 39 LEREAYGAALDACIKRLGLDVPAYERYLDWASNVVRGD 76
>UniRef100_UPI00002926A5 UPI00002926A5 UniRef100 entry
Length = 137
Score = 30.8 bits (68), Expect = 7.7
Identities = 16/38 (42%), Positives = 24/38 (63%), Gaps = 2/38 (5%)
Query: 23 LYSEAYGS--ERCMKRWGRRIPALDAVVDSITDVVGGD 58
L EAYG+ + C+KR G +PA + +D ++VV GD
Sbjct: 39 LEREAYGAALDACIKRLGLDVPAYERYLDWASNVVRGD 76
>UniRef100_Q63KH1 Hypothetical protein [Burkholderia pseudomallei]
Length = 468
Score = 30.8 bits (68), Expect = 7.7
Identities = 10/31 (32%), Positives = 23/31 (73%)
Query: 49 DSITDVVGGDHRVLKILMEELKRKGGGSGGG 79
D+ + + G D +++ +L ++L+++ GG+GGG
Sbjct: 30 DARSQIAGLDQQIMGLLRQQLQKRDGGTGGG 60
>UniRef100_Q97UC0 Hypothetical protein [Sulfolobus solfataricus]
Length = 220
Score = 30.8 bits (68), Expect = 7.7
Identities = 11/35 (31%), Positives = 21/35 (59%)
Query: 21 WSLYSEAYGSERCMKRWGRRIPALDAVVDSITDVV 55
WS Y+E + E + RRIP+LD + +++ ++
Sbjct: 14 WSRYTERFNEEELYVKVVRRIPSLDGLTENVMGII 48
>UniRef100_P31790 Retrovirus-related Gag polyprotein [Mouse intracisternal
a-particle]
Length = 255
Score = 30.8 bits (68), Expect = 7.7
Identities = 25/103 (24%), Positives = 46/103 (44%), Gaps = 6/103 (5%)
Query: 1 EQMKRWKVRGKEKGYLRASF---WSLYSEAYGSERCMKRWGRRIPALDAVVDSITDVVGG 57
+++K VR ++KG L+A W L E C + LD + +S+++V G
Sbjct: 53 DKLKGDLVREQQKGKLKAGIIPLWKLVKSCLTDEDCQQMVEAGQKVLDEIQESLSEVERG 112
Query: 58 DHRVLKILMEELKRKGGGSGGGNLEMEKVFKLAREVYGKVVKK 100
+ ++ LK G +G LE E+ + G++ K+
Sbjct: 113 EKVKVERKQSALKNLGLSTG---LEPEEKRYKGKNALGEIRKR 152
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 191,748,255
Number of Sequences: 2790947
Number of extensions: 7366277
Number of successful extensions: 23203
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 23194
Number of HSP's gapped (non-prelim): 15
length of query: 113
length of database: 848,049,833
effective HSP length: 89
effective length of query: 24
effective length of database: 599,655,550
effective search space: 14391733200
effective search space used: 14391733200
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC140030.9


