

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.7 - phase: 0
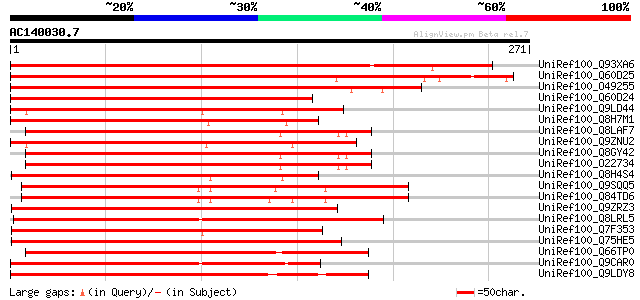
(271 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93XA6 NAC domain protein NAC2 [Phaseolus vulgaris] 407 e-112
UniRef100_Q60D25 Putative NAC domain protein NAC2 [Solanum demis... 317 3e-85
UniRef100_O49255 NAC-domain containing protein 29 [Arabidopsis t... 312 7e-84
UniRef100_Q60D24 Putative NAC domain protein NAC2 [Solanum demis... 258 9e-68
UniRef100_Q9LD44 Putative jasmonic acid regulatory protein [Arab... 241 2e-62
UniRef100_Q8H7M1 Putative NAM (No apical meristem) protein [Oryz... 240 3e-62
UniRef100_Q8LAF7 NAM protein, putative [Arabidopsis thaliana] 239 4e-62
UniRef100_Q9ZNU2 NAC-domain containing protein 18 [Arabidopsis t... 239 4e-62
UniRef100_Q8GY42 Hypothetical protein At1g61110/F11P17_16 [Arabi... 239 6e-62
UniRef100_O22734 F11P17.16 protein [Arabidopsis thaliana] 239 6e-62
UniRef100_Q8H4S4 Putative OsNAC5 protein [Oryza sativa] 236 6e-61
UniRef100_Q9SQQ5 NAM-like protein [Arabidopsis thaliana] 235 8e-61
UniRef100_Q84TD6 At3g04070 [Arabidopsis thaliana] 231 2e-59
UniRef100_Q9ZRZ3 GRAB1 protein [Triticum sp] 229 6e-59
UniRef100_Q8LRL5 Nam-like protein 10 [Petunia hybrida] 228 1e-58
UniRef100_Q7F353 Development regulation gene OsNAC4 [Oryza sativa] 227 3e-58
UniRef100_Q75HE5 Putative NAC-domain protein [Oryza sativa] 226 4e-58
UniRef100_Q66TP0 Jasmonic acid 2 [Solanum tuberosum] 225 1e-57
UniRef100_Q9CAR0 GRAB1-like protein; 10550-11502 [Arabidopsis th... 224 1e-57
UniRef100_Q9LDY8 Putative jasmonic acid regulatory protein [Arab... 224 2e-57
>UniRef100_Q93XA6 NAC domain protein NAC2 [Phaseolus vulgaris]
Length = 256
Score = 407 bits (1045), Expect = e-112
Identities = 195/255 (76%), Positives = 219/255 (85%), Gaps = 5/255 (1%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M+++ SELPPGFRFHPTDEELIV+YLCNQATSKPCPASIIPEVD+YKFDPWELPDK+EF
Sbjct: 1 MDATTPSELPPGFRFHPTDEELIVYYLCNQATSKPCPASIIPEVDLYKFDPWELPDKTEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFFSPR+RKYPNGVRPNRAT+SGYWKATGTDKAI SGSK +GVKKSLVFYKGRPP
Sbjct: 61 GENEWYFFSPRDRKYPNGVRPNRATVSGYWKATGTDKAIYSGSKLVGVKKSLVFYKGRPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 180
KG KTDWIMHEYRL S++ ++ IGSMRLDDWVLCRIYKKK+ GKTL+ KE + Q
Sbjct: 121 KGDKTDWIMHEYRLAESKQPVNRKIGSMRLDDWVLCRIYKKKNTGKTLEHKETHPKVQMT 180
Query: 181 DSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPILSDG---STLDFQINNSNIGIDPYV 237
+ I NND E +MMNL R+ SLTYLLDMNY GPILSDG ST DFQI+N+NIGIDP+V
Sbjct: 181 NLIAANND--EQKMMNLPRTWSLTYLLDMNYLGPILSDGSYCSTFDFQISNANIGIDPFV 238
Query: 238 KPQPVEMTNHYEADS 252
QPVEM N+Y +DS
Sbjct: 239 NSQPVEMANNYVSDS 253
>UniRef100_Q60D25 Putative NAC domain protein NAC2 [Solanum demissum]
Length = 281
Score = 317 bits (811), Expect = 3e-85
Identities = 165/275 (60%), Positives = 204/275 (74%), Gaps = 13/275 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M SS+LPPGFRFHPTDEELI++YL QATS+PCP SIIPE+D+YKFDPWELP+K+EF
Sbjct: 1 MVGKISSDLPPGFRFHPTDEELIMYYLRYQATSRPCPVSIIPEIDVYKFDPWELPEKAEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFF+PR+RKYPNGVRPNRA +SGYWKATGTDKAI S +K +G+KK+LVFYKG+PP
Sbjct: 61 GENEWYFFTPRDRKYPNGVRPNRAAVSGYWKATGTDKAIYSANKYVGIKKALVFYKGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQT-SKHIGSMRLDDWVLCRIYKKKHMGKTLQ--QKEDYSTH 177
KGVKTDWIMHEYRL S+ QT SK GSMRLDDWVLCRIYKKK++GKT++ + E+
Sbjct: 121 KGVKTDWIMHEYRLSDSKSQTYSKQSGSMRLDDWVLCRIYKKKNLGKTIEMMKVEEEELE 180
Query: 178 QFNDSIITNNDDGELEMMNLTRSCSLTYLLDMNYFGPI---LSDGSTLD---FQINN-SN 230
N SI + G + M L R CSL++LL+++YFG I LSD D + +NN SN
Sbjct: 181 AQNVSINNAIEVGGPQTMKLPRICSLSHLLELDYFGSIPQLLSDNLLYDDQSYTMNNVSN 240
Query: 231 IGIDPYVKPQPVEMTNHYEADSHSSITN--QPIFV 263
V Q + TN+ +++ + N QP+FV
Sbjct: 241 TSNVDQVSSQQ-QNTNNITSNNCNIFFNYQQPLFV 274
>UniRef100_O49255 NAC-domain containing protein 29 [Arabidopsis thaliana]
Length = 268
Score = 312 bits (799), Expect = 7e-84
Identities = 147/225 (65%), Positives = 174/225 (77%), Gaps = 10/225 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ME ++ S LPPGFRFHPTDEELIV+YL NQ SKPCP SIIPEVDIYKFDPW+LP+K+EF
Sbjct: 1 MEVTSQSTLPPGFRFHPTDEELIVYYLRNQTMSKPCPVSIIPEVDIYKFDPWQLPEKTEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFFSPRERKYPNGVRPNRA +SGYWKATGTDKAI SGS +GVKK+LVFYKGRPP
Sbjct: 61 GENEWYFFSPRERKYPNGVRPNRAAVSGYWKATGTDKAIHSGSSNVGVKKALVFYKGRPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTH--- 177
KG+KTDWIMHEYRL S+K ++K GSMRLD+WVLCRIYKK+ K L ++E +
Sbjct: 121 KGIKTDWIMHEYRLHDSRKASTKRNGSMRLDEWVLCRIYKKRGASKLLNEQEGFMDEVLM 180
Query: 178 QFNDSIITNNDDGELE-------MMNLTRSCSLTYLLDMNYFGPI 215
+ ++ N + E M L R+CSL +LL+M+Y GP+
Sbjct: 181 EDETKVVVNEAERRTEEEIMMMTSMKLPRTCSLAHLLEMDYMGPV 225
>UniRef100_Q60D24 Putative NAC domain protein NAC2 [Solanum demissum]
Length = 163
Score = 258 bits (660), Expect = 9e-68
Identities = 117/159 (73%), Positives = 138/159 (86%), Gaps = 1/159 (0%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M SS+LPPGFRFHPTDEELI++YL QATS+PCP SIIPE+D+YKFDPWELP+K+EF
Sbjct: 1 MVGKISSDLPPGFRFHPTDEELIMYYLRYQATSRPCPVSIIPEIDVYKFDPWELPEKAEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFF+PR+RKYPNGVRPNRA +SGYWKATGTDKAI S +K +G+KK+LVFYKG+PP
Sbjct: 61 GENEWYFFTPRDRKYPNGVRPNRAAVSGYWKATGTDKAIYSANKYVGIKKALVFYKGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQT-SKHIGSMRLDDWVLCRI 158
KGVKTDWIMHEYRL S+ QT SK GSMR+ ++CR+
Sbjct: 121 KGVKTDWIMHEYRLSDSKSQTYSKQSGSMRVRTSLICRL 159
>UniRef100_Q9LD44 Putative jasmonic acid regulatory protein [Arabidopsis thaliana]
Length = 364
Score = 241 bits (614), Expect = 2e-62
Identities = 115/191 (60%), Positives = 138/191 (72%), Gaps = 17/191 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL+VHYL +A S P P +II EVD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVVHYLKRKAASAPLPVAIIAEVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKK 110
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + G++++GVKK
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVLASDGNQKVGVKK 120
Query: 111 SLVFYKGRPPKGVKTDWIMHEYRLIGSQKQT-------SKHIGSMRLDDWVLCRIYKKKH 163
+LVFY G+PPKGVK+DWIMHEYRLI ++ S+RLDDWVLCRIYKK +
Sbjct: 121 ALVFYSGKPPKGVKSDWIMHEYRLIENKPNNRPPGCDFGNKKNSLRLDDWVLCRIYKKNN 180
Query: 164 MGKTLQQKEDY 174
+ + +D+
Sbjct: 181 ASRHVDNDKDH 191
>UniRef100_Q8H7M1 Putative NAM (No apical meristem) protein [Oryza sativa]
Length = 392
Score = 240 bits (612), Expect = 3e-62
Identities = 114/173 (65%), Positives = 129/173 (73%), Gaps = 12/173 (6%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M S + LPPGFRFHPTDEELIVHYL N+A S PCP SII +VDIYKFDPW+LP K +
Sbjct: 1 MVLSNPAMLPPGFRFHPTDEELIVHYLRNRAASSPCPVSIIADVDIYKFDPWDLPSKENY 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSG-----SKQIGVKKSLVFY 115
+ EWYFFSPR+RKYPNG+RPNRA SGYWKATGTDK I S ++ +GVKK+LVFY
Sbjct: 61 GDREWYFFSPRDRKYPNGIRPNRAAGSGYWKATGTDKPIHSSGGAATNESVGVKKALVFY 120
Query: 116 KGRPPKGVKTDWIMHEYRLIGSQKQTSK-------HIGSMRLDDWVLCRIYKK 161
KGRPPKG KT+WIMHEYRL + + SMRLDDWVLCRIYKK
Sbjct: 121 KGRPPKGTKTNWIMHEYRLAAADAHAANTYRPMKFRNTSMRLDDWVLCRIYKK 173
>UniRef100_Q8LAF7 NAM protein, putative [Arabidopsis thaliana]
Length = 323
Score = 239 bits (611), Expect = 4e-62
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 16 LPPGFRFHPTDEELVVHYLKKKAASVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 75
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 76 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 135
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 136 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 195
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 196 EGMLSKSSANSSSTSVLDNNDN 217
>UniRef100_Q9ZNU2 NAC-domain containing protein 18 [Arabidopsis thaliana]
Length = 320
Score = 239 bits (611), Expect = 4e-62
Identities = 118/197 (59%), Positives = 139/197 (69%), Gaps = 16/197 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL++HYL +A S P P +II +VD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVIHYLKRKADSVPLPVAIIADVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS----GSKQIGV 108
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + S GSK++GV
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVISTGGGGSKKVGV 120
Query: 109 KKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQTSKHIG----SMRLDDWVLCRIYKKKHM 164
KK+LVFY G+PPKGVK+DWIMHEYRL ++ G S+RLDDWVLCRIYKK +
Sbjct: 121 KKALVFYSGKPPKGVKSDWIMHEYRLTDNKPTHICDFGNKKNSLRLDDWVLCRIYKKNNS 180
Query: 165 GKTLQQKEDYSTHQFND 181
+ + H ND
Sbjct: 181 TASRHHHHLHHIHLDND 197
>UniRef100_Q8GY42 Hypothetical protein At1g61110/F11P17_16 [Arabidopsis thaliana]
Length = 323
Score = 239 bits (610), Expect = 6e-62
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 16 LPPGFRFHPTDEELVVHYLKKKADSVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 75
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 76 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 135
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 136 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 195
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 196 EGMLSKSSANSSSTSVLDNNDN 217
>UniRef100_O22734 F11P17.16 protein [Arabidopsis thaliana]
Length = 300
Score = 239 bits (610), Expect = 6e-62
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 13 LPPGFRFHPTDEELVVHYLKKKADSVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 72
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 73 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 132
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 133 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 192
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 193 EGMLSKSSANSSSTSVLDNNDN 214
>UniRef100_Q8H4S4 Putative OsNAC5 protein [Oryza sativa]
Length = 425
Score = 236 bits (601), Expect = 6e-61
Identities = 111/179 (62%), Positives = 131/179 (73%), Gaps = 19/179 (10%)
Query: 2 ESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE 61
+ ++ ELPPGFRFHPTDEEL+VHYL +A S P P +II EVD+YKFDPW+LP+K+ F
Sbjct: 22 QPGSAPELPPGFRFHPTDEELVVHYLKKKAASVPLPVTIIAEVDLYKFDPWDLPEKANFG 81
Query: 62 ENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGS---KQIGVKKSLVFYKGR 118
E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK I S +++GVKK+LVFY+G+
Sbjct: 82 EQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPIMSSGSTREKVGVKKALVFYRGK 141
Query: 119 PPKGVKTDWIMHEYRLIGSQKQT----------------SKHIGSMRLDDWVLCRIYKK 161
PPKGVKT+WIMHEYRL + SK S+RLDDWVLCRIYKK
Sbjct: 142 PPKGVKTNWIMHEYRLTDTSSSAAAVATTRRPPPPITGGSKGAVSLRLDDWVLCRIYKK 200
>UniRef100_Q9SQQ5 NAM-like protein [Arabidopsis thaliana]
Length = 343
Score = 235 bits (600), Expect = 8e-61
Identities = 121/220 (55%), Positives = 145/220 (65%), Gaps = 18/220 (8%)
Query: 7 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWY 66
S LPPGFRFHPTDEELI+HYL + +S P P SII +VDIYK DPW+LP K+ F E EWY
Sbjct: 8 SSLPPGFRFHPTDEELILHYLRKKVSSSPVPLSIIADVDIYKSDPWDLPAKAPFGEKEWY 67
Query: 67 FFSPRERKYPNGVRPNRATLSGYWKATGTDK--AIKSGS---KQIGVKKSLVFYKGRPPK 121
FFSPR+RKYPNG RPNRA SGYWKATGTDK A+ +G + IG+KK+LVFY+G+PPK
Sbjct: 68 FFSPRDRKYPNGARPNRAAASGYWKATGTDKLIAVPNGEGFHENIGIKKALVFYRGKPPK 127
Query: 122 GVKTDWIMHEYRLIGS---QKQTSKHIGSMRLDDWVLCRIYKKKH---------MGKTLQ 169
GVKT+WIMHEYRL S ++ S G LDDWVLCRIYKK H + + Q
Sbjct: 128 GVKTNWIMHEYRLADSLSPKRINSSRSGGSELDDWVLCRIYKKSHASLSSPDVALVTSNQ 187
Query: 170 QKEDYSTHQFND-SIITNNDDGELEMMNLTRSCSLTYLLD 208
+ E+ F D N + + SCS + LLD
Sbjct: 188 EHEENDNEPFVDRGTFLPNLQNDQPLKRQKSSCSFSNLLD 227
>UniRef100_Q84TD6 At3g04070 [Arabidopsis thaliana]
Length = 359
Score = 231 bits (589), Expect = 2e-59
Identities = 123/236 (52%), Positives = 147/236 (62%), Gaps = 34/236 (14%)
Query: 7 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWY 66
S LPPGFRFHPTDEELI+HYL + +S P P SII +VDIYK DPW+LP K+ F E EWY
Sbjct: 8 SSLPPGFRFHPTDEELILHYLRKKVSSSPVPLSIIADVDIYKSDPWDLPAKAPFGEKEWY 67
Query: 67 FFSPRERKYPNGVRPNRATLSGYWKATGTDK--AIKSGS---KQIGVKKSLVFYKGRPPK 121
FFSPR+RKYPNG RPNRA SGYWKATGTDK A+ +G + IG+KK+LVFY+G+PPK
Sbjct: 68 FFSPRDRKYPNGARPNRAAASGYWKATGTDKLIAVPNGEGFHENIGIKKALVFYRGKPPK 127
Query: 122 GVKTDWIMHEYRL--------IGSQKQTSKHIG-----------SMRLDDWVLCRIYKKK 162
GVKT+WIMHEYRL I S + + SMRLDDWVLCRIYKK
Sbjct: 128 GVKTNWIMHEYRLADSLSPKRINSSRSGGSEVNNNFGDRNSKEYSMRLDDWVLCRIYKKS 187
Query: 163 H---------MGKTLQQKEDYSTHQFND-SIITNNDDGELEMMNLTRSCSLTYLLD 208
H + + Q+ E+ F D N + + SCS + LLD
Sbjct: 188 HASLSSPDVALVTSNQEHEENDNEPFVDRGTFLPNLQNDQPLKRQKSSCSFSNLLD 243
>UniRef100_Q9ZRZ3 GRAB1 protein [Triticum sp]
Length = 287
Score = 229 bits (584), Expect = 6e-59
Identities = 97/170 (57%), Positives = 129/170 (75%)
Query: 2 ESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE 61
++ A LPPGFRFHPTDEEL+ YLC +A + P II E+D+Y+FDPWELP+++ F
Sbjct: 10 DAEAELNLPPGFRFHPTDEELVADYLCARAAGRAPPVPIIAELDLYRFDPWELPERALFG 69
Query: 62 ENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPK 121
EWYFF+PR+RKYPNG RPNRA GYWKATG D+ + + +G+KK+LVFY GRP
Sbjct: 70 AREWYFFTPRDRKYPNGSRPNRAAGGGYWKATGADRPVARAGRTVGIKKALVFYHGRPSA 129
Query: 122 GVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQK 171
GVKTDWIMHEYRL G+ + +K+ G++RLD+WVLCR+Y KK+ + +Q++
Sbjct: 130 GVKTDWIMHEYRLAGADGRAAKNGGTLRLDEWVLCRLYNKKNQWEKMQRQ 179
>UniRef100_Q8LRL5 Nam-like protein 10 [Petunia hybrida]
Length = 304
Score = 228 bits (582), Expect = 1e-58
Identities = 104/194 (53%), Positives = 137/194 (70%), Gaps = 2/194 (1%)
Query: 3 SSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEE 62
++A +LPPGFRFHPTDEEL++HYLC + S+P II E+D+YK+DPW+LPD + + E
Sbjct: 2 TTAELQLPPGFRFHPTDEELVMHYLCRKCASQPIAVPIIAEIDLYKYDPWDLPDLALYGE 61
Query: 63 NEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKG 122
EWYFFSPR+RKYPNG RPNRA +GYWKATG DK I K +G+KK+LVFY G+ PKG
Sbjct: 62 KEWYFFSPRDRKYPNGSRPNRAAGTGYWKATGADKPI-GHPKAVGIKKALVFYAGKAPKG 120
Query: 123 VKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK-HMGKTLQQKEDYSTHQFND 181
KT+WIMHEYRL + K+ S+RLDDWVLCRIY KK + K + + D
Sbjct: 121 EKTNWIMHEYRLADVDRSARKNNNSLRLDDWVLCRIYNKKGSIEKNQLNNKKIMNTSYMD 180
Query: 182 SIITNNDDGELEMM 195
+++ +D + E++
Sbjct: 181 MTVSSEEDRKPEIL 194
>UniRef100_Q7F353 Development regulation gene OsNAC4 [Oryza sativa]
Length = 316
Score = 227 bits (578), Expect = 3e-58
Identities = 98/164 (59%), Positives = 123/164 (74%), Gaps = 2/164 (1%)
Query: 2 ESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE 61
++ A LPPGFRFHPTDEEL+VHYLC + +P P II EVD+YK DPW+LP+K+ F
Sbjct: 12 DAEAELNLPPGFRFHPTDEELVVHYLCRKVARQPLPVPIIAEVDLYKLDPWDLPEKALFG 71
Query: 62 ENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKKSLVFYKGRP 119
EWYFF+PR+RKYPNG RPNRA GYWKATG DK + K ++ +G+KK+LVFY G+
Sbjct: 72 RKEWYFFTPRDRKYPNGSRPNRAAGRGYWKATGADKPVAPKGSARTVGIKKALVFYSGKA 131
Query: 120 PKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKH 163
P+GVKTDWIMHEYRL + + GS +LD+WVLCR+Y KK+
Sbjct: 132 PRGVKTDWIMHEYRLADADRAPGGKKGSQKLDEWVLCRLYNKKN 175
>UniRef100_Q75HE5 Putative NAC-domain protein [Oryza sativa]
Length = 316
Score = 226 bits (577), Expect = 4e-58
Identities = 99/173 (57%), Positives = 130/173 (74%), Gaps = 1/173 (0%)
Query: 2 ESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE 61
++ A LPPGFRFHPTD+EL+ HYLC +A + P II EVD+YKFDPW+LP+++ F
Sbjct: 10 DAEAELNLPPGFRFHPTDDELVEHYLCRKAAGQRLPVPIIAEVDLYKFDPWDLPERALFG 69
Query: 62 ENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPK 121
EWYFF+PR+RKYPNG RPNRA +GYWKATG DK + + +G+KK+LVFY G+ P+
Sbjct: 70 AREWYFFTPRDRKYPNGSRPNRAAGNGYWKATGADKPVAPRGRTLGIKKALVFYAGKAPR 129
Query: 122 GVKTDWIMHEYRLIGSQKQTS-KHIGSMRLDDWVLCRIYKKKHMGKTLQQKED 173
GVKTDWIMHEYRL + + + GS+RLDDWVLCR+Y KK+ + +QQ ++
Sbjct: 130 GVKTDWIMHEYRLADAGRAAAGAKKGSLRLDDWVLCRLYNKKNEWEKMQQGKE 182
>UniRef100_Q66TP0 Jasmonic acid 2 [Solanum tuberosum]
Length = 349
Score = 225 bits (573), Expect = 1e-57
Identities = 104/179 (58%), Positives = 129/179 (71%), Gaps = 3/179 (1%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRF+PTDEEL+V YLC + P II E+D+YKFDPW LP K+ F E EWYFF
Sbjct: 14 LPPGFRFYPTDEELLVQYLCKKVAGHDFPLQIIGEIDLYKFDPWVLPSKATFGEKEWYFF 73
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDWI 128
SPR+RKYPNG RPNR SGYWKATGTDK I S +++G+KK+LVFY G+ PKG KT+WI
Sbjct: 74 SPRDRKYPNGSRPNRVAGSGYWKATGTDKIITSQGRKVGIKKALVFYVGKAPKGSKTNWI 133
Query: 129 MHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSIITNN 187
MHEYRL S K K+ GS +LD+WVLCRIYKK G +S+++++ T++
Sbjct: 134 MHEYRLFESSK---KNNGSSKLDEWVLCRIYKKNSSGPKPLMSGLHSSNEYSHGSSTSS 189
>UniRef100_Q9CAR0 GRAB1-like protein; 10550-11502 [Arabidopsis thaliana]
Length = 253
Score = 224 bits (572), Expect = 1e-57
Identities = 101/162 (62%), Positives = 123/162 (75%), Gaps = 2/162 (1%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M+S A + PPGFRFHPTDEEL++ YLC + S+P PA II E+D+Y++DPW+LPD + +
Sbjct: 2 MKSGADLQFPPGFRFHPTDEELVLMYLCRKCASQPIPAPIITELDLYRYDPWDLPDMALY 61
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
E EWYFFSPR+RKYPNG RPNRA +GYWKATG DK I K +G+KK+LVFY G+PP
Sbjct: 62 GEKEWYFFSPRDRKYPNGSRPNRAAGTGYWKATGADKPI-GRPKPVGIKKALVFYSGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 162
G KT+WIMHEYRL + K S+RLDDWVLCRIY KK
Sbjct: 121 NGEKTNWIMHEYRLADVDRSVRKK-NSLRLDDWVLCRIYNKK 161
>UniRef100_Q9LDY8 Putative jasmonic acid regulatory protein [Arabidopsis thaliana]
Length = 317
Score = 224 bits (571), Expect = 2e-57
Identities = 106/187 (56%), Positives = 132/187 (69%), Gaps = 8/187 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
++ A LPPGFRF+PTDEEL+V YLC +A +I E+D+YKFDPW LP K+ F
Sbjct: 6 LDPLAQLSLPPGFRFYPTDEELMVEYLCRKAAGHDFSLQLIAEIDLYKFDPWVLPSKALF 65
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
E EWYFFSPR+RKYPNG RPNR SGYWKATGTDK I + +++G+KK+LVFY G+ P
Sbjct: 66 GEKEWYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKVISTEGRRVGIKKALVFYIGKAP 125
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 180
KG KT+WIMHEYRLI + S+ GS +LDDWVLCRIYKK +T QK+ Y+ +
Sbjct: 126 KGTKTNWIMHEYRLI----EPSRRNGSTKLDDWVLCRIYKK----QTSAQKQAYNNLMTS 177
Query: 181 DSIITNN 187
+NN
Sbjct: 178 GREYSNN 184
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 509,715,523
Number of Sequences: 2790947
Number of extensions: 22917661
Number of successful extensions: 47812
Number of sequences better than 10.0: 318
Number of HSP's better than 10.0 without gapping: 260
Number of HSP's successfully gapped in prelim test: 58
Number of HSP's that attempted gapping in prelim test: 47031
Number of HSP's gapped (non-prelim): 331
length of query: 271
length of database: 848,049,833
effective HSP length: 125
effective length of query: 146
effective length of database: 499,181,458
effective search space: 72880492868
effective search space used: 72880492868
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC140030.7


