

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.11 + phase: 0
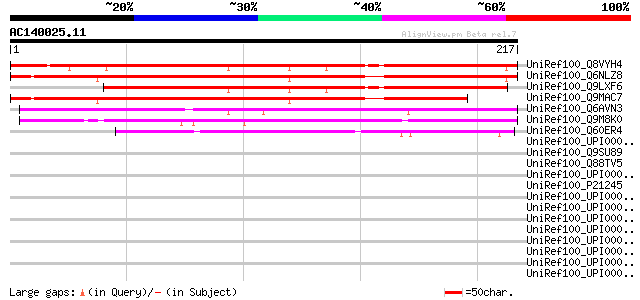
(217 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VYH4 AT5g15260/F8M21_150 [Arabidopsis thaliana] 207 1e-52
UniRef100_Q6NLZ8 Hypothetical protein At3g01170 [Arabidopsis tha... 202 4e-51
UniRef100_Q9LXF6 Hypothetical protein F8M21_150 [Arabidopsis tha... 197 2e-49
UniRef100_Q9MAC7 T4P13.14 protein [Arabidopsis thaliana] 174 2e-42
UniRef100_Q6AVN3 Hypothetical protein OJ1119_H02.10 [Oryza sativa] 142 6e-33
UniRef100_Q9M8K0 F28L1.12 protein [Arabidopsis thaliana] 116 4e-25
UniRef100_Q60ER4 Hypothetical protein OJ1764_D01.18 [Oryza sativa] 84 3e-15
UniRef100_UPI000033A117 UPI000033A117 UniRef100 entry 38 0.21
UniRef100_Q9SU89 Hypothetical protein T16L4.70 [Arabidopsis thal... 36 0.61
UniRef100_Q88TV5 Tyrosine--tRNA ligase [Lactobacillus plantarum] 35 1.0
UniRef100_UPI000026B0CB UPI000026B0CB UniRef100 entry 35 1.4
UniRef100_P21245 Peroxisomal membrane protein PMP47A [Candida bo... 34 2.3
UniRef100_UPI0000317AEA UPI0000317AEA UniRef100 entry 33 4.0
UniRef100_UPI0000315683 UPI0000315683 UniRef100 entry 33 4.0
UniRef100_UPI00002A8E9A UPI00002A8E9A UniRef100 entry 33 4.0
UniRef100_UPI0000299118 UPI0000299118 UniRef100 entry 33 4.0
UniRef100_UPI000028EDE7 UPI000028EDE7 UniRef100 entry 33 4.0
UniRef100_UPI000027E737 UPI000027E737 UniRef100 entry 33 4.0
UniRef100_UPI0000302FD8 UPI0000302FD8 UniRef100 entry 33 5.2
UniRef100_UPI00002CE91B UPI00002CE91B UniRef100 entry 33 5.2
>UniRef100_Q8VYH4 AT5g15260/F8M21_150 [Arabidopsis thaliana]
Length = 234
Score = 207 bits (528), Expect = 1e-52
Identities = 125/238 (52%), Positives = 155/238 (64%), Gaps = 25/238 (10%)
Query: 1 MVNFHSLISSVDQSSSSMANSIDL--------AKSRNRKTLNSLQIPQ----CERSRSAV 48
MV F + IS VDQS MA+S D S NRK+ +S C+ SRSA
Sbjct: 1 MVYFQNSISLVDQSPI-MAHSPDFHHHSKQSRTSSSNRKSSSSSCSSNFSVCCDGSRSAA 59
Query: 49 VDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVA--VAPVIYVSLGVSVS 106
+DVVI IAV+ + GFL+FPYI+F+ +S ++ LVKEE+ P++Y + +S+S
Sbjct: 60 IDVVILIAVITSSGFLIFPYIKFITIKSVEIFSELSCLVKEEILRNPDPIVYGLIALSIS 119
Query: 107 CAVVATWFFIAY--TSRKCSNPNCKGLKNA-AEFDIQLETEDCVKNSPSLGKDGGGIKKG 163
C ++ W + + +C PNCKGL+ A AEFDIQLETEDCVK+S S GK KKG
Sbjct: 120 CTALSAWMIVILLCSRHRCGKPNCKGLRKANAEFDIQLETEDCVKSSNS-GKSVS--KKG 176
Query: 164 LFKIPCDHHRELEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVPGPKK----NRKIKK 217
LF++P DHHRELEAELKKMAP NGRAVLV R KCGCSVGRLEVPGPKK RK+KK
Sbjct: 177 LFELPRDHHRELEAELKKMAPPNGRAVLVFRAKCGCSVGRLEVPGPKKQQLQQRKVKK 234
>UniRef100_Q6NLZ8 Hypothetical protein At3g01170 [Arabidopsis thaliana]
Length = 215
Score = 202 bits (515), Expect = 4e-51
Identities = 114/224 (50%), Positives = 146/224 (64%), Gaps = 16/224 (7%)
Query: 1 MVNFHSLISSVDQSSSSMANSIDLAKSRNRKTLNSL-----QIPQCERSRSAVVDVVIFI 55
MV HS IS V S M S KSRN + +S + P C+ S+SA +DVVI I
Sbjct: 1 MVYLHSSIS-VCNSVDPMIMSHSSPKSRNTRNKSSSSPTCSKFPVCDGSQSAAIDVVILI 59
Query: 56 AVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAPVIYVSLGVSVSCAVVATWFF 115
AV+ A GFL FPY++ + +S ++ LVK+E+ P++Y SL +S+ CA ++TW
Sbjct: 60 AVITACGFLFFPYVKLITLKSIEVFSDLSLLVKQEILQNPIVYGSLALSIFCAAISTWLV 119
Query: 116 IAY-TSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIPCDHHRE 174
I T ++C PNCKGL+ A EFDIQLETE+CVK+S + K+G+F++P HHRE
Sbjct: 120 ILLCTMQRCGKPNCKGLRKAVEFDIQLETEECVKSSSN--------KRGMFELPRVHHRE 171
Query: 175 LEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVPGPKK-NRKIKK 217
LEAELKKMAP NGRAVLV R +CGCSV RL V GPKK RKIKK
Sbjct: 172 LEAELKKMAPPNGRAVLVFRARCGCSVRRLVVSGPKKQQRKIKK 215
>UniRef100_Q9LXF6 Hypothetical protein F8M21_150 [Arabidopsis thaliana]
Length = 294
Score = 197 bits (501), Expect = 2e-49
Identities = 103/178 (57%), Positives = 129/178 (71%), Gaps = 8/178 (4%)
Query: 41 CERSRSAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVA--VAPVIY 98
C+ SRSA +DVVI IAV+ + GFL+FPYI+F+ +S ++ LVKEE+ P++Y
Sbjct: 36 CDGSRSAAIDVVILIAVITSSGFLIFPYIKFITIKSVEIFSELSCLVKEEILRNPDPIVY 95
Query: 99 VSLGVSVSCAVVATWFFIAY--TSRKCSNPNCKGLKNA-AEFDIQLETEDCVKNSPSLGK 155
+ +S+SC ++ W + + +C PNCKGL+ A AEFDIQLETEDCVK+S S GK
Sbjct: 96 GLIALSISCTALSAWMIVILLCSRHRCGKPNCKGLRKANAEFDIQLETEDCVKSSNS-GK 154
Query: 156 DGGGIKKGLFKIPCDHHRELEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVPGPKKNR 213
KKGLF++P DHHRELEAELKKMAP NGRAVLV R KCGCSVGRLEVPGPKK +
Sbjct: 155 SVS--KKGLFELPRDHHRELEAELKKMAPPNGRAVLVFRAKCGCSVGRLEVPGPKKQQ 210
>UniRef100_Q9MAC7 T4P13.14 protein [Arabidopsis thaliana]
Length = 596
Score = 174 bits (440), Expect = 2e-42
Identities = 97/202 (48%), Positives = 129/202 (63%), Gaps = 15/202 (7%)
Query: 1 MVNFHSLISSVDQSSSSMANSIDLAKSRNRKTLNSL-----QIPQCERSRSAVVDVVIFI 55
MV HS IS V S M S KSRN + +S + P C+ S+SA +DVVI I
Sbjct: 1 MVYLHSSIS-VCNSVDPMIMSHSSPKSRNTRNKSSSSPTCSKFPVCDGSQSAAIDVVILI 59
Query: 56 AVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAPVIYVSLGVSVSCAVVATWFF 115
AV+ A GFL FPY++ + +S ++ LVK+E+ P++Y SL +S+ CA ++TW
Sbjct: 60 AVITACGFLFFPYVKLITLKSIEVFSDLSLLVKQEILQNPIVYGSLALSIFCAAISTWLV 119
Query: 116 IAY-TSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIPCDHHRE 174
I T ++C PNCKGL+ A EFDIQLETE+CVK+S + K+G+F++P HHRE
Sbjct: 120 ILLCTMQRCGKPNCKGLRKAVEFDIQLETEECVKSSSN--------KRGMFELPRVHHRE 171
Query: 175 LEAELKKMAPVNGRAVLVLRGK 196
LEAELKKMAP NGRAVLV R +
Sbjct: 172 LEAELKKMAPPNGRAVLVFRAR 193
>UniRef100_Q6AVN3 Hypothetical protein OJ1119_H02.10 [Oryza sativa]
Length = 241
Score = 142 bits (358), Expect = 6e-33
Identities = 90/224 (40%), Positives = 118/224 (52%), Gaps = 14/224 (6%)
Query: 5 HSLISSVDQSSSSMANSIDLAKSRNRKTLNSLQIPQCERSRSAVVDVVIFIAVVIALGFL 64
H+ +S D ++ A + S R+ L C+ S A VD V +AVV AL FL
Sbjct: 21 HAAAASTDSAACGTAPARPRRGSSPRRGRRRLFTSLCDHSAMAAVDAVALLAVVSALAFL 80
Query: 65 VFPYIQFVMSESFKLCGLFMDLVKEEVA---VAPVIYVSLGVSVSC-AVVATWFFIAYTS 120
V PY++ V +E + GL DL VA AP G +++ A V W + + +
Sbjct: 81 VTPYVRMVAAE---VGGLVSDLDAAGVASSYYAPFAAAGAGAAIAAVAGVVAWDAVGHRA 137
Query: 121 RKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIPC-------DHHR 173
R+C P C+GL+ A EFDIQLETE+CV+ G + L D HR
Sbjct: 138 RRCGKPRCRGLRKAVEFDIQLETEECVRGQQQRLLPLPGGRAALLAAAGARPVQLGDAHR 197
Query: 174 ELEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVPGPKKNRKIKK 217
ELEAEL+KMAP NGR VL+ R CGC GR+EV G KK R+IKK
Sbjct: 198 ELEAELRKMAPPNGRTVLIFRSPCGCPKGRMEVWGAKKVRRIKK 241
>UniRef100_Q9M8K0 F28L1.12 protein [Arabidopsis thaliana]
Length = 241
Score = 116 bits (291), Expect = 4e-25
Identities = 79/221 (35%), Positives = 121/221 (54%), Gaps = 13/221 (5%)
Query: 5 HSLISSVDQSSSSMANSIDLAKSRNRKTLNSLQIPQCERSRSAVVDVVIFIAVVIALGFL 64
HS +S+ SSS+ ++ + + S + + +SL + C+ S SA +D++I I V+ + FL
Sbjct: 12 HSTTNSMPPSSSASRSASNHSSSSSSSS-SSLHL--CKHSPSATLDLLILILVLFSGAFL 68
Query: 65 VFPYIQFV------MSESF-KLCGLFMDLVKEEVAVAPVIYV-SLGVSVSCAVVATWFFI 116
+ Y ++ +S F L L ++ ++ P Y + V + ++
Sbjct: 69 LSSYFSYLFHSFSLLSSHFPSLSSLIFSDDEDLSSIPPASYFFAFAVFFAASIAFLDLCC 128
Query: 117 AYTSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIPCDHHRELE 176
SRKC NP CKGLK A EFD+QL+TE+CVK+ + D K G P + L
Sbjct: 129 GPRSRKCRNPKCKGLKKAMEFDLQLQTEECVKSGATKEIDRLPWKGGSESNP--DYECLR 186
Query: 177 AELKKMAPVNGRAVLVLRGKCGCSVGRLEVPGPKKNRKIKK 217
AEL++MAP NGRAVL+ R +CGC V +LE GPK+ R+ KK
Sbjct: 187 AELRRMAPPNGRAVLLFRSRCGCPVAKLEGWGPKRGRRHKK 227
>UniRef100_Q60ER4 Hypothetical protein OJ1764_D01.18 [Oryza sativa]
Length = 255
Score = 83.6 bits (205), Expect = 3e-15
Identities = 59/184 (32%), Positives = 89/184 (48%), Gaps = 17/184 (9%)
Query: 46 SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAPVIYVSLGVSV 105
SA +D++I + V+ ++ FL+ + V L L +A A LG +
Sbjct: 68 SATLDLLILLLVLFSVAFLLASSLAHVSRSLTPL--LASPPAAAALASAAAAMPYLGAAA 125
Query: 106 SCAVVATWFFIAYTSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLF 165
+ A R+C NP C+GL A EFD+QL+TE+ V+ G GG ++
Sbjct: 126 ALAGATFLSCSRLPRRRCRNPRCRGLVKALEFDVQLQTEEAVR--AGTGSTSGGADAAMW 183
Query: 166 K----IPCD--------HHRELEAELKKMAPVNGRAVLVLRGKCGCSVGRLEVPG-PKKN 212
+ +P + L AEL++MAP NGRAVL+ R +CGC + +LE G PK
Sbjct: 184 REIEALPWKGGQGGNNPDYECLRAELRRMAPPNGRAVLLFRNRCGCPIAKLEGWGVPKSK 243
Query: 213 RKIK 216
R+ K
Sbjct: 244 RRSK 247
>UniRef100_UPI000033A117 UPI000033A117 UniRef100 entry
Length = 249
Score = 37.7 bits (86), Expect = 0.21
Identities = 20/73 (27%), Positives = 43/73 (58%), Gaps = 9/73 (12%)
Query: 51 VVIFIAVV----IALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAPVIYVSLGVSVS 106
+ IF+A++ +++G FP+I +M +F + GL +K+++ + P+ V+L + +
Sbjct: 73 IAIFLAIIAIILLSVGIESFPFISLIMGGTFAIYGL----IKKKIELDPLYSVTLEILLL 128
Query: 107 CAVVATW-FFIAY 118
C + + FFI+Y
Sbjct: 129 CPIALGFIFFISY 141
>UniRef100_Q9SU89 Hypothetical protein T16L4.70 [Arabidopsis thaliana]
Length = 493
Score = 36.2 bits (82), Expect = 0.61
Identities = 23/71 (32%), Positives = 38/71 (53%), Gaps = 6/71 (8%)
Query: 11 VDQSSSSMANSIDLAKSRNRKTLNSLQIPQCERSRSAVVDVVIFIAVV------IALGFL 64
+DQ+S+ ++S A NR + SL QC+++ S V V+FI + L FL
Sbjct: 25 LDQASNLFSSSSSAAAPINRSSFVSLLKKQCDQNSSPVTKKVLFIETLPNMVQSKILSFL 84
Query: 65 VFPYIQFVMSE 75
+F Y +F +S+
Sbjct: 85 LFEYQRFCVSD 95
>UniRef100_Q88TV5 Tyrosine--tRNA ligase [Lactobacillus plantarum]
Length = 418
Score = 35.4 bits (80), Expect = 1.0
Identities = 23/75 (30%), Positives = 34/75 (44%), Gaps = 7/75 (9%)
Query: 129 KGLKNAAEFDIQLETE-------DCVKNSPSLGKDGGGIKKGLFKIPCDHHRELEAELKK 181
+G K D+ E + D K PS + IK G +I + ++EAE+
Sbjct: 335 QGFKGVPSADVSAEKQNIVLWLVDATKFEPSRRQAREDIKNGAIRINGEKVTDVEAEIDP 394
Query: 182 MAPVNGRAVLVLRGK 196
A NG+ V+V RGK
Sbjct: 395 SAAFNGKFVIVRRGK 409
>UniRef100_UPI000026B0CB UPI000026B0CB UniRef100 entry
Length = 284
Score = 35.0 bits (79), Expect = 1.4
Identities = 26/92 (28%), Positives = 46/92 (49%), Gaps = 4/92 (4%)
Query: 26 KSRNRKTLNSLQIPQCERSRSAVVDVVIFIAVVIALGFLVFPYIQFVMSE-SFKLCGLFM 84
K+ N K L ++++ S ++ D +I+I ++ +G V + V S KL +
Sbjct: 134 KNHNYKFLKNIKLEN-SISNPSIKDPIIYILIISLIGGFVLNLMPCVFPVLSIKLMSVLS 192
Query: 85 DLVKEEVAVAPVIYVSLGVSVSCAVVATWFFI 116
+ K+ IY SLG+ S ++AT+FFI
Sbjct: 193 N--KQGTIRLSFIYTSLGILFSFLILATFFFI 222
>UniRef100_P21245 Peroxisomal membrane protein PMP47A [Candida boidinii]
Length = 423
Score = 34.3 bits (77), Expect = 2.3
Identities = 32/117 (27%), Positives = 55/117 (46%), Gaps = 10/117 (8%)
Query: 53 IFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDLVKEEVAVAPVIYVSLGV-SVSCAVVA 111
+F +V AL ++ P IQ+ + E K F+ +K+ + PV + LG A +
Sbjct: 203 LFTGIVPALFLVLNPIIQYTIFEQLKS---FIVKIKKR-NITPVDALLLGAFGKLIATII 258
Query: 112 TWFFIAYTSRKCSNPNCKGLKNAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIP 168
T+ +I SR + K + +E D++ E D V++ P G D +K+ K P
Sbjct: 259 TYPYITLRSRM----HVKSMTEISE-DVEKERTDSVQSLPEDGSDEDNLKENSAKSP 310
>UniRef100_UPI0000317AEA UPI0000317AEA UniRef100 entry
Length = 198
Score = 33.5 bits (75), Expect = 4.0
Identities = 30/95 (31%), Positives = 43/95 (44%), Gaps = 14/95 (14%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S V+D F+ + GFLVF + S K +F DL
Sbjct: 58 SLNPLSDFQCSTDKSRGLLSRVIDTTNFVTTWVISGFLVFEIFMYFTSLDLK---IFFDL 114
Query: 87 VKEEVAVAPVIYVSLGVSVSC---AVVATWFFIAY 118
+ P+I + G C VVAT++ Y
Sbjct: 115 ---WLPFVPMIAILFGFLPGCGPQVVVATFYLNGY 146
>UniRef100_UPI0000315683 UPI0000315683 UniRef100 entry
Length = 353
Score = 33.5 bits (75), Expect = 4.0
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 8/92 (8%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S VVD F+ + GFLVF + S K +F DL
Sbjct: 243 SLNPLSDFQCSTDKSRGLLSRVVDTTNFVTTWVISGFLVFEIFMYFTSLDLK---IFFDL 299
Query: 87 VKEEVAVAPVIYVSLGVSVSCAVVATWFFIAY 118
V + +++ L VVAT++ Y
Sbjct: 300 WLPFVPLVAILFGFLPGCGPQVVVATFYLNGY 331
>UniRef100_UPI00002A8E9A UPI00002A8E9A UniRef100 entry
Length = 356
Score = 33.5 bits (75), Expect = 4.0
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 8/92 (8%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S VVD F+ + GFLVF + S K +F DL
Sbjct: 243 SLNPLSDFQCSTDKSRGLLSRVVDTTNFVTTWVISGFLVFEIFMYFTSLDLK---IFFDL 299
Query: 87 VKEEVAVAPVIYVSLGVSVSCAVVATWFFIAY 118
V + +++ L VVAT++ Y
Sbjct: 300 WLPFVPLVAILFGFLPGCGPQVVVATFYLNGY 331
>UniRef100_UPI0000299118 UPI0000299118 UniRef100 entry
Length = 382
Score = 33.5 bits (75), Expect = 4.0
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 8/92 (8%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S VVD F+ + GFLVF + S K +F DL
Sbjct: 242 SLNPLSDFQCSTDKSRGLLSRVVDTTNFVTTWVISGFLVFEIFMYFTSLDLK---IFFDL 298
Query: 87 VKEEVAVAPVIYVSLGVSVSCAVVATWFFIAY 118
V + +++ L VVAT++ Y
Sbjct: 299 WLPFVPLVAILFGFLPGCGPQVVVATFYLNGY 330
>UniRef100_UPI000028EDE7 UPI000028EDE7 UniRef100 entry
Length = 144
Score = 33.5 bits (75), Expect = 4.0
Identities = 31/95 (32%), Positives = 43/95 (44%), Gaps = 14/95 (14%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S VVD F+ + GFLVF + + K LF DL
Sbjct: 3 SLNPLSDFQCSTDKSRGFVSRVVDTTNFVTTWVISGFLVFEIFMYFTNIDLK---LFFDL 59
Query: 87 VKEEVAVAPVIYVSLGVSVSCA---VVATWFFIAY 118
+ P+I + G C VVAT++ Y
Sbjct: 60 ---WLPFVPLIAILFGFLPGCGPQIVVATFYLNGY 91
>UniRef100_UPI000027E737 UPI000027E737 UniRef100 entry
Length = 297
Score = 33.5 bits (75), Expect = 4.0
Identities = 29/92 (31%), Positives = 42/92 (45%), Gaps = 8/92 (8%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S VVD F+ + GFLVF + S K +F DL
Sbjct: 157 SLNPLSDFQCSTDKSRGFLSRVVDTTNFVTTWVISGFLVFEIFMYFTSLDLK---IFFDL 213
Query: 87 VKEEVAVAPVIYVSLGVSVSCAVVATWFFIAY 118
V + +++ L VVAT++ Y
Sbjct: 214 WLPFVPLVAILFGFLPGCGPQVVVATFYLNGY 245
>UniRef100_UPI0000302FD8 UPI0000302FD8 UniRef100 entry
Length = 200
Score = 33.1 bits (74), Expect = 5.2
Identities = 30/91 (32%), Positives = 43/91 (46%), Gaps = 14/91 (15%)
Query: 32 TLNSLQIPQC--ERSR---SAVVDVVIFIAVVIALGFLVFPYIQFVMSESFKLCGLFMDL 86
+LN L QC ++SR S VVD F++ + GFLVF F S K +F D+
Sbjct: 60 SLNPLSDFQCSTDKSRGFLSRVVDTTNFVSTWVICGFLVFEIFMFFSSIDLK---VFFDI 116
Query: 87 VKEEVAVAPVIYVSLGVSVSC---AVVATWF 114
+ P+I + G C VVAT++
Sbjct: 117 ---WLPFVPLIAIFFGFLPGCGPQVVVATFY 144
>UniRef100_UPI00002CE91B UPI00002CE91B UniRef100 entry
Length = 258
Score = 33.1 bits (74), Expect = 5.2
Identities = 24/75 (32%), Positives = 32/75 (42%), Gaps = 3/75 (4%)
Query: 133 NAAEFDIQLETEDCVKNSPSLGKDGGGIKKGLFKIPCDHHRELEAELKKMAPVNGRAVLV 192
N D++ D +N S G G G + G+ + P HR AE K+ RA
Sbjct: 31 NKRRSDVEPAARDGERNKASRGDAGRGTRGGIVRPPSHTHR---AEAKRAKRGGERATQR 87
Query: 193 LRGKCGCSVGRLEVP 207
R + VGR EVP
Sbjct: 88 PRRRIARHVGREEVP 102
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 329,408,018
Number of Sequences: 2790947
Number of extensions: 12344935
Number of successful extensions: 28186
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 30
Number of HSP's that attempted gapping in prelim test: 28161
Number of HSP's gapped (non-prelim): 38
length of query: 217
length of database: 848,049,833
effective HSP length: 122
effective length of query: 95
effective length of database: 507,554,299
effective search space: 48217658405
effective search space used: 48217658405
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC140025.11


