

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.11 + phase: 0
(381 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
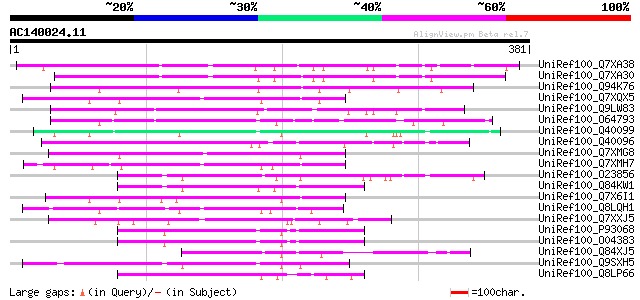
UniRef100_Q7XA38 Putative receptor protein kinase [Solanum bulbo... 188 2e-46
UniRef100_Q7XA30 Putative receptor protein kinase [Solanum bulbo... 178 2e-43
UniRef100_Q94K76 Hypothetical protein At5g18470 [Arabidopsis tha... 135 2e-30
UniRef100_Q7XQX5 OSJNBb0108J11.18 protein [Oryza sativa] 102 2e-20
UniRef100_Q9LW83 Receptor kinase 1 [Arabidopsis thaliana] 100 5e-20
UniRef100_O64793 T1F15.1 protein [Arabidopsis thaliana] 98 3e-19
UniRef100_Q40099 Secreted glycoprotein 2 [Ipomoea trifida] 94 6e-18
UniRef100_Q40096 Receptor protein kinase [Ipomoea trifida] 89 2e-16
UniRef100_Q7XMG8 OSJNBa0028I23.14 protein [Oryza sativa] 85 3e-15
UniRef100_Q7XMH7 OSJNBa0028I23.5 protein [Oryza sativa] 85 4e-15
UniRef100_O23856 S glycoprotein [Brassica campestris] 83 1e-14
UniRef100_Q84KW1 S-locus receptor kinase [Brassica oleracea] 82 2e-14
UniRef100_Q7X6I1 OSJNBb0108J11.20 protein [Oryza sativa] 82 2e-14
UniRef100_Q8LQH1 Putative S-receptor kinase [Oryza sativa] 82 2e-14
UniRef100_Q7XXJ5 OSJNBa0094O15.5 protein [Oryza sativa] 82 3e-14
UniRef100_P93068 Receptor-like kinase [Brassica oleracea] 80 7e-14
UniRef100_O04383 Serine /threonine kinase precursor [Brassica ol... 80 7e-14
UniRef100_Q84XJ5 S-receptor kinase 13-8 [Arabidopsis lyrata] 80 7e-14
UniRef100_Q9SXH5 SRK13 [Brassica oleracea] 80 9e-14
UniRef100_Q8LP66 S-locus-related I [Brassica amplexicaulis] 79 2e-13
>UniRef100_Q7XA38 Putative receptor protein kinase [Solanum bulbocastanum]
Length = 761
Score = 188 bits (477), Expect = 2e-46
Identities = 142/396 (35%), Positives = 208/396 (51%), Gaps = 36/396 (9%)
Query: 6 IKKQVVLIYLWLWWNTTA--NICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNS 63
+ + ++ L L+++ T N+ A SLKPGD+LN+ L S+ GKF L F + S +
Sbjct: 4 LANSIFILILTLFFSHTCIINLLSVAAITSLKPGDELNHSQVLDSEGGKFKLGFFSISQT 63
Query: 64 DFQCLFISVNAD-YGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQP 122
+ L I D K +W+ + N + N+ +L++D +G LKI S K ++ + P
Sbjct: 64 NNYYLGIWYAGDPQDKKLWIANPNTPLLNNSGLLTIDTTGTLKITSGG-KTVVNITPPLL 122
Query: 123 TNNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSL--- 179
T +++A + +GN VLQ N + LWQSFD+P++ L P MKLG N T NW+L
Sbjct: 123 TRSSIARLQGSGNLVLQDETQNRT---LWQSFDHPTNTLFPGMKLGYNLTTKQNWTLTSW 179
Query: 180 ------VSDKFNLEWEPKQG--ELNIKKSGKVYWKSGKLKSNGLFENIPA--NVQSRYQY 229
S F L E Q +L I++ G+VYW SG + N F + A + +RYQY
Sbjct: 180 LSSYIPASGAFTLSLESIQDAFQLVIRRRGEVYWISGAWR-NQSFPLLTALHDSSNRYQY 238
Query: 230 --IIVSNKDEDSFTFEVKDGKFAQWELSSKGKLV--GDDG--YIANADMCYGYNSDGGCQ 283
+VS KD F F+ DG F EL+ G +V G+D Y + CYGY S GC
Sbjct: 239 NLNLVSEKDGVFFQFDAPDGSFPSLELNFNGAIVGGGEDSRVYALYNEFCYGYESQDGCV 298
Query: 284 KWEDIPTCREPGEMFQKKAGRPSID---NSTTYEFDVTYSYSDCKIRCWKNCSCNGFQLY 340
+P CR+ G+ F++K+G ID NS +Y+ + + S DC RCW++CSC GF
Sbjct: 299 S-NQLPECRKDGDKFEQKSG-DFIDRSKNSNSYD-NASTSLGDCMKRCWEHCSCVGFTT- 354
Query: 341 YSNMTGCVFLSWNSTQYVDMVPD--KFYTLVKTTKS 374
SN TGC+ + N VD + K Y LV + S
Sbjct: 355 TSNGTGCIIWNGNGEFQVDESGNTVKKYVLVSSKSS 390
>UniRef100_Q7XA30 Putative receptor protein kinase [Solanum bulbocastanum]
Length = 711
Score = 178 bits (452), Expect = 2e-43
Identities = 125/353 (35%), Positives = 180/353 (50%), Gaps = 29/353 (8%)
Query: 34 LKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNAD-YGKVVWVYDINHSIDFN 92
+KPGD LN+ L S+ G+F L F + ++ L I D K +W+ + N I N
Sbjct: 1 MKPGDVLNHSQVLDSEGGRFKLGFFSIPQTNKTYLGIWYAGDPVEKKLWIANPNTPILNN 60
Query: 93 TSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLPNGSMSVLWQ 152
+ +L+LD +G L+I S K ++ ++P T + +A + D+GNFV+Q N + LWQ
Sbjct: 61 SGLLTLDSTGALRITSGG-KTVVNIATPLLTGSLIARLQDSGNFVVQDETRNRT---LWQ 116
Query: 153 SFDYPSDVLIPMMKLGVNRKTGHNWSLVS----------DKFNLEWEPKQG--ELNIKKS 200
SFD+P+ L+P MKLG N T NW+L S F L E Q +L + +
Sbjct: 117 SFDHPTSCLLPGMKLGYNLTTRQNWTLTSWLVSSAVPAPGAFTLSLEAIQDAFQLVVSRR 176
Query: 201 GKVYWKSGKLKSNGLFENIPA--NVQSRYQY--IIVSNKDEDSFTFEVKDGKFAQWELSS 256
G+VYW SG + G F +P+ + + YQY +VS D F FE G F EL S
Sbjct: 177 GEVYWTSGAWNNQG-FPFLPSFRDSATTYQYNLNLVSGTDGMFFQFEATKGSFPSLELFS 235
Query: 257 KGKLVGDDG--YIANADMCYGYNSDGGCQKWEDIPTCREPGEMFQKKAGRPSID---NST 311
G + DG Y CYGY D GC +P CR+ G+ F++K G ID +T
Sbjct: 236 DGAIAAGDGSIYTRYNKFCYGYGGDDGCVS-SQLPECRKDGDKFEQKRG-DFIDLSGTTT 293
Query: 312 TYEFDVTYSYSDCKIRCWKNCSCNGFQLYYSNMTGCVFLSWNSTQYVDMVPDK 364
+Y + + S DC +CW++CSC GF SN TGC+ + VD +K
Sbjct: 294 SYYDNASISLGDCMQKCWEHCSCVGFTTLNSNGTGCLISNGKRDFRVDESGEK 346
>UniRef100_Q94K76 Hypothetical protein At5g18470 [Arabidopsis thaliana]
Length = 413
Score = 135 bits (339), Expect = 2e-30
Identities = 104/353 (29%), Positives = 161/353 (45%), Gaps = 44/353 (12%)
Query: 31 SDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSD--------FQCLFISVNADYGKVVWV 82
+D+LKPG +L +L S G F L F +S + L I + +VWV
Sbjct: 30 TDTLKPGQQLRDWEQLISADGIFTLGFFTPKDSSTSELGSAGLRYLGIWPQSIPINLVWV 89
Query: 83 YDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQP----TNNTVATMLDAGNFVL 138
+ S+ ++ LS+D +GVLKI N PI++ P N A +LD GNFV+
Sbjct: 90 GNPTESVSDSSGSLSIDTNGVLKITQANAIPILVNQRPAAQLSLVGNVSAILLDTGNFVV 149
Query: 139 QQFLPNGSMS-VLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS---------DKFNLEW 188
++ P G VLWQSFD+P++ L+P MK+G N +T S+ S F L
Sbjct: 150 REIRPGGVPGRVLWQSFDHPTNTLLPGMKIGFNLRTKKEVSVTSWITDQVPVPGAFRLGL 209
Query: 189 EPK-QGELNIKKSGKVYWKSGKLKSNGLFENIPANVQSR---YQYIIVSNKDEDSFTFEV 244
+P +L + + G++YW SG L +NG ++ V Y++ SNK F++ +
Sbjct: 210 DPSGANQLLVWRRGEIYWSSGILTNNG-SSHLNLEVSRHYIDYEFKFDSNKYMKYFSYSI 268
Query: 245 KDGK---FAQWELSSKGKLVGDDGYIANADMCYGYNSDGGCQK----------WEDIPTC 291
K F+ W L + G++ +N + S C+ E C
Sbjct: 269 KKANSSVFSSWFLDTLGQITVTFSLSSNNSSTWISESSEPCKTDLKNSSAICITEKPTAC 328
Query: 292 REPGEMFQKKAGRPSIDNSTTYEF----DVTYSYSDCKIRCWKNCSCNGFQLY 340
R+ E F+ + G +N+ Y F ++ SDC CW+NCSC FQ +
Sbjct: 329 RKGSEYFEPRRGYMMENNTGYYPFYYDDSLSAGLSDCHGTCWRNCSCIAFQAF 381
>UniRef100_Q7XQX5 OSJNBb0108J11.18 protein [Oryza sativa]
Length = 807
Score = 102 bits (254), Expect = 2e-20
Identities = 80/263 (30%), Positives = 125/263 (47%), Gaps = 29/263 (11%)
Query: 10 VVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQF---GNNSNSDFQ 66
++L+ L L + + A +D+L PG + +L S GKF L F G+ S+ +
Sbjct: 3 ILLVILGLHLCSLHLPAISAAADTLSPGQSIAGDDRLVSSNGKFALGFFNTGSKSSGNDT 62
Query: 67 CLFISVNADYGKV-----VWVYDINHSI-DFNTSVLSLDYSGVLKIESQNRKPIIIYSSP 120
+ + + KV VW+ + + D +S L++ G L I S+ I+ S
Sbjct: 63 LSYWYLGIWFNKVPNKTHVWIANRGSPVTDATSSHLTISPDGNLAIVSRADSSIVWSSQA 122
Query: 121 QPT-NNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSL 179
T NNTVA +LD GN VLQ + S +LW+SFD+P+DV +P K+G+N+ TG N +
Sbjct: 123 NITSNNTVAVLLDTGNLVLQS--SSNSSHILWESFDHPTDVFLPSAKIGLNKITGLNRRI 180
Query: 180 VSDK---------FNLEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQSR---- 226
S + +++E+ PK G + S YW SG+ F IP V
Sbjct: 181 FSRRDLVDQSPSVYSMEFGPKGGYQLVWNSSVEYWSSGEWNGR-YFSRIPEMVVKSPHYT 239
Query: 227 ---YQYIIVSNKDEDSFTFEVKD 246
+Q V+N E FT+ + D
Sbjct: 240 PFIFQIEYVNNDQEVYFTYRIHD 262
>UniRef100_Q9LW83 Receptor kinase 1 [Arabidopsis thaliana]
Length = 805
Score = 100 bits (250), Expect = 5e-20
Identities = 102/371 (27%), Positives = 151/371 (40%), Gaps = 73/371 (19%)
Query: 31 SDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNADY------------GK 78
+D+L G L +L S F L+F N NS L I N Y K
Sbjct: 24 TDTLLQGQYLKDGQELVSAFNIFKLKFFNFENSSNWYLGIWYNNFYLSGGNKKYGDIKDK 83
Query: 79 VVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVL 138
VW+ + N+ + + L++D G L+I + ++ SS + T NT +LD+GN L
Sbjct: 84 AVWIANRNNPVLGRSGSLTVDSLGRLRI-LRGASSLLELSSTETTGNTTLKLLDSGNLQL 142
Query: 139 QQFLPNGSMS-VLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLV-----------SDKFNL 186
Q+ +GSM LWQSFDYP+D L+P MKLG N KTG W L S F +
Sbjct: 143 QEMDSDGSMKRTLWQSFDYPTDTLLPGMKLGFNVKTGKRWELTSWLGDTLPASGSFVFGM 202
Query: 187 EWEPKQGELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQYIIVSNKDEDSFTFEVK 245
+ + L I G VYW SG G E + N + + VS + E F +
Sbjct: 203 D-DNITNRLTILWLGNVYWASGLWFKGGFSLEKLNTN---GFIFSFVSTESEHYFMYSGD 258
Query: 246 DG----KFAQWELSSKGKL--VGDDG---YIANADMCYGYNSDGGC-------------- 282
+ F + + +G L + DG ++ + +G + GC
Sbjct: 259 ENYGGPLFPRIRIDQQGSLQKINLDGVKKHVHCSPSVFGEELEYGCYQQNFRNCVPARYK 318
Query: 283 ------------------QKWEDIPTCREPGEMFQKKAGRPSIDNSTTY-EFDVTYSYSD 323
+K D+ C G F++ PS +N + E S D
Sbjct: 319 EVTGSWDCSPFGFGYTYTRKTYDLSYCSRFGYTFRETVS-PSAENGFVFNEIGRRLSSYD 377
Query: 324 CKIRCWKNCSC 334
C ++C +NCSC
Sbjct: 378 CYVKCLQNCSC 388
>UniRef100_O64793 T1F15.1 protein [Arabidopsis thaliana]
Length = 833
Score = 98.2 bits (243), Expect = 3e-19
Identities = 99/351 (28%), Positives = 152/351 (43%), Gaps = 43/351 (12%)
Query: 31 SDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSD-------FQCLFISVNADYGKVVWVY 83
+D+L G L +L S F L+F N NS+ F L+++ ++ + VW+
Sbjct: 24 TDTLHQGQFLKDGQELVSAFKIFKLKFFNFKNSENLYLGIWFNNLYLNTDSQ-DRPVWIA 82
Query: 84 DINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFLP 143
+ N+ I + L++D G LKI + ++ SS + T NT +LD+GN LQ+
Sbjct: 83 NRNNPISDRSGSLTVDSLGRLKI-LRGASTMLELSSIETTRNTTLQLLDSGNLQLQEMDA 141
Query: 144 NGSMS-VLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGE-------- 194
+GSM VLWQSFDYP+D L+P MKLG + KT W L S + + P G
Sbjct: 142 DGSMKRVLWQSFDYPTDTLLPGMKLGFDGKTRKRWELTS--WLGDTLPASGSFVFGMDTN 199
Query: 195 ----LNIKKSGKVYWKSGKLKSNGLFENIPANVQSRYQYIIVSNKDEDSFTFEVKDGKFA 250
L I G +YW SG L + G F N + + + VS K F + G
Sbjct: 200 ITNVLTILWRGNMYWSSG-LWNKGRFSEEELN-ECGFLFSFVSTKSGQYFMY---SGDQD 254
Query: 251 QWELSSKGKLVGDDGYIANADMCYGYNSDGGCQKWEDIPTCREPG-----EMFQKKAGRP 305
++ + G + M N + C G E + + R
Sbjct: 255 DARTFFPTIMIDEQGILRREQMHRQRNRQNYRNR-----NCLAAGYVVRDEPYGFTSFRV 309
Query: 306 SIDNSTTYEFDV--TYSYSDCKIRCWKNCSCNGFQLYYSNMTGCVFLSWNS 354
++ +S + F + T+S DC C +N SC + + TGC WN+
Sbjct: 310 TVSSSASNGFVLSGTFSSVDCSAICLQNSSCLAYASTEPDGTGCEI--WNT 358
>UniRef100_Q40099 Secreted glycoprotein 2 [Ipomoea trifida]
Length = 451
Score = 94.0 bits (232), Expect = 6e-18
Identities = 99/408 (24%), Positives = 157/408 (38%), Gaps = 72/408 (17%)
Query: 18 WWNTTANICVEATS---DSLKPGDKLNYKSKLCSKQGKFCLQF---GNNSNSDFQCLFIS 71
W+ I + T+ D++ P L L S G F L F G NS + ++
Sbjct: 13 WYFLILQILIPTTAIAVDTITPTQPLTQNQTLVSAGGVFQLGFFSPGGNSGGLYVGIWYK 72
Query: 72 VNADYGKVVWVYDINHSIDFN-TSVLSLDYSGVLKIESQNRKPIIIYSSPQPT--NNTVA 128
D +VWV + + + N T L + G + + Q I S+ + NTVA
Sbjct: 73 EIQDR-TIVWVANRDKPLRNNSTGFLKIGEDGNIHLVDQTENSIWSSSNSNQSVPENTVA 131
Query: 129 TMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEW 188
+LD+GN VL++ + LWQ FDYP+D L+P MKLG + KTG N + S +
Sbjct: 132 QLLDSGNLVLRRENDENPENYLWQGFDYPTDTLLPGMKLGWDSKTGRNRYISS--WKTPT 189
Query: 189 EPKQGELNIK------------KSGKVYWKSGKLKSNGLFENIPANVQSRYQYIIVSNKD 236
+P +G++ K K + +SG G + + +V K
Sbjct: 190 DPSEGDITFKLDINGLPEAFLRKKDNIITRSGGWNGIGFSGVTEMQTKEVIDFSLVMTKH 249
Query: 237 EDSFTFEVKDGKFAQWELSSKGKL-----------VGDDGYIANADMCYGYNSDG----- 280
E +TFE+++ +++ ++ + + + A D C Y G
Sbjct: 250 EVYYTFEIRNKTLLSRLVANYTEILERYTWVPENRIWNRFWYAPKDQCDNYGECGTYGIC 309
Query: 281 --------GC---------QKW-----------EDIPTCREPGEMFQKKAGRPSIDNSTT 312
GC Q W D C G + P S+T
Sbjct: 310 DTDKSPVCGCLVGFEPRKQQAWSLRDGSGGCFRHDQLDCETDGFLTMNNMKLP---ESST 366
Query: 313 YEFDVTYSYSDCKIRCWKNCSCNGFQLYYSNMTGCVFLSWNSTQYVDM 360
DVT S +CK C +NCSC + Y + G + W + + +DM
Sbjct: 367 SFVDVTMSLDECKEMCVRNCSCTAYSNYNISNGGSGCVIW-TAELLDM 413
>UniRef100_Q40096 Receptor protein kinase [Ipomoea trifida]
Length = 853
Score = 89.0 bits (219), Expect = 2e-16
Identities = 94/376 (25%), Positives = 151/376 (40%), Gaps = 72/376 (19%)
Query: 24 NICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLFISVNADYGK-VVWV 82
N+ V DS+ P L L S G F L F + SD + I K VVWV
Sbjct: 24 NLAVALAVDSITPTQPLAGNRTLVSSDGLFELGFFTPNGSDQSYVGIWYKEIEPKTVVWV 83
Query: 83 YDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQQFL 142
+ + + + +L + G + + I ++ NTVA +LD+GNFVL++
Sbjct: 84 GNRDGASRGSAGILKIGEDGNIHLVDGGGNFIWSPTNQSAARNTVAQLLDSGNFVLRRED 143
Query: 143 PNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHN-----WSLVSD--------KFNLEWE 189
+ LWQSFDYP+D L+P MKLG + KTG N W ++D K ++
Sbjct: 144 DENPENYLWQSFDYPTDTLLPGMKLGWDSKTGLNRYISAWKSLNDPGEGPISFKLDINGL 203
Query: 190 PKQGELNIKKSGKVYWKSGKLKSNGL-FENIP-ANVQSRYQYIIVSNKDEDSFTFEV--- 244
P E+ ++ K+ ++SG NG+ F +P + + V K+E ++FE+
Sbjct: 204 P---EIFLRNRDKIVYRSGPW--NGVRFSGVPEMKPTATITFSFVMTKNERYYSFELHNK 258
Query: 245 ---------KDGKFAQWELSSKGKLVGDDGYIANADMCYGYNSDG--------------- 280
++G ++ K+ Y A D C Y G
Sbjct: 259 TLYSRLLVTRNGNLERYAWIPTSKIWSKFWY-APKDQCDSYKECGTFGFCDTNMSPVCQC 317
Query: 281 -------------------GCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFDVTYSY 321
GC ++ ++ CR+ G + P D S+++ D T +
Sbjct: 318 LVGFRPKSPQAWDLRDGSDGCVRYHEL-ECRKDGFLTMNFMKLP--DTSSSF-VDTTMNL 373
Query: 322 SDCKIRCWKNCSCNGF 337
+C C NCSC +
Sbjct: 374 DECMKMCKNNCSCTAY 389
>UniRef100_Q7XMG8 OSJNBa0028I23.14 protein [Oryza sativa]
Length = 802
Score = 85.1 bits (209), Expect = 3e-15
Identities = 76/238 (31%), Positives = 111/238 (45%), Gaps = 22/238 (9%)
Query: 29 ATSDSLKPGDKLNYKSKLCSKQGKFCLQF-GNNSNSDFQCLFISVNADYGKV-----VWV 82
A D++ P L KL S GKF L F S S L + Y K+ VWV
Sbjct: 22 AAMDTMTPAQALFGNGKLISSNGKFALGFFQTGSKSSHNTLNWYLGIWYNKIPKLTPVWV 81
Query: 83 YDINHSI-DFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQ-PTNNTVATMLDAGNFVLQQ 140
+ ++ + D N S L++ G L I ++ + I+ + TN+TVA +L++GN VLQ
Sbjct: 82 ANGDNPVTDPNNSELTISGDGGLVILDRSNRSIVWSTRINITTNDTVAMLLNSGNLVLQN 141
Query: 141 FLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNIKKS 200
FL S LWQSFDYP+ +P KLG ++ +G N LVS K +++ P + + + S
Sbjct: 142 FL--NSSDALWQSFDYPTHTFLPGAKLGWSKISGLNSRLVSRKNSIDLAPGKYSVELDPS 199
Query: 201 GKVYWKSGKLKS----------NG-LFENIPANV-QSRYQYIIVSNKDEDSFTFEVKD 246
G + L S NG F +IP + V N E FT+ + D
Sbjct: 200 GANQYIFTLLNSSTPYLTSGVWNGQYFPSIPEMAGPFIVNFTFVDNDQEKYFTYSLLD 257
>UniRef100_Q7XMH7 OSJNBa0028I23.5 protein [Oryza sativa]
Length = 677
Score = 84.7 bits (208), Expect = 4e-15
Identities = 83/262 (31%), Positives = 125/262 (47%), Gaps = 34/262 (12%)
Query: 11 VLIYLWLWWNTTANICVEATS---DSLKPGDKLNYKSKLCSKQGKFCLQFGN------NS 61
+LIY+ L ++ +C+ A + D++ G+ L K KL SK G++ L F +
Sbjct: 3 LLIYVVLLFS----LCISANAAMTDTISMGNALGRKDKLVSKNGRYALGFFETERVEVSQ 58
Query: 62 NSDFQCLFISVNADYGKVV--WVYDINHSIDFNTSV-LSLDYSGVLKIESQNRKPIIIYS 118
S L I N K+ WV + ++ I+ TS+ L++ + G L I +++ K II S
Sbjct: 59 KSSKWYLGIWFN-QVPKITPAWVANRDNPINDPTSLELTIFHDGNLVILNRSAKTIIWSS 117
Query: 119 SPQPTNN-TVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNW 177
TNN T A +L +GN +L P+ S VLWQSFDYP+D L P KLG ++ TG N
Sbjct: 118 QANITNNNTSAMLLSSGNLILTN--PSNSSEVLWQSFDYPTDTLFPRAKLGWDKVTGLNR 175
Query: 178 SLVSDK---------FNLEWEPK---QGELNIKKSGKVYWKSGKLKSNGLFENIPANV-Q 224
++S K + E +P Q L S YW SG + F +P
Sbjct: 176 RIISWKNSKDLAAGVYCKELDPSGVDQSLLTPLNSFTPYWSSGPWNGD-YFAAVPEMASH 234
Query: 225 SRYQYIIVSNKDEDSFTFEVKD 246
+ + V N E FT+ + D
Sbjct: 235 TVFNSTFVHNDQERYFTYTLVD 256
>UniRef100_O23856 S glycoprotein [Brassica campestris]
Length = 430
Score = 83.2 bits (204), Expect = 1e-14
Identities = 87/334 (26%), Positives = 140/334 (41%), Gaps = 74/334 (22%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN-----TVATMLDAG 134
VWV + ++ + + L + ++ ++ N+ ++S+ + N VA +L G
Sbjct: 76 VWVANRDNPLSSSIGTLKISNMNLVLLDHSNKS---VWSTNRTRGNERSSPVVAELLANG 132
Query: 135 NFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGE 194
NFV++ + NG+ LWQSFDYP+D L+P MKLG + KTG N L S K + +P G+
Sbjct: 133 NFVMRDYNNNGASGFLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTSWKSS--DDPSSGD 190
Query: 195 ----LNIKKSGKVYWKSGKLKS------NGL-FENIPANVQSRYQ-YIIVSNKDEDSFTF 242
L ++ + Y SG + NG+ F IP + + Y Y N +E ++TF
Sbjct: 191 FLYKLQNRRLPEFYLSSGVFRLYRSGPWNGIGFSGIPEDEKLSYMVYNFTENSEEVAYTF 250
Query: 243 EVKDGK-FAQWELSSKG---KLVGD------------------DGYIANADMCY------ 274
+ + +++ LSSKG +L D D YI Y
Sbjct: 251 RMTNNSIYSRLTLSSKGDFQRLTWDPSLEIWNMFWSSPVDPQCDSYIMCGAYAYCDVNTS 310
Query: 275 -------GYNS-----------DGGCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFD 316
G+N GGC + + +C G K P +T D
Sbjct: 311 PVCNCIQGFNPRNIQRWDQRVWAGGCVRRTQL-SCNGDGFTRMKNMKLP---ETTMAIVD 366
Query: 317 VTYSYSDCKIRCWKNCSCNGFQL--YYSNMTGCV 348
+ +C+ RC +C+C F + TGCV
Sbjct: 367 RSIGEKECEKRCLSDCNCTAFANADIRNGGTGCV 400
>UniRef100_Q84KW1 S-locus receptor kinase [Brassica oleracea]
Length = 440
Score = 82.4 bits (202), Expect = 2e-14
Identities = 63/199 (31%), Positives = 101/199 (50%), Gaps = 23/199 (11%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN----TVATMLDAGN 135
VWV + + + L + S ++ ++ N+ ++S+ N VA +L GN
Sbjct: 74 VWVANRDSPLSNAIGTLKISGSNLVLLDHSNKS---VWSTNLTRGNERSPVVAELLANGN 130
Query: 136 FVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS---------DKFNL 186
FV++ F NG+ LWQSFDYP+D L+P MKLG + KTG N L S +F+
Sbjct: 131 FVIRYFSNNGASGFLWQSFDYPTDTLLPEMKLGYDHKTGLNRLLTSWRSSDDPSRGEFSY 190
Query: 187 EWEPKQG--ELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQ-YIIVSNKDEDSFTF 242
+ + ++G E I K G +SG NG+ F IP + + Y Y N +E ++TF
Sbjct: 191 QLDTQRGMPEFFIMKEGSQGQRSGPW--NGVQFSGIPEDRKLSYMVYNFTENNEEVAYTF 248
Query: 243 EVKDGKF-AQWELSSKGKL 260
V + F ++ ++S +G L
Sbjct: 249 RVTNNSFYSRLKISPEGVL 267
>UniRef100_Q7X6I1 OSJNBb0108J11.20 protein [Oryza sativa]
Length = 849
Score = 82.4 bits (202), Expect = 2e-14
Identities = 73/247 (29%), Positives = 109/247 (43%), Gaps = 28/247 (11%)
Query: 27 VEATSDSLKPGDKLNYKSKLCSKQGKFCLQF---GNNSNSDFQCLFISVNADYGKV---- 79
++ T D++ PG +L KL S G+F L F +N +S I + + V
Sbjct: 67 IQPTLDAISPGQELAAGDKLVSSNGRFALGFFQTDSNKSSSNSTPNIYLGIWFNTVPKFT 126
Query: 80 -VWVYDINHSI-DFNTSVLSLDYSGVLKIESQN--RKPIIIYSSPQ--PTNNTVATMLDA 133
VWV + + + D + L + G L I + + +++SS PTN T A +LD
Sbjct: 127 PVWVANGENPVADLASCKLLVSSDGNLAIVATTHAKNSSMVWSSKANIPTNTTHAVLLDD 186
Query: 134 GNFVLQQF-LPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQ 192
GN VL+ N S ++LWQSFD+P+D ++ K+G N TG N LVS K ++ P
Sbjct: 187 GNLVLRSTSTTNASSTILWQSFDHPTDTVLQGGKIGWNNATGVNRRLVSRKNTVDQAPGM 246
Query: 193 GELNIK------------KSGKVYWKSGKLKSNGLFENIPANV-QSRYQYIIVSNKDEDS 239
+ S YW SG F NIP V Q+ SN+ E
Sbjct: 247 YSFELLGHNGPTSMVSTFNSSNPYWSSGDWNGR-YFSNIPETVGQTWLSLNFTSNEQEKY 305
Query: 240 FTFEVKD 246
+ + D
Sbjct: 306 IEYAIAD 312
>UniRef100_Q8LQH1 Putative S-receptor kinase [Oryza sativa]
Length = 826
Score = 82.4 bits (202), Expect = 2e-14
Identities = 79/262 (30%), Positives = 128/262 (48%), Gaps = 34/262 (12%)
Query: 10 VVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSD---FQ 66
++LI LW + A + A D++ G L+ + L S+ GKF L F NS +
Sbjct: 25 MMLISCLLWLHREAAPSLAA--DTVTVGRPLSGRQVLVSRGGKFALGFFQPDNSSQRWYM 82
Query: 67 CLFISVNADYGKVVWVYDINHSI-DFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPT-- 123
++ + D+ KV WV + + D +TS L++ G + + + R P+ +S+ T
Sbjct: 83 GIWYNKIPDHTKV-WVANRRAPLSDPDTSRLAISADGNMVLLDRARPPV--WSTNVTTGV 139
Query: 124 --NNTVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVS 181
N+TV +LD GN VL + + VLWQSFD+ D +P +LG N+ TG LV
Sbjct: 140 AANSTVGVILDTGNLVLAD--ASNTSVVLWQSFDHFGDTWLPGGRLGRNKLTGEVTRLVG 197
Query: 182 DK---------FNLEWEP---KQGELNIKKSGKVYWKSGKLKSNGLFENIP------ANV 223
K F+LE +P Q ++ S ++YW SG + G+F ++P A+
Sbjct: 198 WKGYDDPTPGMFSLELDPGGASQYVMSWNGSSRLYWSSGNW-TGGMFSSVPEMMASNADP 256
Query: 224 QSRYQYIIVSNKDEDSFTFEVK 245
S Y + V ++E F ++VK
Sbjct: 257 LSLYTFNYVDGENESYFFYDVK 278
>UniRef100_Q7XXJ5 OSJNBa0094O15.5 protein [Oryza sativa]
Length = 813
Score = 81.6 bits (200), Expect = 3e-14
Identities = 90/302 (29%), Positives = 131/302 (42%), Gaps = 58/302 (19%)
Query: 29 ATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNS-----NSDFQCLFISVNADYGK----- 78
A +D+LK G L+ KL S+ GKF L F N S +SD + + K
Sbjct: 22 AATDTLKAGQVLSAGDKLVSRNGKFALGFFNPSANISKSSDNISSSWYIGIWFNKIPVFT 81
Query: 79 VVWVYDINHSI---DFNTSVLSLDYSGVLKIESQNRKPII----IYSSPQPTNNTVATML 131
VVWV + SI DF + L + G L I + + II I + + + NT +
Sbjct: 82 VVWVANRERSIAEPDFKLTQLKISQDGNLAIVNHANESIIWSTRIVNRTEASMNTSVLLH 141
Query: 132 DAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPK 191
D+GN V+Q + S +VLWQSFDYP+DV +P K+G N+ TG N VS K ++
Sbjct: 142 DSGNLVIQ----STSNAVLWQSFDYPTDVALPNAKIGWNKVTGLNRVGVSKKSLIDMGTG 197
Query: 192 QGELNIKKSGK------------VYW-----KSGKLKSNGLFENIPANVQSR-------- 226
+ + +G YW +SG +K L + + N Q+R
Sbjct: 198 SYSVQLYTNGTRRVTLEHRNPSIEYWYWSPDESG-MKIPALKQLLYMNPQTRGLVTPAYV 256
Query: 227 ----YQYIIVSNKDEDSFTFEVKD----GKFAQWELSSKGKLVGDDGYIANADMCYGYNS 278
+Y ++ DE S TF + D KF W S+ K Y D C Y++
Sbjct: 257 NSSEEEYYSYNSSDESSSTFLLLDINGQIKFNVW---SQDKHSWQSLYTQPVDPCRSYDT 313
Query: 279 DG 280
G
Sbjct: 314 CG 315
>UniRef100_P93068 Receptor-like kinase [Brassica oleracea]
Length = 847
Score = 80.5 bits (197), Expect = 7e-14
Identities = 61/199 (30%), Positives = 99/199 (49%), Gaps = 23/199 (11%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRK--PIIIYSSPQPTNNTVATMLDAGNFV 137
VWV + +H + +T L + S ++ ++ + + + VA +LD GNFV
Sbjct: 76 VWVANRDHPLSTSTGTLKISDSNLVVVDGSDTAVWSTNLTGGGDVRSPVVAELLDNGNFV 135
Query: 138 LQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNI 197
L+ N VLWQSFD+P+D L+P MKLG + KTG NW L S + +P G+ +
Sbjct: 136 LRDSNNNDPDIVLWQSFDFPTDTLLPEMKLGWDLKTGFNWFLRS--WKSPDDPSSGDYSF 193
Query: 198 K-------------KSGKVYWKSGKLKSNGL-FENIPANVQSRY-QYIIVSNKDEDSFTF 242
K K+ +VY +SG NG+ F +P Y ++ ++ E +++F
Sbjct: 194 KLKTRGFPEAFLWNKASQVY-RSGPW--NGIRFSGVPEMQPFDYIEFNFTTSNQEVTYSF 250
Query: 243 EV-KDGKFAQWELSSKGKL 260
+ KD +++ LSS G L
Sbjct: 251 HITKDNMYSRLSLSSTGSL 269
>UniRef100_O04383 Serine /threonine kinase precursor [Brassica oleracea]
Length = 847
Score = 80.5 bits (197), Expect = 7e-14
Identities = 61/199 (30%), Positives = 99/199 (49%), Gaps = 23/199 (11%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRK--PIIIYSSPQPTNNTVATMLDAGNFV 137
VWV + +H + +T L + S ++ ++ + + + VA +LD GNFV
Sbjct: 76 VWVANRDHPLSTSTGTLKISDSNLVVVDGSDTAVWSTNLTGGGDVRSPVVAELLDNGNFV 135
Query: 138 LQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGELNI 197
L+ N VLWQSFD+P+D L+P MKLG + KTG NW L S + +P G+ +
Sbjct: 136 LRDSNNNDPDIVLWQSFDFPTDTLLPEMKLGWDLKTGFNWFLRS--WKSPDDPSSGDYSF 193
Query: 198 K-------------KSGKVYWKSGKLKSNGL-FENIPANVQSRY-QYIIVSNKDEDSFTF 242
K K+ +VY +SG NG+ F +P Y ++ ++ E +++F
Sbjct: 194 KLKTRGFPEAFLWNKASQVY-RSGPW--NGIRFSGVPEMQPFDYIEFNFTTSNQEVTYSF 250
Query: 243 EV-KDGKFAQWELSSKGKL 260
+ KD +++ LSS G L
Sbjct: 251 HITKDNMYSRLSLSSTGSL 269
>UniRef100_Q84XJ5 S-receptor kinase 13-8 [Arabidopsis lyrata]
Length = 312
Score = 80.5 bits (197), Expect = 7e-14
Identities = 74/229 (32%), Positives = 104/229 (45%), Gaps = 49/229 (21%)
Query: 127 VATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNL 186
VA +L GNFVL+ NG LWQSFDYP+D L+P MKLG++RKTG+N L S K +
Sbjct: 21 VAELLPNGNFVLRDSKTNGQDGFLWQSFDYPTDTLLPHMKLGLDRKTGNNRFLTSWKNS- 79
Query: 187 EWEPKQGELNIK------------KSGKVYWKSGKLKSNGL-FENIPANVQSRY---QYI 230
++P G L+ K KSG ++SG +G+ F IP + ++ Y
Sbjct: 80 -YDPSSGSLSYKLEIQGLPEFFVSKSGVPVFRSGPW--DGIQFSGIPEMQRWKHVNISYN 136
Query: 231 IVSNKDEDSFTFEVKDGKFAQWELSSKGKLVGDDGYIANADMCYGYNSDGGCQKWEDIPT 290
NK+E +FT+ V I +A +S+G Q + IPT
Sbjct: 137 FTENKEEVAFTYRVT---------------------IPDAYAGMTMDSEGLLQLFTWIPT 175
Query: 291 CREPGEMFQKKAGRPSIDNSTTYEFDVTYSYSDCKIRCWKNCSC-NGFQ 338
E + AG I Y+ Y+Y C NC+C GF+
Sbjct: 176 TLEWNMFWLSSAGECDI-----YQRCSPYTY--CDRNKTPNCNCIKGFE 217
>UniRef100_Q9SXH5 SRK13 [Brassica oleracea]
Length = 854
Score = 80.1 bits (196), Expect = 9e-14
Identities = 66/257 (25%), Positives = 124/257 (47%), Gaps = 26/257 (10%)
Query: 10 VVLIYLWLWWNTTANICVEATSDSLKPGDKLNYKSKLCSKQGKFCLQFGNNSNSDFQCLF 69
+++ ++W+ + +I ++++SL ++ L S F L F ++S L
Sbjct: 17 LLVFFVWILFRPAFSINTLSSTESLT----ISSNRTLVSPGNVFELGFFKTTSSSRWYLG 72
Query: 70 ISVNA-DYGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN--- 125
I Y VWV + ++ + + L + + ++ ++ N+ ++S+ N
Sbjct: 73 IWYKKFPYRTYVWVANRDNPLSNDIGTLKISGNNLVLLDHSNKS---VWSTNVTRGNERS 129
Query: 126 -TVATMLDAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKF 184
VA +LD GNFV++ N + LWQSFDYP+D L+P MKLG + KTG N L S +
Sbjct: 130 PVVAELLDNGNFVMRDSNSNNASQFLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTS--W 187
Query: 185 NLEWEPKQGELNIK----KSGKVYWKSGKLKS------NGL-FENIPANVQSRYQ-YIIV 232
+P G+ + K + + Y G +++ +G+ F IP + + Y Y
Sbjct: 188 RSSDDPSSGDYSYKLEPGRLPEFYLWKGNIRTHRSGPWSGIQFSGIPEDQRLSYMVYNFT 247
Query: 233 SNKDEDSFTFEVKDGKF 249
N++E ++TF++ + F
Sbjct: 248 ENREEVAYTFQMTNNSF 264
>UniRef100_Q8LP66 S-locus-related I [Brassica amplexicaulis]
Length = 419
Score = 79.3 bits (194), Expect = 2e-13
Identities = 61/200 (30%), Positives = 95/200 (47%), Gaps = 27/200 (13%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQ 139
VWV + ++ + +T L + +S + ++ N + T+ A +L GNFVL+
Sbjct: 75 VWVANRDNPLHSSTGTLKISHSNLFLLDQFNTPVWSTNITETVTSPLTAELLSNGNFVLR 134
Query: 140 QFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDK---------FNLEWEP 190
+ LWQSFD+P D L+P MKLG N KTGH+ L S K ++ + E
Sbjct: 135 DSKTKDTNQFLWQSFDFPVDTLLPEMKLGRNLKTGHDRVLTSWKSPTDPSSGDYSFKLET 194
Query: 191 KQGEL-----NIKKSGKVYWKSGKLKSNGLFENIPANVQSRYQYII---VSNKDEDSFTF 242
QG L +K KVY ++ F IP + YI+ + NK+E S+ F
Sbjct: 195 HQGSLLHEFYLLKNELKVY------RTGPWFNAIPK--MQNWSYIVNSFIDNKEEVSYAF 246
Query: 243 EVKDGKF--AQWELSSKGKL 260
+V + K ++ +SS G L
Sbjct: 247 KVNNHKMIHTRFRMSSTGLL 266
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 718,101,417
Number of Sequences: 2790947
Number of extensions: 32850261
Number of successful extensions: 68469
Number of sequences better than 10.0: 473
Number of HSP's better than 10.0 without gapping: 357
Number of HSP's successfully gapped in prelim test: 117
Number of HSP's that attempted gapping in prelim test: 67505
Number of HSP's gapped (non-prelim): 946
length of query: 381
length of database: 848,049,833
effective HSP length: 129
effective length of query: 252
effective length of database: 488,017,670
effective search space: 122980452840
effective search space used: 122980452840
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC140024.11


