

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.4 - phase: 0
(205 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
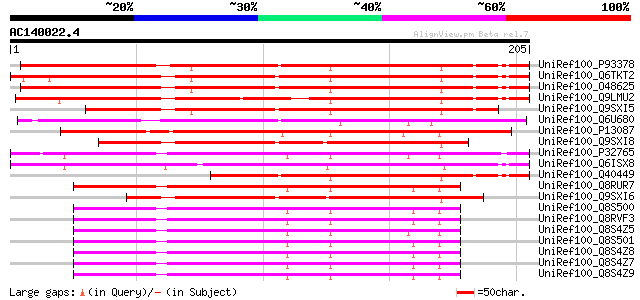
UniRef100_P93378 Tumor-related protein [Nicotiana tabacum] 192 3e-48
UniRef100_Q6TKT2 Miraculin-like protein [Solanum brevidens] 173 3e-42
UniRef100_O48625 Lemir [Lycopersicon esculentum] 171 8e-42
UniRef100_Q9LMU2 Hypothetical protein F2H15.9 (Hypothetical prot... 166 3e-40
UniRef100_Q9SXI5 Miraculin homologue [Solanum melongena] 159 4e-38
UniRef100_Q6U680 Kunitz trypsin inhibitor 4 [Populus balsamifera... 149 4e-35
UniRef100_P13087 Miraculin precursor [Richadella dulcifica] 141 1e-32
UniRef100_Q9SXI8 Miraculin homologue [Youngia japonica] 139 5e-32
UniRef100_P32765 21 kDa seed protein precursor [Theobroma cacao] 135 7e-31
UniRef100_Q6ISX8 Pathogen-inducible trypsin-inhibitor-like prote... 130 2e-29
UniRef100_Q40449 Tumor-related protein [Nicotiana glauca x Nicot... 124 2e-27
UniRef100_Q8RUR7 Trypsin inhibitor [Theobroma obovatum] 120 3e-26
UniRef100_Q9SXI6 Miraculin homologue [Taraxacum officinale] 119 4e-26
UniRef100_Q8S500 Trypsin inhibitor [Theobroma subincanum] 119 6e-26
UniRef100_Q8RVF3 Trypsin inhibitor [Theobroma grandiflorum] 118 1e-25
UniRef100_Q8S4Z5 Trypsin inhibitor [Theobroma cacao] 116 3e-25
UniRef100_Q8S501 Trypsin inhibitor [Theobroma angustifolium] 115 5e-25
UniRef100_Q8S4Z8 Trypsin inhibitor [Theobroma mammosum] 115 5e-25
UniRef100_Q8S4Z7 Trypsin inhibitor [Theobroma mammosum] 115 5e-25
UniRef100_Q8S4Z9 Trypsin inhibitor [Theobroma mammosum] 115 7e-25
>UniRef100_P93378 Tumor-related protein [Nicotiana tabacum]
Length = 210
Score = 192 bits (489), Expect = 3e-48
Identities = 103/205 (50%), Positives = 145/205 (70%), Gaps = 13/205 (6%)
Query: 5 FLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGN 64
FL F+I F +F+ + AP V+DI+GKK+ TG+ YYILPV+RG+GGGLT+ +
Sbjct: 10 FLIFTISFNSFLSSSAEAPPAVVDIAGKKLRTGIDYYILPVVRGRGGGLTLDST-----G 64
Query: 65 NNTCPL-YVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLN 123
N +CPL V+QE+ E+KNG +TFTP N KKGVI STDLNIK + + C Q+ +WKL+
Sbjct: 65 NESCPLDAVVQEQQEIKNGLPLTFTPVNPKKGVIRESTDLNIK-FSAASICVQTTLWKLD 123
Query: 124 KV--LSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQTLCRELGLY 180
+G +F+ GG EGNPG +TI NWFKIEK ++DY +CP+VC C+ +C+++G++
Sbjct: 124 DFDETTGKYFITIGGNEGNPGRETISNWFKIEKFERDYKLVYCPTVCNFCKVICKDVGIF 183
Query: 181 VYDHGKKHLALSDQVPSFRVVFKRA 205
+ D G + LALSD VP F+V+FK+A
Sbjct: 184 IQD-GIRRLALSD-VP-FKVMFKKA 205
>UniRef100_Q6TKT2 Miraculin-like protein [Solanum brevidens]
Length = 209
Score = 173 bits (438), Expect = 3e-42
Identities = 101/214 (47%), Positives = 140/214 (65%), Gaps = 18/214 (8%)
Query: 1 MKTS--FLAFSIIFLA---FICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTV 55
MKT+ FL F I+ ++ F+ +P V+DI GK + TGV YYILPV+RG+GGGLT+
Sbjct: 5 MKTNQLFLPFLILVISSNSFLSSAAESPPEVVDIDGKILRTGVDYYILPVVRGRGGGLTM 64
Query: 56 VNENNLNGNNNTCPL-YVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTC 114
+ + CPL V+QE E+ G +TFTP N KKGVI STDLNI + + C
Sbjct: 65 DSIGD-----KICPLDAVVQEHQEIDQGLPLTFTPVNPKKGVIRESTDLNI-IFSANSIC 118
Query: 115 AQSQVWKLNKV--LSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQ 171
Q+ WKL+ +G +F+ GG +GNPG +TI NWFKIEK D+DY +CP+VC C+
Sbjct: 119 VQTTQWKLDDFDETTGQYFITLGGNQGNPGRETISNWFKIEKFDRDYKLLYCPTVCDFCK 178
Query: 172 TLCRELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
+C+E+G+++ D G + LALSD VP F+V+FK+A
Sbjct: 179 VICKEIGIFIQD-GVRRLALSD-VP-FKVMFKKA 209
>UniRef100_O48625 Lemir [Lycopersicon esculentum]
Length = 205
Score = 171 bits (434), Expect = 8e-42
Identities = 96/205 (46%), Positives = 134/205 (64%), Gaps = 13/205 (6%)
Query: 5 FLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGN 64
FL +I F + + +P V+DI GK + TGV YYILPV+RG+GGGLT+ + +
Sbjct: 10 FLILAISFNSLLSSAAESPPEVVDIDGKILRTGVDYYILPVVRGRGGGLTMDSIGD---- 65
Query: 65 NNTCPL-YVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLN 123
CPL V+QE E+ G +TFTP + KKGVI STDLNI + + C Q+ WKL+
Sbjct: 66 -KMCPLDAVVQEHNEIDQGLPLTFTPVDPKKGVIRESTDLNI-IFSANSICVQTTQWKLD 123
Query: 124 KV--LSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQTLCRELGLY 180
+G +F+ GG +GNPG +TI NWFKIEK D+DY +CP+VC C+ +CR++G++
Sbjct: 124 DFDETTGQYFITLGGDQGNPGVETISNWFKIEKYDRDYKLLYCPTVCDFCKVICRDIGIF 183
Query: 181 VYDHGKKHLALSDQVPSFRVVFKRA 205
+ D G + LALSD VP F+V+FK+A
Sbjct: 184 IQD-GVRRLALSD-VP-FKVMFKKA 205
>UniRef100_Q9LMU2 Hypothetical protein F2H15.9 (Hypothetical protein At1g17860)
(At1g17860/F2H15_8) (Putative lemir (Miraculin) protein)
[Arabidopsis thaliana]
Length = 196
Score = 166 bits (421), Expect = 3e-40
Identities = 100/210 (47%), Positives = 131/210 (61%), Gaps = 23/210 (10%)
Query: 3 TSFLAFSIIFLAFICK----TFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNE 58
+S L ++ FI T AA EPV DI+GK + TGV YYILPVIRG+GGGLT+ N
Sbjct: 2 SSLLYIFLLLAVFISHRGVTTEAAVEPVKDINGKSLLTGVNYYILPVIRGRGGGLTMSNL 61
Query: 59 NNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQ 118
TCP V+Q++ EV G V F+PY+ K I STD+NIK T
Sbjct: 62 KT-----ETCPTSVIQDQFEVSQGLPVKFSPYD-KSRTIPVSTDVNIKFSPTS------- 108
Query: 119 VWKLNKV--LSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQTLCR 175
+W+L + WF++T GVEGNPG T+ NWFKI+K +KDY FCP+VC C+ +CR
Sbjct: 109 IWELANFDETTKQWFISTCGVEGNPGQKTVDNWFKIDKFEKDYKIRFCPTVCNFCKVICR 168
Query: 176 ELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
++G++V D GK+ LALSD VP +V+FKRA
Sbjct: 169 DVGVFVQD-GKRRLALSD-VP-LKVMFKRA 195
>UniRef100_Q9SXI5 Miraculin homologue [Solanum melongena]
Length = 160
Score = 159 bits (402), Expect = 4e-38
Identities = 83/167 (49%), Positives = 115/167 (68%), Gaps = 11/167 (6%)
Query: 31 GKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPL-YVLQEKLEVKNGQAVTFTP 89
GK + TG+ YYILPV+RG+GGGLT+ + N TCPL V+QE+ EVK G +TFTP
Sbjct: 1 GKILRTGIDYYILPVVRGRGGGLTMDSIGN-----KTCPLDAVVQEQEEVKQGLPLTFTP 55
Query: 90 YNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPGFDTIF 147
+N KKGVI STDLNI + + C Q+ WKL+ +G +F+ GG +GNPG +TI
Sbjct: 56 FNPKKGVIRESTDLNI-IFSANSICVQTTQWKLDNFDETTGKYFITLGGNQGNPGRETIS 114
Query: 148 NWFKIEKADKDYVFSFCPSVCK-CQTLCRELGLYVYDHGKKHLALSD 193
NWFKIEK ++DY +CP+VC C+ +C+++G+++ D G + LALSD
Sbjct: 115 NWFKIEKFERDYKLVYCPTVCDFCKVICKDIGIFIQD-GVRRLALSD 160
>UniRef100_Q6U680 Kunitz trypsin inhibitor 4 [Populus balsamifera subsp. trichocarpa
x Populus deltoides]
Length = 212
Score = 149 bits (376), Expect = 4e-35
Identities = 86/205 (41%), Positives = 120/205 (57%), Gaps = 14/205 (6%)
Query: 4 SFLAFSIIFLAFICKTFAAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNG 63
SFL F+ F+ + A EPVLDI G+++ G +Y I + G GGG +++
Sbjct: 10 SFLLFA--FVLSVPSIEAYTEPVLDIQGEELKAGTEYIIGSIFFGAGGG-------DVSA 60
Query: 64 NNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLN 123
N TCP V+Q ++ G VTF+P ++ VI STDLNIK + K C S VWK+
Sbjct: 61 TNKTCPDDVIQYSSDLLQGLPVTFSPASSDDDVIRVSTDLNIK-FSIKKACDHSSVWKIQ 119
Query: 124 KVLSGV--WFLATGGVEGNPGFDTIFNWFKIEKAD-KDYVFSFCP-SVCKCQTLCRELGL 179
K + WF+ TGG EGNPG DT+ NWFKIEKA Y CP +C C LCR++G+
Sbjct: 120 KSSNSEVQWFVTTGGEEGNPGIDTLTNWFKIEKAGILGYKLVSCPEGICHCGVLCRDIGI 179
Query: 180 YVYDHGKKHLALSDQVPSFRVVFKR 204
Y + G++ L+LSD++ F V+FK+
Sbjct: 180 YRENDGRRILSLSDKLSPFLVLFKK 204
>UniRef100_P13087 Miraculin precursor [Richadella dulcifica]
Length = 220
Score = 141 bits (355), Expect = 1e-32
Identities = 81/184 (44%), Positives = 113/184 (61%), Gaps = 8/184 (4%)
Query: 21 AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVK 80
+AP PVLDI G+K+ TG YYI+PV+R GGGLT V+ NG CP V+Q + EV
Sbjct: 31 SAPNPVLDIDGEKLRTGTNYYIVPVLRDHGGGLT-VSATTPNG-TFVCPPRVVQTRKEVD 88
Query: 81 NGQAVTFTPYNAKKGVILTSTDLNIK-SYVTKTTCAQSQVWKLNKV--LSGVWFLATGGV 137
+ + + F P N K+ V+ STDLNI S S VW+L+K +G +F+ GGV
Sbjct: 89 HDRPLAFFPENPKEDVVRVSTDLNINFSAFMPCRWTSSTVWRLDKYDESTGQYFVTIGGV 148
Query: 138 EGNPGFDTIFNWFKIEK--ADKDYVFSFCPSVC-KCQTLCRELGLYVYDHGKKHLALSDQ 194
+GNPG +TI +WFKIE+ Y FCP+VC C+ C ++G+Y+ G++ LALSD+
Sbjct: 149 KGNPGPETISSWFKIEEFCGSGFYKLVFCPTVCGSCKVKCGDVGIYIDQKGRRRLALSDK 208
Query: 195 VPSF 198
+F
Sbjct: 209 PFAF 212
>UniRef100_Q9SXI8 Miraculin homologue [Youngia japonica]
Length = 142
Score = 139 bits (350), Expect = 5e-32
Identities = 73/147 (49%), Positives = 95/147 (63%), Gaps = 8/147 (5%)
Query: 36 TGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKG 95
+G +YYILPV RG GGGLT+ + N +TCPL V+Q LEV NG +TFTP + KKG
Sbjct: 3 SGTEYYILPVFRGMGGGLTLASTRN-----DTCPLDVVQADLEVDNGLPLTFTPVDPKKG 57
Query: 96 VILTSTDLNIKSYVTKTTCAQSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKA 155
VI STDLNI + + C QS VW L + G ++ GV GNPG +TI NWFKIEK
Sbjct: 58 VIRESTDLNI-IFSASSICIQSNVWNLEE-YDGQLIVSAHGVAGNPGRETISNWFKIEKY 115
Query: 156 DKDYVFSFCPSVCK-CQTLCRELGLYV 181
+ DY FCP+VC C+ +C ++G+ +
Sbjct: 116 EDDYKIVFCPTVCDFCRPVCGDIGVLI 142
>UniRef100_P32765 21 kDa seed protein precursor [Theobroma cacao]
Length = 221
Score = 135 bits (340), Expect = 7e-31
Identities = 84/216 (38%), Positives = 123/216 (56%), Gaps = 18/216 (8%)
Query: 1 MKTSFLAFSIIFLAFICKTF------AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLT 54
MKT+ ++F AF K++ AA PVLD G ++ TGV+YY+L I G GGG
Sbjct: 1 MKTATAVVLLLF-AFTSKSYFFGVANAANSPVLDTDGDELQTGVQYYVLSSISGAGGGGL 59
Query: 55 VVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSY-VTKTT 113
+ +CP V+Q + ++ NG V F+ ++K V+ STD+NI+ +
Sbjct: 60 ALGR----ATGQSCPEIVVQRRSDLDNGTPVIFSNADSKDDVVRVSTDVNIEFVPIRDRL 115
Query: 114 CAQSQVWKLNKV--LSGVWFLATGGVEGNPGFDTIFNWFKIEKAD-KDYVFSFCPSVC-K 169
C+ S VW+L+ +G W++ T GV+G PG +T+ +WFKIEKA Y F FCPSVC
Sbjct: 116 CSTSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNTLCSWFKIEKAGVLGYKFRFCPSVCDS 175
Query: 170 CQTLCRELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
C TLC ++G + D G+ LALSD + +FK+A
Sbjct: 176 CTTLCSDIGRHSDDDGQIRLALSDN--EWAWMFKKA 209
>UniRef100_Q6ISX8 Pathogen-inducible trypsin-inhibitor-like protein [Medicago
truncatula]
Length = 213
Score = 130 bits (328), Expect = 2e-29
Identities = 88/217 (40%), Positives = 118/217 (53%), Gaps = 16/217 (7%)
Query: 1 MKTSFLAFSIIFLAFICKTF-----AAPEPVLDISGKKVTTGVKYYILPVIRGKGGGLTV 55
MK + LAF +F + A+ E V+D GKK+ YYI+PV K G
Sbjct: 1 MKNTLLAFFFLFTFLSSQPLLGAAEASNEQVVDTLGKKLRADANYYIIPVPIYKCGPYGK 60
Query: 56 VNENN----LNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIKSYVTK 111
+ L TCPL V+ ++ +TF P N KKGVI STDLNIK
Sbjct: 61 CRSSGSSLALASIGKTCPLDVVV--VDRYQALPLTFIPVNPKKGVIRVSTDLNIKFSSRA 118
Query: 112 TTCAQSQVWKLNK--VLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK 169
T S VWKL++ V WF+ GGV GNPG++TI NWFKIEK Y FCPSV +
Sbjct: 119 TCLHHSMVWKLDRFNVSKRQWFITIGGVAGNPGWETINNWFKIEKYGDAYKLVFCPSVVQ 178
Query: 170 C-QTLCRELGLYVYDHGKKHLALSDQVPSFRVVFKRA 205
+ +C+++G++V ++G K LALSD VP +V F++A
Sbjct: 179 SFKHMCKDVGVFVDENGNKRLALSD-VP-LKVKFQQA 213
>UniRef100_Q40449 Tumor-related protein [Nicotiana glauca x Nicotiana langsdorffii]
Length = 125
Score = 124 bits (311), Expect = 2e-27
Identities = 65/129 (50%), Positives = 92/129 (70%), Gaps = 7/129 (5%)
Query: 80 KNGQAVTFTPYNAKKGVILTSTDLNIKSYVTKTTCAQSQVWKLNKV--LSGVWFLATGGV 137
+NG +TFTP N KKGVI STDLNIK + + C Q+ +WKL+ +G +F+ GG
Sbjct: 1 ENGLPLTFTPVNPKKGVIRESTDLNIK-FSAASICVQTTLWKLDDFDETTGKYFITIGGN 59
Query: 138 EGNPGFDTIFNWFKIEKADKDYVFSFCPSVCK-CQTLCRELGLYVYDHGKKHLALSDQVP 196
EGNPG +TI NWFKIEK ++DY +CP+VC C+ +C+++G+++ D G + LALSD VP
Sbjct: 60 EGNPGRETISNWFKIEKFERDYKLVYCPTVCNFCKVICKDVGIFIQD-GIRRLALSD-VP 117
Query: 197 SFRVVFKRA 205
F+V+FK+A
Sbjct: 118 -FKVMFKKA 125
>UniRef100_Q8RUR7 Trypsin inhibitor [Theobroma obovatum]
Length = 154
Score = 120 bits (300), Expect = 3e-26
Identities = 65/158 (41%), Positives = 96/158 (60%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YY++ VI G GGG + + CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYVVSVIWGAGGGGLALGR----ATDQKCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ TGGV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGVKGEPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A + Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGNVYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q9SXI6 Miraculin homologue [Taraxacum officinale]
Length = 136
Score = 119 bits (299), Expect = 4e-26
Identities = 62/142 (43%), Positives = 88/142 (61%), Gaps = 8/142 (5%)
Query: 47 RGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAVTFTPYNAKKGVILTSTDLNIK 106
RG+GGG+T+ CPL V+Q E+ NG +TFTP + KKGVI STDLNI
Sbjct: 2 RGRGGGVTLAPTRT-----ELCPLDVVQANQELDNGLPLTFTPVDPKKGVIRESTDLNI- 55
Query: 107 SYVTKTTCAQSQVWKLNKVLSGVWFLATGGVEGNPGFDTIFNWFKIEKADKDYVFSFCPS 166
+ + C QS VW + + G ++ GV+GNPG +T+ NWFKIEK D DY FCP+
Sbjct: 56 IFSASSICIQSNVWMIEEY-DGSLIVSAHGVQGNPGQETLSNWFKIEKFDDDYKIVFCPA 114
Query: 167 VCK-CQTLCRELGLYVYDHGKK 187
VC C+ LC ++G+ + ++G++
Sbjct: 115 VCDVCRPLCGDIGVEIDENGRR 136
>UniRef100_Q8S500 Trypsin inhibitor [Theobroma subincanum]
Length = 154
Score = 119 bits (297), Expect = 6e-26
Identities = 64/158 (40%), Positives = 95/158 (59%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YY++ I G GGG + + CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYVMSAIWGAGGGGLALGR----ATDQKCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ TGGV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGVKGEPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A + Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGNVYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8RVF3 Trypsin inhibitor [Theobroma grandiflorum]
Length = 154
Score = 118 bits (295), Expect = 1e-25
Identities = 64/158 (40%), Positives = 95/158 (59%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YY+L + G GGG + + CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYVLSALWGAGGGGLDLGR----ATDQKCPEIVIQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ TGGV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGVKGEPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A + Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGNVYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8S4Z5 Trypsin inhibitor [Theobroma cacao]
Length = 154
Score = 116 bits (291), Expect = 3e-25
Identities = 64/158 (40%), Positives = 93/158 (58%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YY+L I G GGG + +CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELQTGVQYYVLSSISGAGGGGLALGR----ATGQSCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ ++K V+ STD+NI+ + C+ S VW+L+ +G W++ T GV+G PG
Sbjct: 57 IFSNADSKDDVVRVSTDVNIEFVPIRDRLCSTSTVWRLDNYDNSAGKWWVTTDGVKGEPG 116
Query: 143 FDTIFNWFKIEKAD-KDYVFSFCPSVC-KCQTLCRELG 178
+T+ +WFKIEKA Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLCSWFKIEKAGVLGYKFRFCPSVCDSCTTLCSDIG 154
>UniRef100_Q8S501 Trypsin inhibitor [Theobroma angustifolium]
Length = 154
Score = 115 bits (289), Expect = 5e-25
Identities = 64/158 (40%), Positives = 93/158 (58%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YY++ I G GGG + N CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYVVSTIWGAGGGGLDLGR----ATNQKCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ T GV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTDGVKGEPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGTLYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8S4Z8 Trypsin inhibitor [Theobroma mammosum]
Length = 154
Score = 115 bits (289), Expect = 5e-25
Identities = 65/158 (41%), Positives = 94/158 (59%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YYI+ I G GGG + N CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYIVSAIWGAGGGGLDLGR----ATNQKCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ T GV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTDGVKGVPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A + Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGNIYKFRFCPSVCDSCTTLCSDIG 154
>UniRef100_Q8S4Z7 Trypsin inhibitor [Theobroma mammosum]
Length = 154
Score = 115 bits (289), Expect = 5e-25
Identities = 65/158 (41%), Positives = 94/158 (59%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YYI+ I G GGG + N CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYIVSAIWGAGGGGLDLGR----ATNQKCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ T GV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTDGVKGVPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A + Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGNIYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8S4Z9 Trypsin inhibitor [Theobroma mammosum]
Length = 154
Score = 115 bits (288), Expect = 7e-25
Identities = 64/158 (40%), Positives = 94/158 (58%), Gaps = 9/158 (5%)
Query: 26 VLDISGKKVTTGVKYYILPVIRGKGGGLTVVNENNLNGNNNTCPLYVLQEKLEVKNGQAV 85
VLD G ++ TGV+YY++ I G GGG + N CP V+Q + ++ NG V
Sbjct: 1 VLDTDGDELRTGVQYYVVSAIWGAGGGGLDLGR----ATNQKCPEIVVQRRSDLDNGTPV 56
Query: 86 TFTPYNAKKGVILTSTDLNIKSY-VTKTTCAQSQVWKLNKV--LSGVWFLATGGVEGNPG 142
F+ +++ V+ STD+NI+ + C+ S VWKL+ +G W++ T GV+G PG
Sbjct: 57 IFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTDGVKGVPG 116
Query: 143 FDTIFNWFKIEKADKD-YVFSFCPSVC-KCQTLCRELG 178
+T+ NWFKIE+A + Y F FCPSVC C TLC ++G
Sbjct: 117 PNTLSNWFKIEEAGGNIYKFRFCPSVCDSCTTLCSDIG 154
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.139 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 370,801,214
Number of Sequences: 2790947
Number of extensions: 16154741
Number of successful extensions: 37911
Number of sequences better than 10.0: 174
Number of HSP's better than 10.0 without gapping: 87
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 37634
Number of HSP's gapped (non-prelim): 178
length of query: 205
length of database: 848,049,833
effective HSP length: 121
effective length of query: 84
effective length of database: 510,345,246
effective search space: 42869000664
effective search space used: 42869000664
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 72 (32.3 bits)
Medicago: description of AC140022.4


