

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.13 + phase: 0
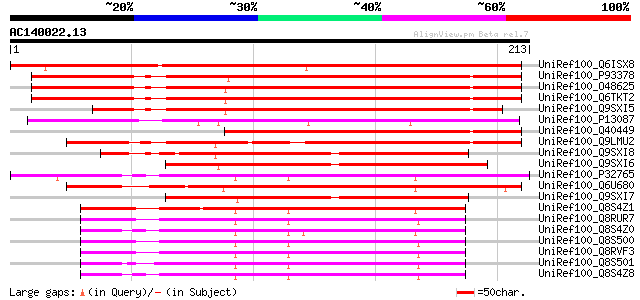
(213 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6ISX8 Pathogen-inducible trypsin-inhibitor-like prote... 233 2e-60
UniRef100_P93378 Tumor-related protein [Nicotiana tabacum] 213 3e-54
UniRef100_O48625 Lemir [Lycopersicon esculentum] 205 7e-52
UniRef100_Q6TKT2 Miraculin-like protein [Solanum brevidens] 203 2e-51
UniRef100_Q9SXI5 Miraculin homologue [Solanum melongena] 177 1e-43
UniRef100_P13087 Miraculin precursor [Richadella dulcifica] 176 4e-43
UniRef100_Q40449 Tumor-related protein [Nicotiana glauca x Nicot... 173 3e-42
UniRef100_Q9LMU2 Hypothetical protein F2H15.9 (Hypothetical prot... 163 3e-39
UniRef100_Q9SXI8 Miraculin homologue [Youngia japonica] 155 9e-37
UniRef100_Q9SXI6 Miraculin homologue [Taraxacum officinale] 137 1e-31
UniRef100_P32765 21 kDa seed protein precursor [Theobroma cacao] 136 3e-31
UniRef100_Q6U680 Kunitz trypsin inhibitor 4 [Populus balsamifera... 135 5e-31
UniRef100_Q9SXI7 Miraculin homologue [Youngia japonica] 132 6e-30
UniRef100_Q8S4Z1 Trypsin inhibitor [Theobroma sylvestre] 125 1e-27
UniRef100_Q8RUR7 Trypsin inhibitor [Theobroma obovatum] 124 2e-27
UniRef100_Q8S4Z0 Trypsin inhibitor [Theobroma bicolor] 124 2e-27
UniRef100_Q8S500 Trypsin inhibitor [Theobroma subincanum] 123 3e-27
UniRef100_Q8RVF3 Trypsin inhibitor [Theobroma grandiflorum] 121 1e-26
UniRef100_Q8S501 Trypsin inhibitor [Theobroma angustifolium] 120 2e-26
UniRef100_Q8S4Z8 Trypsin inhibitor [Theobroma mammosum] 120 2e-26
>UniRef100_Q6ISX8 Pathogen-inducible trypsin-inhibitor-like protein [Medicago
truncatula]
Length = 213
Score = 233 bits (595), Expect = 2e-60
Identities = 122/212 (57%), Positives = 149/212 (69%), Gaps = 3/212 (1%)
Query: 1 MKSICILLAVLFA-LSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
MK+ + LF LS+QPLLG A+AS EQVVDT GKK+RA +YYI PVP C G
Sbjct: 1 MKNTLLAFFFLFTFLSSQPLLGAAEASNEQVVDTLGKKLRADANYYIIPVPIYKCGPYGK 60
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTS 119
C SG L S +TCPL+VVVV+ ++ +TF PVNPKKGVIRVSTDLNIK S +
Sbjct: 61 CR-SSGSSLALASIGKTCPLDVVVVDRYQALPLTFIPVNPKKGVIRVSTDLNIKFSSRAT 119
Query: 120 C-EESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNF 178
C S +W LD F+ S QWF+T GGV GNPG +T++NWFKIEKY D YK VFCP+V
Sbjct: 120 CLHHSMVWKLDRFNVSKRQWFITIGGVAGNPGWETINNWFKIEKYGDAYKLVFCPSVVQS 179
Query: 179 CKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQ 210
K MC++VG+F D NGN+R+AL+DVP KV+FQ
Sbjct: 180 FKHMCKDVGVFVDENGNKRLALSDVPLKVKFQ 211
>UniRef100_P93378 Tumor-related protein [Nicotiana tabacum]
Length = 210
Score = 213 bits (542), Expect = 3e-54
Identities = 108/204 (52%), Positives = 143/204 (69%), Gaps = 14/204 (6%)
Query: 10 VLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI 69
++F +S L + +P VVD GKK+R G+DYYI PV GRG G + +
Sbjct: 11 LIFTISFNSFLSSSAEAPPAVVDIAGKKLRTGIDYYILPVVR----GRG------GGLTL 60
Query: 70 ARSPNETCPLNVVVVEGFR---GQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIW 126
+ NE+CPL+ VV E G +TFTPVNPKKGVIR STDLNIK S + C ++T+W
Sbjct: 61 DSTGNESCPLDAVVQEQQEIKNGLPLTFTPVNPKKGVIRESTDLNIKFSAASICVQTTLW 120
Query: 127 TLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNV 186
LDDFD +TG++F+T GG GNPG++T+ NWFKIEK+E DYK V+CPTVCNFCKV+C++V
Sbjct: 121 KLDDFDETTGKYFITIGGNEGNPGRETISNWFKIEKFERDYKLVYCPTVCNFCKVICKDV 180
Query: 187 GIFRDSNGNQRVALTDVPYKVRFQ 210
GIF +G +R+AL+DVP+KV F+
Sbjct: 181 GIF-IQDGIRRLALSDVPFKVMFK 203
>UniRef100_O48625 Lemir [Lycopersicon esculentum]
Length = 205
Score = 205 bits (521), Expect = 7e-52
Identities = 107/204 (52%), Positives = 142/204 (69%), Gaps = 14/204 (6%)
Query: 10 VLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI 69
++ A+S LL A SP +VVD +GK +R GVDYYI PV GRG G + +
Sbjct: 11 LILAISFNSLLSSAAESPPEVVDIDGKILRTGVDYYILPVVR----GRG------GGLTM 60
Query: 70 ARSPNETCPLNVVVVEGF---RGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIW 126
++ CPL+ VV E +G +TFTPV+PKKGVIR STDLNI S N+ C ++T W
Sbjct: 61 DSIGDKMCPLDAVVQEHNEIDQGLPLTFTPVDPKKGVIRESTDLNIIFSANSICVQTTQW 120
Query: 127 TLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNV 186
LDDFD +TGQ+F+T GG GNPG +T+ NWFKIEKY+ DYK ++CPTVC+FCKV+CR++
Sbjct: 121 KLDDFDETTGQYFITLGGDQGNPGVETISNWFKIEKYDRDYKLLYCPTVCDFCKVICRDI 180
Query: 187 GIFRDSNGNQRVALTDVPYKVRFQ 210
GIF +G +R+AL+DVP+KV F+
Sbjct: 181 GIF-IQDGVRRLALSDVPFKVMFK 203
>UniRef100_Q6TKT2 Miraculin-like protein [Solanum brevidens]
Length = 209
Score = 203 bits (517), Expect = 2e-51
Identities = 104/204 (50%), Positives = 141/204 (68%), Gaps = 14/204 (6%)
Query: 10 VLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI 69
++ +S+ L A SP +VVD +GK +R GVDYYI PV GRG G + +
Sbjct: 15 LILVISSNSFLSSAAESPPEVVDIDGKILRTGVDYYILPVVR----GRG------GGLTM 64
Query: 70 ARSPNETCPLNVVVVEGF---RGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIW 126
++ CPL+ VV E +G +TFTPVNPKKGVIR STDLNI S N+ C ++T W
Sbjct: 65 DSIGDKICPLDAVVQEHQEIDQGLPLTFTPVNPKKGVIRESTDLNIIFSANSICVQTTQW 124
Query: 127 TLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNV 186
LDDFD +TGQ+F+T GG GNPG++T+ NWFKIEK++ DYK ++CPTVC+FCKV+C+ +
Sbjct: 125 KLDDFDETTGQYFITLGGNQGNPGRETISNWFKIEKFDRDYKLLYCPTVCDFCKVICKEI 184
Query: 187 GIFRDSNGNQRVALTDVPYKVRFQ 210
GIF +G +R+AL+DVP+KV F+
Sbjct: 185 GIF-IQDGVRRLALSDVPFKVMFK 207
>UniRef100_Q9SXI5 Miraculin homologue [Solanum melongena]
Length = 160
Score = 177 bits (450), Expect = 1e-43
Identities = 91/171 (53%), Positives = 121/171 (70%), Gaps = 14/171 (8%)
Query: 35 GKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGF---RGQG 91
GK +R G+DYYI PV GRG G + + N+TCPL+ VV E +G
Sbjct: 1 GKILRTGIDYYILPVVR----GRG------GGLTMDSIGNKTCPLDAVVQEQEEVKQGLP 50
Query: 92 VTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPGK 151
+TFTP NPKKGVIR STDLNI S N+ C ++T W LD+FD +TG++F+T GG GNPG+
Sbjct: 51 LTFTPFNPKKGVIRESTDLNIIFSANSICVQTTQWKLDNFDETTGKYFITLGGNQGNPGR 110
Query: 152 DTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALTD 202
+T+ NWFKIEK+E DYK V+CPTVC+FCKV+C+++GIF +G +R+AL+D
Sbjct: 111 ETISNWFKIEKFERDYKLVYCPTVCDFCKVICKDIGIF-IQDGVRRLALSD 160
>UniRef100_P13087 Miraculin precursor [Richadella dulcifica]
Length = 220
Score = 176 bits (446), Expect = 4e-43
Identities = 96/210 (45%), Positives = 126/210 (59%), Gaps = 17/210 (8%)
Query: 8 LAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFV 67
++ L A + PLL AD++P V+D +G+K+R G +YYI PV G G
Sbjct: 14 VSALLAAAANPLLSAADSAPNPVLDIDGEKLRTGTNYYIVPVLRDH---------GGGLT 64
Query: 68 LIARSPNET--CPLNVVVV--EGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCE-- 121
+ A +PN T CP VV E + + F P NPK+ V+RVSTDLNI S C
Sbjct: 65 VSATTPNGTFVCPPRVVQTRKEVDHDRPLAFFPENPKEDVVRVSTDLNINFSAFMPCRWT 124
Query: 122 ESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKY--EDDYKFVFCPTVCNFC 179
ST+W LD +D STGQ+FVT GGV GNPG +T+ +WFKIE++ YK VFCPTVC C
Sbjct: 125 SSTVWRLDKYDESTGQYFVTIGGVKGNPGPETISSWFKIEEFCGSGFYKLVFCPTVCGSC 184
Query: 180 KVMCRNVGIFRDSNGNQRVALTDVPYKVRF 209
KV C +VGI+ D G +R+AL+D P+ F
Sbjct: 185 KVKCGDVGIYIDQKGRRRLALSDKPFAFEF 214
>UniRef100_Q40449 Tumor-related protein [Nicotiana glauca x Nicotiana langsdorffii]
Length = 125
Score = 173 bits (438), Expect = 3e-42
Identities = 78/122 (63%), Positives = 101/122 (81%), Gaps = 1/122 (0%)
Query: 89 GQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGN 148
G +TFTPVNPKKGVIR STDLNIK S + C ++T+W LDDFD +TG++F+T GG GN
Sbjct: 3 GLPLTFTPVNPKKGVIRESTDLNIKFSAASICVQTTLWKLDDFDETTGKYFITIGGNEGN 62
Query: 149 PGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVR 208
PG++T+ NWFKIEK+E DYK V+CPTVCNFCKV+C++VGIF +G +R+AL+DVP+KV
Sbjct: 63 PGRETISNWFKIEKFERDYKLVYCPTVCNFCKVICKDVGIF-IQDGIRRLALSDVPFKVM 121
Query: 209 FQ 210
F+
Sbjct: 122 FK 123
>UniRef100_Q9LMU2 Hypothetical protein F2H15.9 (Hypothetical protein At1g17860)
(At1g17860/F2H15_8) (Putative lemir (Miraculin) protein)
[Arabidopsis thaliana]
Length = 196
Score = 163 bits (412), Expect = 3e-39
Identities = 92/189 (48%), Positives = 122/189 (63%), Gaps = 20/189 (10%)
Query: 24 DASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV 83
+A+ E V D GK + GV+YYI PV GRG G + ++ ETCP +V+
Sbjct: 23 EAAVEPVKDINGKSLLTGVNYYILPV----IRGRG------GGLTMSNLKTETCPTSVIQ 72
Query: 84 --VEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVT 141
E +G V F+P + K I VSTD+NIK S ++IW L +FD +T QWF++
Sbjct: 73 DQFEVSQGLPVKFSPYD-KSRTIPVSTDVNIKFS------PTSIWELANFDETTKQWFIS 125
Query: 142 TGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALT 201
T GV GNPG+ TVDNWFKI+K+E DYK FCPTVCNFCKV+CR+VG+F +G +R+AL+
Sbjct: 126 TCGVEGNPGQKTVDNWFKIDKFEKDYKIRFCPTVCNFCKVICRDVGVF-VQDGKRRLALS 184
Query: 202 DVPYKVRFQ 210
DVP KV F+
Sbjct: 185 DVPLKVMFK 193
>UniRef100_Q9SXI8 Miraculin homologue [Youngia japonica]
Length = 142
Score = 155 bits (391), Expect = 9e-37
Identities = 80/153 (52%), Positives = 107/153 (69%), Gaps = 15/153 (9%)
Query: 38 VRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV--VEGFRGQGVTFT 95
+R+G +YYI PV RG +G G L A + N+TCPL+VV +E G +TFT
Sbjct: 1 LRSGTEYYILPV------FRG---MGGGLTL-ASTRNDTCPLDVVQADLEVDNGLPLTFT 50
Query: 96 PVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVD 155
PV+PKKGVIR STDLNI S ++ C +S +W L+++D GQ V+ GV GNPG++T+
Sbjct: 51 PVDPKKGVIRESTDLNIIFSASSICIQSNVWNLEEYD---GQLIVSAHGVAGNPGRETIS 107
Query: 156 NWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGI 188
NWFKIEKYEDDYK VFCPTVC+FC+ +C ++G+
Sbjct: 108 NWFKIEKYEDDYKIVFCPTVCDFCRPVCGDIGV 140
>UniRef100_Q9SXI6 Miraculin homologue [Taraxacum officinale]
Length = 136
Score = 137 bits (346), Expect = 1e-31
Identities = 66/134 (49%), Positives = 92/134 (68%), Gaps = 5/134 (3%)
Query: 65 GFVLIARSPNETCPLNVVVV--EGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEE 122
G V +A + E CPL+VV E G +TFTPV+PKKGVIR STDLNI S ++ C +
Sbjct: 6 GGVTLAPTRTELCPLDVVQANQELDNGLPLTFTPVDPKKGVIRESTDLNIIFSASSICIQ 65
Query: 123 STIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVM 182
S +W ++++D G V+ GV GNPG++T+ NWFKIEK++DDYK VFCP VC+ C+ +
Sbjct: 66 SNVWMIEEYD---GSLIVSAHGVQGNPGQETLSNWFKIEKFDDDYKIVFCPAVCDVCRPL 122
Query: 183 CRNVGIFRDSNGNQ 196
C ++G+ D NG +
Sbjct: 123 CGDIGVEIDENGRR 136
>UniRef100_P32765 21 kDa seed protein precursor [Theobroma cacao]
Length = 221
Score = 136 bits (343), Expect = 3e-31
Identities = 79/219 (36%), Positives = 127/219 (57%), Gaps = 15/219 (6%)
Query: 1 MKSICILLAVLFALSTQP-LLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
MK+ ++ +LFA +++ G A+A+ V+DT+G +++ GV YY+ + G
Sbjct: 1 MKTATAVVLLLFAFTSKSYFFGVANAANSPVLDTDGDELQTGVQYYV----LSSISG--- 53
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQG--VTFTPVNPKKGVIRVSTDLNIK--TS 115
G G + + R+ ++CP VV G V F+ + K V+RVSTD+NI+
Sbjct: 54 --AGGGGLALGRATGQSCPEIVVQRRSDLDNGTPVIFSNADSKDDVVRVSTDVNIEFVPI 111
Query: 116 LNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPT 174
+ C ST+W LD++D+S G+W+VTT GV G PG +T+ +WFKIEK YKF FCP+
Sbjct: 112 RDRLCSTSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNTLCSWFKIEKAGVLGYKFRFCPS 171
Query: 175 VCNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA 213
VC+ C +C ++G D +G R+AL+D + F+ ++
Sbjct: 172 VCDSCTTLCSDIGRHSDDDGQIRLALSDNEWAWMFKKAS 210
>UniRef100_Q6U680 Kunitz trypsin inhibitor 4 [Populus balsamifera subsp. trichocarpa
x Populus deltoides]
Length = 212
Score = 135 bits (341), Expect = 5e-31
Identities = 77/192 (40%), Positives = 116/192 (60%), Gaps = 17/192 (8%)
Query: 24 DASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV 83
+A E V+D +G++++AG +Y I G G+G ++ + N+TCP +V+
Sbjct: 24 EAYTEPVLDIQGEELKAGTEYII-----------GSIFFGAGGGDVSAT-NKTCPDDVIQ 71
Query: 84 VEG--FRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVT 141
+G VTF+P + VIRVSTDLNIK S+ +C+ S++W + +S QWFVT
Sbjct: 72 YSSDLLQGLPVTFSPASSDDDVIRVSTDLNIKFSIKKACDHSSVWKIQKSSNSEVQWFVT 131
Query: 142 TGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVAL 200
TGG GNPG DT+ NWFKIEK YK V CP C V+CR++GI+R+++G + ++L
Sbjct: 132 TGGEEGNPGIDTLTNWFKIEKAGILGYKLVSCPEGICHCGVLCRDIGIYRENDGRRILSL 191
Query: 201 TD--VPYKVRFQ 210
+D P+ V F+
Sbjct: 192 SDKLSPFLVLFK 203
>UniRef100_Q9SXI7 Miraculin homologue [Youngia japonica]
Length = 136
Score = 132 bits (332), Expect = 6e-30
Identities = 64/126 (50%), Positives = 86/126 (67%), Gaps = 5/126 (3%)
Query: 65 GFVLIARSPNETCPLNVVVVEGFRGQGV--TFTPVNPKKGVIRVSTDLNIKTSLNTSCEE 122
G V + + NE CP +VV + G+ TFTPV+PKKGVIR STDLNI + C +
Sbjct: 6 GGVRLVPTRNELCPPDVVQADQELDNGLPLTFTPVDPKKGVIRESTDLNIIFRAYSICIQ 65
Query: 123 STIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVM 182
S +W L+++D G V+ GV GNPG++T+ NWFKIEKYEDDYK VFCPTVC+ C+ +
Sbjct: 66 SNVWMLEEYD---GSLIVSGHGVAGNPGQETISNWFKIEKYEDDYKLVFCPTVCDTCRPI 122
Query: 183 CRNVGI 188
C ++G+
Sbjct: 123 CGDIGV 128
>UniRef100_Q8S4Z1 Trypsin inhibitor [Theobroma sylvestre]
Length = 153
Score = 125 bits (313), Expect = 1e-27
Identities = 67/163 (41%), Positives = 98/163 (60%), Gaps = 15/163 (9%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G ++R GV+YY+ G G + + R+ ++CP +VV GF
Sbjct: 1 VLDTDGDELRTGVEYYVLSAIWG---------AGGGGLDLGRATGQSCP-EIVVQRGFND 50
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK--TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ + K GV+R+STD+NI+ + C ST+W LD++D STG+W+VTT GV
Sbjct: 51 DGIPVIFSNADGKDGVVRLSTDVNIEFVPIRDRLCLTSTVWKLDNYDHSTGKWWVTTDGV 110
Query: 146 LGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCKVMCRNVG 187
GNPG T+ +WFKIE YKF FCP+VC+ C +C ++G
Sbjct: 111 KGNPGHTTLTSWFKIENAGALGYKFRFCPSVCDSCATLCSDIG 153
>UniRef100_Q8RUR7 Trypsin inhibitor [Theobroma obovatum]
Length = 154
Score = 124 bits (311), Expect = 2e-27
Identities = 66/163 (40%), Positives = 97/163 (59%), Gaps = 14/163 (8%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G ++R GV YY+ V G G + + R+ ++ CP VV
Sbjct: 1 VLDTDGDELRTGVQYYVVSVIWG---------AGGGGLALGRATDQKCPEIVVQRRSDLD 51
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK--TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ + + V+RVSTD+NI+ + C ST+W LDD+D+S G+W+VTTGGV
Sbjct: 52 NGTPVIFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGV 111
Query: 146 LGNPGKDTVDNWFKIEKYEDD-YKFVFCPTVCNFCKVMCRNVG 187
G PG +T+ NWFKIE+ + YKF FCP+VC+ C +C ++G
Sbjct: 112 KGEPGPNTLSNWFKIEEAGGNVYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8S4Z0 Trypsin inhibitor [Theobroma bicolor]
Length = 154
Score = 124 bits (311), Expect = 2e-27
Identities = 71/163 (43%), Positives = 98/163 (59%), Gaps = 14/163 (8%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G+++R GV+YY+ + G G G + RS N++CP VV
Sbjct: 1 VLDTDGEELRTGVEYYV----VSALWG-----AGGGGLAPGRSRNQSCPDIVVQRRSDLD 51
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK-TSLNTS-CEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+PV P IRVSTDLNI+ T L S C + +W LDD+D STG+W+V GGV
Sbjct: 52 YGIPVIFSPVKPNDIFIRVSTDLNIEFTPLRDSLCLTTAVWKLDDYDQSTGKWWVIAGGV 111
Query: 146 LGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCKVMCRNVG 187
G+ G T+ NWFKIEK YKF++CP+VC+ C +C ++G
Sbjct: 112 AGDAGPHTLPNWFKIEKNGVLGYKFIYCPSVCDSCTTLCSDIG 154
>UniRef100_Q8S500 Trypsin inhibitor [Theobroma subincanum]
Length = 154
Score = 123 bits (309), Expect = 3e-27
Identities = 65/163 (39%), Positives = 96/163 (58%), Gaps = 14/163 (8%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G ++R GV YY+ G G + + R+ ++ CP VV
Sbjct: 1 VLDTDGDELRTGVQYYVMSAIWG---------AGGGGLALGRATDQKCPEIVVQRRSDLD 51
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK--TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ + + V+RVSTD+NI+ + C ST+W LDD+D+S G+W+VTTGGV
Sbjct: 52 NGTPVIFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGV 111
Query: 146 LGNPGKDTVDNWFKIEKYEDD-YKFVFCPTVCNFCKVMCRNVG 187
G PG +T+ NWFKIE+ + YKF FCP+VC+ C +C ++G
Sbjct: 112 KGEPGPNTLSNWFKIEEAGGNVYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8RVF3 Trypsin inhibitor [Theobroma grandiflorum]
Length = 154
Score = 121 bits (304), Expect = 1e-26
Identities = 64/163 (39%), Positives = 96/163 (58%), Gaps = 14/163 (8%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G ++R GV YY+ G G + + R+ ++ CP V+
Sbjct: 1 VLDTDGDELRTGVQYYVLSALWG---------AGGGGLDLGRATDQKCPEIVIQRRSDLD 51
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK--TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ + + V+RVSTD+NI+ + C ST+W LDD+D+S G+W+VTTGGV
Sbjct: 52 NGTPVIFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTGGV 111
Query: 146 LGNPGKDTVDNWFKIEKYEDD-YKFVFCPTVCNFCKVMCRNVG 187
G PG +T+ NWFKIE+ + YKF FCP+VC+ C +C ++G
Sbjct: 112 KGEPGPNTLSNWFKIEEAGGNVYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8S501 Trypsin inhibitor [Theobroma angustifolium]
Length = 154
Score = 120 bits (302), Expect = 2e-26
Identities = 67/163 (41%), Positives = 96/163 (58%), Gaps = 14/163 (8%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G ++R GV YY+ V T G G + + R+ N+ CP VV
Sbjct: 1 VLDTDGDELRTGVQYYV--VSTI-------WGAGGGGLDLGRATNQKCPEIVVQRRSDLD 51
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK--TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ + + V+RVSTD+NI+ + C ST+W LDD+D+S G+W+VTT GV
Sbjct: 52 NGTPVIFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTDGV 111
Query: 146 LGNPGKDTVDNWFKIEKYEDD-YKFVFCPTVCNFCKVMCRNVG 187
G PG +T+ NWFKIE+ YKF FCP+VC+ C +C ++G
Sbjct: 112 KGEPGPNTLSNWFKIEEAGGTLYKFRFCPSVCDSCATLCSDIG 154
>UniRef100_Q8S4Z8 Trypsin inhibitor [Theobroma mammosum]
Length = 154
Score = 120 bits (301), Expect = 2e-26
Identities = 67/163 (41%), Positives = 97/163 (59%), Gaps = 14/163 (8%)
Query: 30 VVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGFRG 89
V+DT+G ++R GV YYI + G G G + + R+ N+ CP VV
Sbjct: 1 VLDTDGDELRTGVQYYI----VSAIWG-----AGGGGLDLGRATNQKCPEIVVQRRSDLD 51
Query: 90 QG--VTFTPVNPKKGVIRVSTDLNIK--TSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ + + V+RVSTD+NI+ + C ST+W LDD+D+S G+W+VTT GV
Sbjct: 52 NGTPVIFSNADSEDDVVRVSTDINIEFVPIRDRLCSTSTVWKLDDYDNSAGKWWVTTDGV 111
Query: 146 LGNPGKDTVDNWFKIEKYEDD-YKFVFCPTVCNFCKVMCRNVG 187
G PG +T+ NWFKIE+ + YKF FCP+VC+ C +C ++G
Sbjct: 112 KGVPGPNTLSNWFKIEEAGGNIYKFRFCPSVCDSCTTLCSDIG 154
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 387,962,244
Number of Sequences: 2790947
Number of extensions: 17247053
Number of successful extensions: 37866
Number of sequences better than 10.0: 210
Number of HSP's better than 10.0 without gapping: 78
Number of HSP's successfully gapped in prelim test: 132
Number of HSP's that attempted gapping in prelim test: 37550
Number of HSP's gapped (non-prelim): 222
length of query: 213
length of database: 848,049,833
effective HSP length: 122
effective length of query: 91
effective length of database: 507,554,299
effective search space: 46187441209
effective search space used: 46187441209
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC140022.13


