

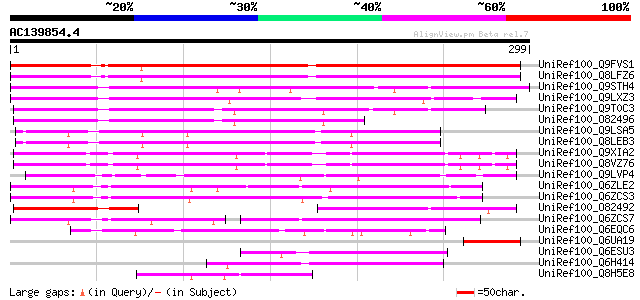
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139854.4 + phase: 0
(299 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FVS1 Hypothetical protein F12K22.16 [Arabidopsis tha... 243 5e-63
UniRef100_Q8LFZ6 Hypothetical protein [Arabidopsis thaliana] 233 5e-60
UniRef100_Q9STH4 Hypothetical protein T4C9.210 [Arabidopsis thal... 219 9e-56
UniRef100_Q9LXZ3 Hypothetical protein T5P19_120 [Arabidopsis tha... 199 1e-49
UniRef100_Q9T0C3 Hypothetical protein T4F9.120 [Arabidopsis thal... 198 1e-49
UniRef100_O82496 T12H20.15 protein [Arabidopsis thaliana] 140 5e-32
UniRef100_Q9LSA5 Gb|AAD43172.1 [Arabidopsis thaliana] 139 1e-31
UniRef100_Q8LEB3 Hypothetical protein [Arabidopsis thaliana] 137 5e-31
UniRef100_Q9XIA2 F13F21.21 protein [Arabidopsis thaliana] 127 4e-28
UniRef100_Q8VZ76 Hypothetical protein At1g49360; F13F21.21 [Arab... 127 4e-28
UniRef100_Q9LVP4 Gb|AAD43172.1 [Arabidopsis thaliana] 127 5e-28
UniRef100_Q6ZLE2 F-box protein family-like [Oryza sativa] 115 1e-24
UniRef100_Q6ZCS3 F-box protein family-like [Oryza sativa] 100 5e-20
UniRef100_O82492 T12H20.2 protein [Arabidopsis thaliana] 87 5e-16
UniRef100_Q6ZCS7 Hypothetical protein P0409A07.24 [Oryza sativa] 76 1e-12
UniRef100_Q6EQC6 Hypothetical protein P0448B03.3 [Oryza sativa] 53 9e-06
UniRef100_Q6UA19 Fiber protein Fb5 [Gossypium barbadense] 53 1e-05
UniRef100_Q6ESU3 Hypothetical protein P0521F09.4 [Oryza sativa] 48 3e-04
UniRef100_Q6H414 Hypothetical protein B1175F05.22 [Oryza sativa] 47 5e-04
UniRef100_Q8H5E8 Hypothetical protein OJ1165_F02.121 [Oryza sativa] 47 5e-04
>UniRef100_Q9FVS1 Hypothetical protein F12K22.16 [Arabidopsis thaliana]
Length = 352
Score = 243 bits (620), Expect = 5e-63
Identities = 129/298 (43%), Positives = 186/298 (62%), Gaps = 14/298 (4%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPE-LDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSL 59
MYFP+ N YDFYDP K Y++ELP+ L G V Y+KDGWLL++++D +H F L
Sbjct: 60 MYFPETKNTYDFYDPSNCKKYTMELPKSLVGFIVRYSKDGWLLMSQED-----SSH-FVL 113
Query: 60 FNPFTRDLITLPKFD--RTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWT 117
FNPFT D++ LP YQ+ FS APTS+ CV+ + V I T PG+ WT
Sbjct: 114 FNPFTMDVVALPFLHLFTYYQLVGFSSAPTSSECVVFTIKDYDPGHVTIRTWSPGQTMWT 173
Query: 118 TVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
++ +++ + +VFSNG+FYCL+ R + VFDP RTW V VPPP+C + K
Sbjct: 174 SMQVESQFLDVDHNNVVFSNGVFYCLNQRNHVAVFDPSLRTWNVLDVPPPRCPDD----K 229
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
+W++GKFM+ +KG+I V+ +DP++FKLDLT W+E +L +T+F S S SRT
Sbjct: 230 SWNEGKFMVGYKGDILVIRTYENKDPLVFKLDLTRGIWEEKDTLGSLTIFVSRKSCESRT 289
Query: 238 YA-TGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPPKD 294
Y G++RNSVYFP++ + K+ + +S D+ RY+ E D + + +N WIEPPK+
Sbjct: 290 YVKDGMLRNSVYFPELCYNEKQSVVYSFDEGRYHLREHDLDWGKQLSSDNIWIEPPKN 347
>UniRef100_Q8LFZ6 Hypothetical protein [Arabidopsis thaliana]
Length = 352
Score = 233 bits (594), Expect = 5e-60
Identities = 126/298 (42%), Positives = 179/298 (59%), Gaps = 14/298 (4%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPE-LDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSL 59
MYFP+ N YDFYDP K Y++ELP+ L G V Y+KDGWLL++++D +H F L
Sbjct: 60 MYFPETKNTYDFYDPSNCKKYTMELPKSLVGFIVRYSKDGWLLMSQED-----SSH-FVL 113
Query: 60 FNPFTRDLITLPKFD--RTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWT 117
FNPFT D++ LP YQ+ FS APTS+ CV+ + V I T PG+ WT
Sbjct: 114 FNPFTMDVVALPFLHLFTYYQLVGFSSAPTSSECVVFTIKDYDPGHVTIRTWSPGQTMWT 173
Query: 118 TVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
++ +++ + +VFSNG+FYCL+ R + VFDP RTW V VPPP+C + K
Sbjct: 174 SMQVESQFLDVDHNNVVFSNGVFYCLNQRNHVAVFDPXLRTWNVLDVPPPRCPDD----K 229
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
+W+ GKFM+ +KG+I + P+ FKLDLT W+E +L +T+F S S SRT
Sbjct: 230 SWNGGKFMVGYKGDILXIRTYENXXPLXFKLDLTRGIWEEKDTLGSLTIFVSRKSCESRT 289
Query: 238 YA-TGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPPKD 294
Y G++RN VYFP++ + K+ + +S D+ RY E D + + +N WIEPPK+
Sbjct: 290 YVKDGMLRNXVYFPELCYNEKQSVVYSFDEGRYXXREHDLDWGKQLSSDNIWIEPPKN 347
>UniRef100_Q9STH4 Hypothetical protein T4C9.210 [Arabidopsis thaliana]
Length = 300
Score = 219 bits (557), Expect = 9e-56
Identities = 122/309 (39%), Positives = 172/309 (55%), Gaps = 19/309 (6%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
MY PK+GN ++ YDP+ +K Y+L LPEL VCY++DGWLL+ + R + F
Sbjct: 1 MYLPKRGNLFELYDPLHQKMYTLNLPELAKSTVCYSRDGWLLMRKTISREM------FFF 54
Query: 61 NPFTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTT-- 118
NPFTR+LI +PK +Y AFSCAPTS CV+L F+ V + STC+P EW T
Sbjct: 55 NPFTRELINVPKCTLSYDAIAFSCAPTSGTCVLLAFKHVSYRITTTSTCHPKATEWVTED 114
Query: 119 VYYDAELSCSMCD--KLVFSNGLFYCLSDRGWLGVFDPLERTWT---VFKVPPPKCLAES 173
+ + + +V++ FYCL +G L FDP R W + +P P
Sbjct: 115 LQFHRRFRSETLNHSNVVYAKRRFYCLDGQGSLYYFDPSSRRWDFSYTYLLPCPYISDRF 174
Query: 174 STAKNWSKGK-FMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRS--LNGVTLFASF 230
S K + F+ KG F + C GE PI+ KL+ + W+E+ S ++G+T+F
Sbjct: 175 SYQYERKKKRIFLAVRKGVFFKIFTCDGEKPIVHKLED--INWEEINSTTIDGLTIFTGL 232
Query: 231 LSSHSRTYATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIE 290
SS R MRNSVYFP++RF KRC+S+SLD+ RYYP +Q +++ + EN WI
Sbjct: 233 YSSEVRLNLPW-MRNSVYFPRLRFNVKRCVSYSLDEERYYPRKQWQEQEDLCPIENLWIR 291
Query: 291 PPKDFTGWM 299
PPK +M
Sbjct: 292 PPKKAVDFM 300
>UniRef100_Q9LXZ3 Hypothetical protein T5P19_120 [Arabidopsis thaliana]
Length = 367
Score = 199 bits (505), Expect = 1e-49
Identities = 115/298 (38%), Positives = 163/298 (54%), Gaps = 25/298 (8%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
+YF K + Y+ YDP +K +L PEL G RVCY+KDGWLL+ + +L F
Sbjct: 76 IYFSKTDDSYELYDPSMQKNCNLHFPELSGFRVCYSKDGWLLMYNPNSYQL------LFF 129
Query: 61 NPFTRDLITLPKFDRTY-QIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTV 119
NPFTRD + +P Y Q AFSCAPTST C++ V + + I T + KEW T
Sbjct: 130 NPFTRDCVPMPTLWMAYDQRMAFSCAPTSTSCLLFTVTSVTWNYITIKTYFANAKEWKTS 189
Query: 120 YYDAEL--SCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
+ L + + +++VFSNG+FYCL++ G L +FDP W V PPK +
Sbjct: 190 VFKNRLQRNFNTFEQIVFSNGVFYCLTNTGCLALFDPSLNYWNVLPGRPPK--------R 241
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
S G FM EH+G IF++++ +P + KLDLT EW E ++L G+T++AS LSS SR
Sbjct: 242 PGSNGCFMTEHQGEIFLIYMYRHMNPTVLKLDLTSFEWAERKTLGGLTIYASALSSESRA 301
Query: 238 ---YATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPP 292
+GI N + + + CI + +D+ SE C + N +EN WI PP
Sbjct: 302 EQQKQSGIW-NCLCLSVFHGFKRTCIYYKVDEE----SEVCFKWKKQNPYENIWIMPP 354
>UniRef100_Q9T0C3 Hypothetical protein T4F9.120 [Arabidopsis thaliana]
Length = 317
Score = 198 bits (504), Expect = 1e-49
Identities = 117/283 (41%), Positives = 155/283 (54%), Gaps = 24/283 (8%)
Query: 3 FPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNP 62
FP + +DP++RK Y+L LPEL G VCY+KDGWLL+ R + FNP
Sbjct: 24 FPNHEDLTFLFDPLERKRYTLNLPELVGTDVCYSKDGWLLMRRSSLVDM------FFFNP 77
Query: 63 FTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTVYYD 122
+TR+LI LPK + +Q AFS APTS CV+L R ++ IS CYPG EW T
Sbjct: 78 YTRELINLPKCELAFQAIAFSSAPTSGTCVVLALRPFTRYIIRISICYPGATEWIT---- 133
Query: 123 AELSCS------MCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTA 176
E SCS M LV++N FYC S G L FD RT + +C +
Sbjct: 134 QEFSCSLRFDPYMHSNLVYANDHFYCFSSGGVLVDFDVASRTMSEQAWNEHRCAYMRNDN 193
Query: 177 KNW---SKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRS--LNGVTLFASFL 231
W K ++ E KG +F+++ C E P+++K L W+E+ S L+GVT+FAS
Sbjct: 194 AEWFNLPKRIYLAEQKGELFLMYTCSSEIPMVYK--LVSSNWEEINSTTLDGVTIFASMY 251
Query: 232 SSHSRTYATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQ 274
SS +R G MRNSVYFPK K C+S+S D+ RYYP +Q
Sbjct: 252 SSETRLDVLG-MRNSVYFPKYGLDCKGCVSYSFDEARYYPRKQ 293
>UniRef100_O82496 T12H20.15 protein [Arabidopsis thaliana]
Length = 705
Score = 140 bits (352), Expect = 5e-32
Identities = 82/211 (38%), Positives = 109/211 (50%), Gaps = 19/211 (9%)
Query: 3 FPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNP 62
FP + +DP++RK Y+L LPEL G VCY+KDGWLL+ R + FNP
Sbjct: 24 FPNHEDLTFLFDPLERKRYTLNLPELVGTDVCYSKDGWLLMRRSSLVDM------FFFNP 77
Query: 63 FTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTVYYD 122
+TR+LI LPK + +Q AFS APTS CV+L R ++ IS CYPG EW T
Sbjct: 78 YTRELINLPKCELAFQAIAFSSAPTSGTCVVLALRPFTRYIIRISICYPGATEWIT---- 133
Query: 123 AELSCS------MCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTA 176
E SCS M LV++N FYC S G L FD RT + +C +
Sbjct: 134 QEFSCSLRFDPYMHSNLVYANDHFYCFSSGGVLVDFDVASRTMSEQAWNEHRCAYMRNDN 193
Query: 177 KNW---SKGKFMIEHKGNIFVVHICCGEDPI 204
W K ++ E KG +F+++ C E P+
Sbjct: 194 AEWFNLPKRIYLAEQKGELFLMYTCSSEIPM 224
>UniRef100_Q9LSA5 Gb|AAD43172.1 [Arabidopsis thaliana]
Length = 380
Score = 139 bits (349), Expect = 1e-31
Identities = 90/253 (35%), Positives = 133/253 (51%), Gaps = 18/253 (7%)
Query: 4 PKKGNCYDFYDPVQRKTYSLELPELDGCR--VCYTKDGWLLLNRQDWRRLDGNHIFSLFN 61
PK+G+ Y ++P + +T+ L+ PEL G R + KDGWLL+ + D + N
Sbjct: 105 PKEGD-YVLFNPSRSQTHHLKFPELTGYRNKLACAKDGWLLVVK------DNPDVVFFLN 157
Query: 62 PFTRDLITLPKFDR--TYQIAAFSCAPTSTGCVILIFRRVGS--SLVAISTCYPGEKEWT 117
PFT + I LP+ + T FS APTST C ++ F ++V + T PGE WT
Sbjct: 158 PFTGERICLPQVPQNSTRDCLTFSAAPTSTSCCVISFTPQSFLYAVVKVDTWRPGESVWT 217
Query: 118 TVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
T ++D + + ++ +FSNG+FYCLS G L VFDP TW V V P +
Sbjct: 218 THHFDQKRYGEVINRCIFSNGMFYCLSTSGRLSVFDPSRETWNVLPVKPCRAFRRKIML- 276
Query: 178 NWSKGKFMIEHKGNIFVV--HICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHS 235
+ FM EH+G+IFVV + FKL+L W+E++ NG+T+F+S +S +
Sbjct: 277 --VRQVFMTEHEGDIFVVTTRRVNNRKLLAFKLNLQGNVWEEMKVPNGLTVFSSDATSLT 334
Query: 236 RTYATGIMRNSVY 248
R RN +Y
Sbjct: 335 RAGLPEEERNILY 347
>UniRef100_Q8LEB3 Hypothetical protein [Arabidopsis thaliana]
Length = 380
Score = 137 bits (344), Expect = 5e-31
Identities = 89/253 (35%), Positives = 132/253 (51%), Gaps = 18/253 (7%)
Query: 4 PKKGNCYDFYDPVQRKTYSLELPELDGCR--VCYTKDGWLLLNRQDWRRLDGNHIFSLFN 61
PK+G+ Y ++P + +T+ L+ PEL G R + KDGWLL+ + D + N
Sbjct: 105 PKEGD-YVLFNPSRSQTHHLKFPELTGYRNKLACAKDGWLLVVK------DNPDVVFFLN 157
Query: 62 PFTRDLITLPKFDR--TYQIAAFSCAPTSTGCVILIFRRVGS--SLVAISTCYPGEKEWT 117
PFT + I LP+ + T FS APTST C ++ F ++V + T PGE WT
Sbjct: 158 PFTGERICLPQVPQNSTRDCLTFSAAPTSTSCCVISFTPQSFLYAVVKVDTWRPGESVWT 217
Query: 118 TVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAK 177
T ++D + + ++ +FSNG+FYCLS G L FDP TW V V P +
Sbjct: 218 THHFDQKRYGEVINRCIFSNGMFYCLSTSGRLSFFDPSRETWNVLPVKPCRAFRRKIML- 276
Query: 178 NWSKGKFMIEHKGNIFVV--HICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHS 235
+ FM EH+G+IFVV + FKL+L W+E++ NG+T+F+S +S +
Sbjct: 277 --VRQVFMTEHEGDIFVVTTRRVNNRKLLAFKLNLQGNVWEEMKVPNGLTVFSSDATSLT 334
Query: 236 RTYATGIMRNSVY 248
R RN +Y
Sbjct: 335 RAGLPEEERNILY 347
>UniRef100_Q9XIA2 F13F21.21 protein [Arabidopsis thaliana]
Length = 481
Score = 127 bits (319), Expect = 4e-28
Identities = 105/314 (33%), Positives = 149/314 (47%), Gaps = 40/314 (12%)
Query: 3 FPKKGNCYDFYDPVQRK-TYSLELPELD-GCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
F KG Y F+DPV++K T + LPEL + Y+KDGWLL+N D L + F F
Sbjct: 160 FQDKGVSYGFFDPVEKKKTKEMNLPELSKSSGILYSKDGWLLMN--DSLSLIADMYF--F 215
Query: 61 NPFTRDLITLPK---FDRTYQIAAFSCAPTSTGCVILIFRRVGSSL-VAISTCYPGEKEW 116
NPFTR+ I LP+ + + AFSCAPT C++ + SS+ + IST PG W
Sbjct: 216 NPFTRERIDLPRNRIMESVHTNFAFSCAPTKKSCLVFGINNISSSVAIKISTWRPGATTW 275
Query: 117 TTVYYDAELSCSM--CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESS 174
+ +++S+GLFY S+ LGVFDP RTW V V P S
Sbjct: 276 LHEDFPNLFPSYFRRLGNILYSDGLFYTASETA-LGVFDPTARTWNVLPVQPIPMAPRSI 334
Query: 175 TAKNWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSH 234
++M E++G+IF+V +P++++L+ W++ +L+G ++F S S
Sbjct: 335 --------RWMTEYEGHIFLVD-ASSLEPMVYRLNRLESVWEKKETLDGSSIFLSDGSCV 385
Query: 235 SRTYATGIMRNSVYFPKVRFYGKR------CISFSLDDRRY--YPSEQCRDKVEPNTFE- 285
TG M N +YF RF +R C +Y Y C D E FE
Sbjct: 386 MTYGLTGSMSNILYFWS-RFINERRSTKSPCPFSRNHPYKYSLYSRSSCEDP-EGYYFEY 443
Query: 286 -------NFWIEPP 292
WIEPP
Sbjct: 444 LTWGQKVGVWIEPP 457
>UniRef100_Q8VZ76 Hypothetical protein At1g49360; F13F21.21 [Arabidopsis thaliana]
Length = 378
Score = 127 bits (319), Expect = 4e-28
Identities = 105/314 (33%), Positives = 149/314 (47%), Gaps = 40/314 (12%)
Query: 3 FPKKGNCYDFYDPVQRK-TYSLELPELD-GCRVCYTKDGWLLLNRQDWRRLDGNHIFSLF 60
F KG Y F+DPV++K T + LPEL + Y+KDGWLL+N D L + F F
Sbjct: 57 FQDKGVSYGFFDPVEKKKTKEMNLPELSKSSGILYSKDGWLLMN--DSLSLIADMYF--F 112
Query: 61 NPFTRDLITLPK---FDRTYQIAAFSCAPTSTGCVILIFRRVGSSL-VAISTCYPGEKEW 116
NPFTR+ I LP+ + + AFSCAPT C++ + SS+ + IST PG W
Sbjct: 113 NPFTRERIDLPRNRIMESVHTNFAFSCAPTKKSCLVFGINNISSSVAIKISTWRPGATTW 172
Query: 117 TTVYYDAELSCSM--CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESS 174
+ +++S+GLFY S+ LGVFDP RTW V V P S
Sbjct: 173 LHEDFPNLFPSYFRRLGNILYSDGLFYTASETA-LGVFDPTARTWNVLPVQPIPMAPRSI 231
Query: 175 TAKNWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSH 234
++M E++G+IF+V +P++++L+ W++ +L+G ++F S S
Sbjct: 232 --------RWMTEYEGHIFLVD-ASSLEPMVYRLNRLESVWEKKETLDGSSIFLSDGSCV 282
Query: 235 SRTYATGIMRNSVYFPKVRFYGKR------CISFSLDDRRY--YPSEQCRDKVEPNTFE- 285
TG M N +YF RF +R C +Y Y C D E FE
Sbjct: 283 MTYGLTGSMSNILYFWS-RFINERRSTKSPCPFSRNHPYKYSLYSRSSCEDP-EGYYFEY 340
Query: 286 -------NFWIEPP 292
WIEPP
Sbjct: 341 LTWGQKVGVWIEPP 354
>UniRef100_Q9LVP4 Gb|AAD43172.1 [Arabidopsis thaliana]
Length = 412
Score = 127 bits (318), Expect = 5e-28
Identities = 89/290 (30%), Positives = 141/290 (47%), Gaps = 25/290 (8%)
Query: 10 YDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNPFTRDLIT 69
Y +DP++ ++Y E P L + K+GWLL+++ D+ +F L NPFTR+ +
Sbjct: 127 YILFDPLRNQSYKQEFPGLGCHKFLSYKNGWLLVSKYDYP----GALFFL-NPFTRERVY 181
Query: 70 LPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTVYYDAELSCSM 129
LP AFS APTST C++ + I T GE WTT + +
Sbjct: 182 LPLLVPC--CGAFSAAPTSTSCLVACV----NYYFEIMTWRLGETVWTTNCFGNHIRGRK 235
Query: 130 CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPP-----PKCLAESSTAKNWSKGKF 184
DK VFSNG+F+CLS G+L VFDP TW + V P CLA + +
Sbjct: 236 WDKCVFSNGMFFCLSTCGYLRVFDPHRATWNILPVKPCILPLEPCLAFRQPSLMGMR-VL 294
Query: 185 MIEHKGNIFVVHICC--GEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRTYATGI 242
M+EH+G+++V+ +FKL+L W+E + L G+T+FA + +S +R +
Sbjct: 295 MMEHEGDVYVISTFSNYNNQASVFKLNLKREVWEEKKELGGLTVFAGYHTSLTRASLLAL 354
Query: 243 MRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQCRDKVEPNTFENFWIEPP 292
RN++Y + + +SL ++ +C + W++PP
Sbjct: 355 DRNNMY---TSHAARSFVYYSLAGNKF---SRCPPGCNYLSDRFAWVDPP 398
>UniRef100_Q6ZLE2 F-box protein family-like [Oryza sativa]
Length = 710
Score = 115 bits (288), Expect = 1e-24
Identities = 83/286 (29%), Positives = 134/286 (46%), Gaps = 22/286 (7%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCY----TKDGWLLLNRQDWRRLDGNHI 56
M+ K+ +DP++ + Y++++ D + +KDGW+ + + G+ I
Sbjct: 407 MHISKQDGMCRLFDPLRGEVYNMQVSIFDTNEDRHIFRSSKDGWVFTSAGIY----GHDI 462
Query: 57 FSLFNPFTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSL----VAISTCYPG 112
F + NPFT D++ P F+R Y S + + C F + SSL + I T
Sbjct: 463 F-IINPFTEDIVEPPMFERRYHYNGVSFSSPNPMCPNCYFFGINSSLSGKFLNIHTWRHE 521
Query: 113 EKEWTT--VYYDAELSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCL 170
E EW YD + ++F G FYCL +G LG FDP TW + P P
Sbjct: 522 ETEWIEQRFEYDVPFPVGYNNPVMFC-GKFYCLGRKGNLGAFDPTSNTWEILDKPEP-IH 579
Query: 171 AESSTAKNWSKGK---FMIEHKGNIFVVHIC-CGEDPIIFKLDLTLMEWKEVRSLNGVTL 226
E +N +G+ ++++ +G + V + E P +FKLDLT M W EV + G L
Sbjct: 580 VEMDLLQNDHRGREFCYLVDLEGELISVLLHNASEAPRVFKLDLTKMSWVEVEDIGGGAL 639
Query: 227 FASFLSSHSRTYATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPS 272
F +SH G N +YFP+ ++ + + +D++ YYPS
Sbjct: 640 FLDHRTSHGVGSPDGGHGNRIYFPRYS-VDRKPVFYDMDNKMYYPS 684
Score = 42.4 bits (98), Expect = 0.015
Identities = 52/200 (26%), Positives = 84/200 (42%), Gaps = 30/200 (15%)
Query: 85 APTSTGCVILIFRRVGSSLVA-ISTCYPGEKEWTTV---YYDAELSCSMCDKLVFSNGLF 140
+P S C ILI + + C PGE EWT + + D LS MC+ + +
Sbjct: 104 SPDSPKCTILIASSPEVEEESYLLYCRPGEDEWTKLVSPFNDIHLSAFMCN---YEGKIC 160
Query: 141 YCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAKNWSKGKFMIEHKGNIFVVHI--- 197
S+ + + D + V + + A S + ++E G +F++ I
Sbjct: 161 SACSNLVVIDMVDGKIQLQRVGTIKDEEKYARGSGCYH------VVESCGKLFLLWIEEL 214
Query: 198 -CCGEDPI-----IFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRTYA----TGIMR-NS 246
C G D + +F LDL LM W+ V S+ +FL S + T++ G+++ N
Sbjct: 215 GCFGNDGLLTAIDVFCLDLELMSWERVESIGS---DRTFLISENYTFSCPSIEGVLQGNC 271
Query: 247 VYFPKVRFYGKRCISFSLDD 266
VY +R F LDD
Sbjct: 272 VYLVWSSCDSERLYKFCLDD 291
>UniRef100_Q6ZCS3 F-box protein family-like [Oryza sativa]
Length = 712
Score = 100 bits (249), Expect = 5e-20
Identities = 75/287 (26%), Positives = 130/287 (45%), Gaps = 24/287 (8%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCRVCY----TKDGWLLLNRQDWRRLDGNHI 56
M+ K+ +DP++ + Y++++ D + +KDGW+L + + GN I
Sbjct: 409 MHISKQDGTCKLFDPLRSENYNIQVTIFDTNEDRHIFRSSKDGWVLASAGIY----GNDI 464
Query: 57 FSLFNPFTRDLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSS----LVAISTCYPG 112
F + NPFT +++ P Y S + ++ C+ F + SS ++ T G
Sbjct: 465 F-IINPFTEEIVEPPMLAFLYNYNGVSFSSSNPMCLDCAFFGINSSDSGKFLSTFTWQHG 523
Query: 113 EKEWTTVYYDAELSCSM-CDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPP---- 167
E W ++ +S + + V +G FYCL +G LGVFDP TW + P P
Sbjct: 524 EPHWIEQEFEYNVSFPVGYNNPVMFDGKFYCLGRKGNLGVFDPTSNTWRILDKPEPIHVE 583
Query: 168 -KCLAESSTAKNWSKGKFMIEHKGNIFVVHI-CCGEDPIIFKLDLTLMEWKEVRSLNGVT 225
E + + ++++ G + V + E P +FKL+ T + W EV + G
Sbjct: 584 MDLFEEDHIGREFC---YLVDMDGELISVFLRNANELPRVFKLNRTEISWVEVEDIGGGA 640
Query: 226 LFASFLSSHSRTYATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPS 272
LF + SS+ G N +YFP+ GK + + ++ + Y PS
Sbjct: 641 LFLDYRSSYGVASPDGGNGNRIYFPRYSKDGKP-VFYDMNKKTYSPS 686
Score = 36.2 bits (82), Expect = 1.1
Identities = 73/290 (25%), Positives = 115/290 (39%), Gaps = 52/290 (17%)
Query: 6 KGNCYDFYDPVQRKTYSLELPELDG--CRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNPF 63
+G+ F D + +PE+ G C C D WLL+ D D + NP
Sbjct: 25 RGDTLTFVDVSDLSLHETVVPEVRGKTCLGCMHGD-WLLM--LDESTADCFLLRITTNPR 81
Query: 64 TR----DLITLPKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCY-----PGEK 114
T+ L P F TY++ +P S C IL V SS A CY PG++
Sbjct: 82 TKIQLPPLHQPPLFLSTYEMLE---SPESANCTIL----VASSTEAEEECYLLHCHPGDE 134
Query: 115 EWTTVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLG-VFDPLERTWTVFKVPPPKCLAES 173
WT S S D + FS+ + R + G ++D + + L +
Sbjct: 135 MWTK-------SVSPYDDISFSSLM------RNYGGKIYDFASNLIAIDVIDGKIELQQL 181
Query: 174 STAKNWSK-----GKF-MIEHKGNIFVVHI----CCGEDPI-----IFKLDLTLMEWKEV 218
T K+ + G++ +IE G +F++ I C +D + +F L+L + W+ V
Sbjct: 182 GTIKDEEEDSRRCGRYHIIESCGKLFLLWIDDLGCFYDDGLLTAIRVFCLNLETLSWERV 241
Query: 219 RSL-NGVTLFASFLSSHSRTYATGIMR-NSVYFPKVRFYGKRCISFSLDD 266
+ N S + S G+++ N VY +R F LDD
Sbjct: 242 EGIGNDRAFLISGTYAFSCPSIEGVLQGNCVYLVWSSCDSERLYKFCLDD 291
>UniRef100_O82492 T12H20.2 protein [Arabidopsis thaliana]
Length = 247
Score = 87.0 bits (214), Expect = 5e-16
Identities = 48/121 (39%), Positives = 70/121 (57%), Gaps = 7/121 (5%)
Query: 178 NWSKGKFMIEHKGNIFVVHICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRT 237
N K ++ E KG +F+++ C E P+++KL + +E +L+GVT+FAS SS +R
Sbjct: 119 NLPKRIYLAEQKGELFLMYTCSSEIPMVYKLVSSNLEEMNSTTLDGVTIFASMYSSETRL 178
Query: 238 YATGIMRNSVYFPKVRFYGKRCISFSLDDRRYYPSEQ------CRDKVEPNTFENFWIEP 291
G MRNSVYFPK K C+S+S D+ RYYP +Q + + E + WIEP
Sbjct: 179 DVLG-MRNSVYFPKYGLDCKGCVSYSFDEARYYPRKQFPKPTMWQMQKELCPLRSLWIEP 237
Query: 292 P 292
P
Sbjct: 238 P 238
Score = 70.9 bits (172), Expect = 4e-11
Identities = 34/72 (47%), Positives = 45/72 (62%), Gaps = 6/72 (8%)
Query: 3 FPKKGNCYDFYDPVQRKTYSLELPELDGCRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNP 62
FP + +DP++RK Y+L LPE+ G VCY+KDGWLL+ R + FNP
Sbjct: 24 FPNHEDMTFLFDPLERKRYTLNLPEVVGTDVCYSKDGWLLMRRSSLVDM------FFFNP 77
Query: 63 FTRDLITLPKFD 74
+TR+LI LPKFD
Sbjct: 78 YTRELINLPKFD 89
>UniRef100_Q6ZCS7 Hypothetical protein P0409A07.24 [Oryza sativa]
Length = 760
Score = 76.3 bits (186), Expect = 1e-12
Identities = 44/139 (31%), Positives = 68/139 (48%), Gaps = 2/139 (1%)
Query: 134 VFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAKNWSKGKFMIEHKGNIF 193
V+ G FY L RG L VF+P W + P P A+ + + +M+E +G +
Sbjct: 599 VYFRGEFYFLGQRGNLSVFNPGNNEWRILDKPEP-IRADLTPYDEGKEACYMVELRGELI 657
Query: 194 VV-HICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRTYATGIMRNSVYFPKV 252
V H E P + KLD + MEW E+ + G LF + +S + + N +YFPK
Sbjct: 658 AVFHRNANEPPRVLKLDESKMEWVEIEDIGGGALFLDYRASIALPSSEAGHGNRIYFPKF 717
Query: 253 RFYGKRCISFSLDDRRYYP 271
GK+ I + L+ ++Y P
Sbjct: 718 SEDGKKAIFYDLEAKKYSP 736
Score = 49.7 bits (117), Expect = 1e-04
Identities = 35/132 (26%), Positives = 64/132 (47%), Gaps = 15/132 (11%)
Query: 1 MYFPKKGNCYDFYDPVQRKTYSLELPELDGCR---VCYTKDGWLLLNRQDWRRLDGNHIF 57
M+ ++ +DP+ YS+++ D ++KDGW+++ + D ++IF
Sbjct: 405 MHCVRQDGACKMFDPLCGVEYSMKVGPFDANERQAFRFSKDGWVIVTQSD------DNIF 458
Query: 58 SLFNPFTRDLITLPKFDRTYQIA--AFSCAPTSTGCVIL-IFRRVGSSLVAISTCYPGEK 114
+ NPFT++++ L Y+ +FS PTS CV L + + + TC P E+
Sbjct: 459 -VINPFTKEIVKLSMASGWYRFTGISFSSVPTSPDCVFLGVCSSPKGDGIKVWTCRPNEE 517
Query: 115 EW--TTVYYDAE 124
E +YY+ E
Sbjct: 518 ETEDNEIYYEEE 529
Score = 40.0 bits (92), Expect = 0.076
Identities = 69/271 (25%), Positives = 106/271 (38%), Gaps = 28/271 (10%)
Query: 12 FYDPVQRKTYSLELPELDG--CRVCYTKDGWLLLNRQDWRRLDGNHIFSLFNPFTRDLIT 69
F D + R + +P+L G C C WLL+ + G +F L N T L
Sbjct: 35 FVDILDRSLHVRVVPDLQGKLCLGCVHGGDWLLMVDEI---TGGCFLFCLSNSSTISLPP 91
Query: 70 L--PKFDRTYQIAAFSCAPTSTGCVILIFRRVGSSLVAISTCYPGEKEWTTVYYDAELSC 127
L P D + S +P + C ++I + C+PG++EWT + S
Sbjct: 92 LREPLGDMGACVVLGS-SPLNRDCTVVITSLPEPEESFLLHCHPGDEEWTKLMVPLG-SD 149
Query: 128 SMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESSTAKNWSKGKFMIE 187
+ KLV G Y LS L D ++ ESS N+ +++E
Sbjct: 150 RLFGKLVNCAGQLYSLSSFRKLLTIDVID---DALHAKILNIEWESSCGHNFE--PYIVE 204
Query: 188 HKGNIFVV--------HICCGEDPIIFKLDLTLMEWKEVRSLNGVTLFASFLSSH---SR 236
G +FVV + C +++LD K+V + T A +S H S
Sbjct: 205 SCGKLFVVLASLYGYPYNCPLNGVSVYRLDRAESMLKKVDDIG--TDRAFLISGHYGFSC 262
Query: 237 TYATGIMR-NSVYFPKVRFYGKRCISFSLDD 266
T G+++ N VY + +R F LDD
Sbjct: 263 TAMEGLVQGNCVYIVWSGYDCERIYKFCLDD 293
>UniRef100_Q6EQC6 Hypothetical protein P0448B03.3 [Oryza sativa]
Length = 384
Score = 53.1 bits (126), Expect = 9e-06
Identities = 61/237 (25%), Positives = 102/237 (42%), Gaps = 35/237 (14%)
Query: 36 TKDGWLLLNRQDWRRLDGNHIFSLFNPFTRDLITLP----KFDRT-YQIAAFSCAPTSTG 90
T GWLL+ +D +H L+NPFT I+LP +F RT Y SC PT
Sbjct: 124 TPQGWLLMVHRD------SHETFLWNPFTSQRISLPFDQDRFLRTNYTRCLLSCKPTDIN 177
Query: 91 CVILIFRRVGSSLVAISTCYPGEKEWTTVYYDAELSCSMCDKLVFSNGLFYCLSDRGWLG 150
CV+L+ + + I CYPG +W Y + ++ L + + + G
Sbjct: 178 CVVLV---LSLNDTVIWYCYPGGTQWFKHEYQSRRFHRHRGSVIGYMALLTVVGGKFYTG 234
Query: 151 VFDPLERTWTVFKVPPPK-----CLAESSTAKNWSKGKFMIEHKGNIFVVH----ICCGE 201
+ D + T+ P PK A + N+S+ +++E G +F + + C +
Sbjct: 235 LGDSV---ITLDFSPNPKFDIIPIKAVQNPMYNFSR-LYLLESSGELFSLFFYPPMTCPK 290
Query: 202 ---DPIIFKLDLTLMEWKEVRSLNGVTLFAS----FLSSHSRTYATGIMRNSVYFPK 251
+ ++KLD+ W +V +L F + F +S S A + N +YF +
Sbjct: 291 RIAEIEVYKLDIPRRAWVKVYTLGDRAFFVNSTKCFGASVSAKEAC-LEENCIYFSR 346
>UniRef100_Q6UA19 Fiber protein Fb5 [Gossypium barbadense]
Length = 37
Score = 52.8 bits (125), Expect = 1e-05
Identities = 20/33 (60%), Positives = 26/33 (78%)
Query: 262 FSLDDRRYYPSEQCRDKVEPNTFENFWIEPPKD 294
+S+DDRRYYP ++C D E + FEN WIEPP+D
Sbjct: 1 YSVDDRRYYPRKECHDWGEQDPFENIWIEPPRD 33
>UniRef100_Q6ESU3 Hypothetical protein P0521F09.4 [Oryza sativa]
Length = 724
Score = 48.1 bits (113), Expect = 3e-04
Identities = 35/122 (28%), Positives = 55/122 (44%), Gaps = 10/122 (8%)
Query: 134 VFSNGLFYCLSDRGWLGVFDPL--ERTWTVFKVPPPKCLAESSTAKNWSKGKFMIEHKGN 191
VF G+ Y L G L ++DP ER + + P S + +G
Sbjct: 540 VFHGGMLYLLGKDGRLALYDPCNHERGFEIHDKP-------ESFGFETDDSYLVESEQGE 592
Query: 192 IFVVHICCGEDPI-IFKLDLTLMEWKEVRSLNGVTLFASFLSSHSRTYATGIMRNSVYFP 250
+ + + P+ I KL+ M+W+EV SL G TLF L++ R+ M+N V+ P
Sbjct: 593 LMAILVGRRGTPVHIVKLNEEAMKWEEVESLQGRTLFTGTLTTMMRSVKIKWMQNKVFLP 652
Query: 251 KV 252
K+
Sbjct: 653 KL 654
>UniRef100_Q6H414 Hypothetical protein B1175F05.22 [Oryza sativa]
Length = 383
Score = 47.4 bits (111), Expect = 5e-04
Identities = 39/141 (27%), Positives = 59/141 (41%), Gaps = 8/141 (5%)
Query: 114 KEWTTVYYDAE---LSCSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCL 170
++WT YDA L S VF G Y L + G L V+DP + + P
Sbjct: 168 EDWTLTEYDANGPRLRPSPVTNPVFHRGSIYLLGEHGRLAVYDPCKHAEGFKILDKPM-- 225
Query: 171 AESSTAKNWSKGKFMIEHKGNIFVVHICCGEDPI-IFKLDLTLMEWKEVRSLNGVTLFAS 229
S + + + +G + V P+ + L+ MEW++V SL G TLF
Sbjct: 226 --SFGFEQYHDSYLVESDQGELMAVLFGRRGTPVHVIVLNKERMEWEQVESLQGRTLFTG 283
Query: 230 FLSSHSRTYATGIMRNSVYFP 250
L+S + M+N V+ P
Sbjct: 284 TLTSMVKKTKFKWMQNRVFLP 304
>UniRef100_Q8H5E8 Hypothetical protein OJ1165_F02.121 [Oryza sativa]
Length = 625
Score = 47.4 bits (111), Expect = 5e-04
Identities = 31/108 (28%), Positives = 46/108 (41%), Gaps = 8/108 (7%)
Query: 74 DRTYQIAAFSCAPTSTGCVILIFRRVGSSL-----VAISTCYPGEKEWTTVYYD--AELS 126
D + S APTS C+ + + V ++ G+++WT D
Sbjct: 482 DNQFDGICLSAAPTSPDCIAFSVEKDRNPTGRNRSVYVTLWRAGDEQWTMQRIDDHTPFR 541
Query: 127 CSMCDKLVFSNGLFYCLSDRGWLGVFDPLERTWTVFKVPPPKCLAESS 174
+ C+ VF +G FYCL RG L VF+P TW V P L + +
Sbjct: 542 TAYCNP-VFYDGEFYCLGTRGGLAVFNPNNTTWRVLDKPEAPVLVDGN 588
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.142 0.476
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 570,197,314
Number of Sequences: 2790947
Number of extensions: 23898741
Number of successful extensions: 51235
Number of sequences better than 10.0: 43
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 51133
Number of HSP's gapped (non-prelim): 50
length of query: 299
length of database: 848,049,833
effective HSP length: 126
effective length of query: 173
effective length of database: 496,390,511
effective search space: 85875558403
effective search space used: 85875558403
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC139854.4


