

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.9 + phase: 0
(500 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
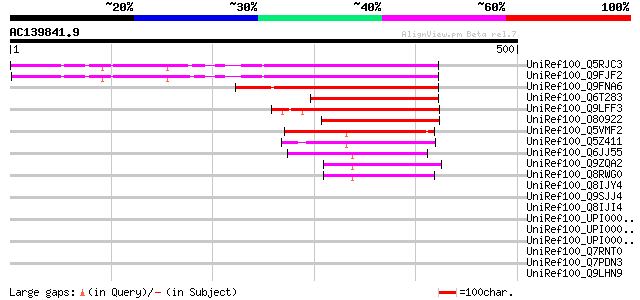
UniRef100_Q5RJC3 At5g59830 [Arabidopsis thaliana] 308 2e-82
UniRef100_Q9FJF2 Arabidopsis thaliana genomic DNA, chromosome 5,... 308 2e-82
UniRef100_Q9FNA6 Arabidopsis thaliana genomic DNA, chromosome 5,... 183 1e-44
UniRef100_Q6T283 Predicted protein [Populus alba x Populus tremula] 148 4e-34
UniRef100_Q9LFF3 Hypothetical protein F4P12_380 [Arabidopsis tha... 142 3e-32
UniRef100_O80922 Hypothetical protein At2g37520 [Arabidopsis tha... 130 9e-29
UniRef100_Q5VMF2 PHD zinc finger protein-like [Oryza sativa] 97 8e-19
UniRef100_Q5Z411 PHD zinc finger protein-like [Oryza sativa] 94 1e-17
UniRef100_Q6JJ55 Putative PHD zinc finger protein [Ipomoea trifida] 90 2e-16
UniRef100_Q9ZQA2 Putative PHD-type zinc finger protein [Arabidop... 75 3e-12
UniRef100_Q8RWG0 Putative PHD-type zinc finger protein [Arabidop... 74 1e-11
UniRef100_Q8IJY4 Regulator of nonsense transcripts, putative [Pl... 45 0.004
UniRef100_Q9SJJ4 Hypothetical protein At2g27980 [Arabidopsis tha... 41 0.070
UniRef100_Q8IJI4 10b antigen, putative [Plasmodium falciparum] 40 0.12
UniRef100_UPI00004671EF UPI00004671EF UniRef100 entry 40 0.20
UniRef100_UPI00002EA680 UPI00002EA680 UniRef100 entry 39 0.27
UniRef100_UPI00003C203D UPI00003C203D UniRef100 entry 38 0.59
UniRef100_Q7RNT0 Plant adhesion molecule 1-related [Plasmodium y... 37 1.0
UniRef100_Q7PDN3 Erythrocyte membrane protein PFEMP3 [Plasmodium... 37 1.3
UniRef100_Q9LHN9 Emb|CAB68186.1 [Arabidopsis thaliana] 37 1.7
>UniRef100_Q5RJC3 At5g59830 [Arabidopsis thaliana]
Length = 425
Score = 308 bits (790), Expect = 2e-82
Identities = 185/428 (43%), Positives = 249/428 (57%), Gaps = 38/428 (8%)
Query: 1 MVKGSGHVSDREPAFDNPSKIEPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNV 60
++K + H S+ + +D+ ++ + KRPH W +D++ + PNKKQA++D G SNV
Sbjct: 10 VMKNNEHTSEEDSVYDHSTRDDSKRPHPWFVDSSRSEMFPNKKQAVQDPVV--GLGKSNV 67
Query: 61 NFTPWENNHNFHSDPSHQNQLIDRLFGSET--RPVNFTEKNTYVSGDGSDVRSKMIANHY 118
WE++ F S NQ +DRL G+E RP+ F +++ G ++K IA Y
Sbjct: 68 GLPLWESSSVFQSV---SNQFMDRLLGAEMPPRPLLFGDRDR-TEGCSHHHQNKSIAESY 123
Query: 119 GDGASFGLSISHSTEDSEPCMNFGGIKKVKVNQVNP--SDVQAPEQHNFDRQSTGDLHHV 176
+ S LSIS+ E + C G +K+ V++V S A E H+ + + +
Sbjct: 124 MEDTSVELSISNGVEVAGGCFGGDGNRKLPVSRVKETMSTHVALEGHSQRKIESSSIQAC 183
Query: 177 YHGEVETRSGSIGLAYGSGDARIRPFGTPYGKVDNTVLSIAESYNKDETNIISFGGFPDE 236
E S I A G PYG D+ I+FG DE
Sbjct: 184 ---SWENESSYINFALA---------GHPYGNEDSQG--------------ITFGEINDE 217
Query: 237 RGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASN-SDVVASTPLVTTKKPESVSKNK 295
GV S +Y Q Y Q + +++E +S S V S V S+ K K
Sbjct: 218 HGVGSASNVVGNY-QSYVQDPIGTLDIVYDQETGSSQTSSGVVSEQQVAKPSLGSLPKTK 276
Query: 296 QDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKGTTYLCGCQSCNY 355
+ KS++KE+ +FP+NVRSLISTGMLDGVPVKYVSV+REELRG+IKG+ YLCGCQ+C++
Sbjct: 277 AEAKSSKKEASTSFPSNVRSLISTGMLDGVPVKYVSVSREELRGVIKGSGYLCGCQTCDF 336
Query: 356 AKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPIN 415
K LNA+ FE+HAGCK+KHPNNHIYFENGKTIYQIVQELR+TPES LFD IQT+FG+PIN
Sbjct: 337 TKVLNAYAFERHAGCKTKHPNNHIYFENGKTIYQIVQELRNTPESILFDVIQTVFGSPIN 396
Query: 416 QKAFRIWK 423
QKAFRIWK
Sbjct: 397 QKAFRIWK 404
>UniRef100_Q9FJF2 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MMN10
[Arabidopsis thaliana]
Length = 415
Score = 308 bits (790), Expect = 2e-82
Identities = 185/427 (43%), Positives = 248/427 (57%), Gaps = 38/427 (8%)
Query: 2 VKGSGHVSDREPAFDNPSKIEPKRPHQWLIDATEGDFLPNKKQAIEDANERSSSGFSNVN 61
+K + H S+ + +D+ ++ + KRPH W +D++ + PNKKQA++D G SNV
Sbjct: 1 MKNNEHTSEEDSVYDHSTRDDSKRPHPWFVDSSRSEMFPNKKQAVQDPVV--GLGKSNVG 58
Query: 62 FTPWENNHNFHSDPSHQNQLIDRLFGSET--RPVNFTEKNTYVSGDGSDVRSKMIANHYG 119
WE++ F S NQ +DRL G+E RP+ F +++ G ++K IA Y
Sbjct: 59 LPLWESSSVFQSV---SNQFMDRLLGAEMPPRPLLFGDRDR-TEGCSHHHQNKSIAESYM 114
Query: 120 DGASFGLSISHSTEDSEPCMNFGGIKKVKVNQVNP--SDVQAPEQHNFDRQSTGDLHHVY 177
+ S LSIS+ E + C G +K+ V++V S A E H+ + + +
Sbjct: 115 EDTSVELSISNGVEVAGGCFGGDGNRKLPVSRVKETMSTHVALEGHSQRKIESSSIQACS 174
Query: 178 HGEVETRSGSIGLAYGSGDARIRPFGTPYGKVDNTVLSIAESYNKDETNIISFGGFPDER 237
E S I A G PYG D+ I+FG DE
Sbjct: 175 R---ENESSYINFALA---------GHPYGNEDSQG--------------ITFGEINDEH 208
Query: 238 GVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASN-SDVVASTPLVTTKKPESVSKNKQ 296
GV S +Y Q Y Q + +++E +S S V S V S+ K K
Sbjct: 209 GVGSTSNVVGNY-QSYVQDPIGTLDIVYDQETGSSQTSSGVVSEQQVAKPSLGSLPKTKA 267
Query: 297 DIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKGTTYLCGCQSCNYA 356
+ KS++KE+ +FP+NVRSLISTGMLDGVPVKYVSV+REELRG+IKG+ YLCGCQ+C++
Sbjct: 268 EAKSSKKEASTSFPSNVRSLISTGMLDGVPVKYVSVSREELRGVIKGSGYLCGCQTCDFT 327
Query: 357 KGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQ 416
K LNA+ FE+HAGCK+KHPNNHIYFENGKTIYQIVQELR+TPES LFD IQT+FG+PINQ
Sbjct: 328 KVLNAYAFERHAGCKTKHPNNHIYFENGKTIYQIVQELRNTPESILFDVIQTVFGSPINQ 387
Query: 417 KAFRIWK 423
KAFRIWK
Sbjct: 388 KAFRIWK 394
>UniRef100_Q9FNA6 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MSH12
[Arabidopsis thaliana]
Length = 536
Score = 183 bits (464), Expect = 1e-44
Identities = 94/202 (46%), Positives = 128/202 (62%), Gaps = 3/202 (1%)
Query: 223 DETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVHVSTTAHEKELEASNSDVVASTPL 282
D + +SFG E + S R + +YE + ++ + E E + S + P
Sbjct: 314 DRSETLSFGDCQKETAMGSSVRVSNNYENFSHDPAI--TKDPLHIEAEENMSFECRNPPY 371
Query: 283 VTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREE-LRGII 341
+ + + +D K+ +K S NTFP+NV+SL+STG+ DGV VKY S +RE L+G+I
Sbjct: 372 ASPRVDTLLVPKIKDTKTAKKGSTNTFPSNVKSLLSTGIFDGVTVKYYSWSRERNLKGMI 431
Query: 342 KGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESS 401
KGT YLCGC +C K LNA+EFE+HA CK+KHPNNHIYFENGKTIY +VQEL++TP+
Sbjct: 432 KGTGYLCGCGNCKLNKVLNAYEFEQHANCKTKHPNNHIYFENGKTIYGVVQELKNTPQEK 491
Query: 402 LFDTIQTIFGAPINQKAFRIWK 423
LFD IQ + G+ IN K F WK
Sbjct: 492 LFDAIQNVTGSDINHKNFNTWK 513
>UniRef100_Q6T283 Predicted protein [Populus alba x Populus tremula]
Length = 868
Score = 148 bits (373), Expect = 4e-34
Identities = 66/128 (51%), Positives = 95/128 (73%), Gaps = 1/128 (0%)
Query: 297 DIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVARE-ELRGIIKGTTYLCGCQSCNY 355
++ ++K PN +PTNV+ L++TG+LD VKY+ + E EL GII G YLCGC SC++
Sbjct: 206 ELNMSKKVVPNNYPTNVKKLLATGILDRARVKYICFSSERELDGIIDGGGYLCGCSSCSF 265
Query: 356 AKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPIN 415
+K L+A+EFE+HAG K++HPNNHIY ENGK IY I+QEL++ P S + I+ + G+ IN
Sbjct: 266 SKVLSAYEFEQHAGAKTRHPNNHIYLENGKPIYSIIQELKTAPLSMIDGVIKDVAGSSIN 325
Query: 416 QKAFRIWK 423
++ FR+WK
Sbjct: 326 EEFFRVWK 333
Score = 39.7 bits (91), Expect = 0.20
Identities = 28/108 (25%), Positives = 48/108 (43%), Gaps = 7/108 (6%)
Query: 288 PESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKGTTYL 347
P S +K K+ +S ++ N + + G+ DG + Y ++ L G +G +
Sbjct: 410 PGSATKQKKTAESGVRKRDNDLHRLL--FMPNGLPDGTELAYYVKGQKILGGYKQGNGIV 467
Query: 348 CGCQSCNYAKGLNAFEFEKHAGCKS-KHPNNHIYFENGKTIYQIVQEL 394
C C ++ +FE HAG + + P HIY N T++ I L
Sbjct: 468 CSCCEIE----ISPSQFESHAGMSARRQPYRHIYTSNRLTLHDIAISL 511
>UniRef100_Q9LFF3 Hypothetical protein F4P12_380 [Arabidopsis thaliana]
Length = 839
Score = 142 bits (357), Expect = 3e-32
Identities = 72/174 (41%), Positives = 107/174 (61%), Gaps = 9/174 (5%)
Query: 259 HVSTTAHEK---ELEASNSDVVASTPLVTTKK----PESVSKNKQDIKSTRKESPNTFPT 311
H+STT K +L N D P++ P +V+ + +K +K F +
Sbjct: 126 HLSTTGITKITFKLSKRNEDF-CDLPMIQEHTWEGYPSNVASSTLGVKMLKKIDSTNFLS 184
Query: 312 NVRSLISTGMLDGVPVKYVSV-AREELRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGC 370
NV+ L+ TG+LDG VKY+S A EL+GII YLCGC +C+++K L A+EFE+HAG
Sbjct: 185 NVKKLLGTGILDGARVKYLSTSAARELQGIIHSGGYLCGCTACDFSKVLGAYEFERHAGG 244
Query: 371 KSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWKG 424
K+KHPNNHIY ENG+ +Y ++QELR P L + I+ + G+ ++++ F+ WKG
Sbjct: 245 KTKHPNNHIYLENGRPVYNVIQELRIAPPDVLEEVIRKVAGSALSEEGFQAWKG 298
>UniRef100_O80922 Hypothetical protein At2g37520 [Arabidopsis thaliana]
Length = 825
Score = 130 bits (327), Expect = 9e-29
Identities = 56/118 (47%), Positives = 85/118 (71%), Gaps = 1/118 (0%)
Query: 308 TFPTNVRSLISTGMLDGVPVKYVSVAR-EELRGIIKGTTYLCGCQSCNYAKGLNAFEFEK 366
++P+NV+ L+ TG+L+G VKY+S +L GII YLCGC +CN++K L+A+EFE+
Sbjct: 162 SYPSNVKKLLETGILEGARVKYISTPPVRQLLGIIHSGGYLCGCTTCNFSKVLSAYEFEQ 221
Query: 367 HAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWKG 424
HAG K++HPNNHI+ EN + +Y IVQEL++ P L + I+ + G+ +N++ R WKG
Sbjct: 222 HAGAKTRHPNNHIFLENRRAVYNIVQELKTAPRVVLEEVIRNVAGSALNEEGLRAWKG 279
>UniRef100_Q5VMF2 PHD zinc finger protein-like [Oryza sativa]
Length = 1025
Score = 97.4 bits (241), Expect = 8e-19
Identities = 51/151 (33%), Positives = 92/151 (60%), Gaps = 4/151 (2%)
Query: 272 SNSDVVASTPLVTTKKPESVSKNKQDIKSTRKESPNT-FPTNVRSLISTGMLDGVPVKYV 330
S ++++ S + + S S K ++K ++K + T P+N+R L++TG+L+G+PV+Y+
Sbjct: 345 STNNLLQSKEAIDSTSDSSRSVKKMEMKMSKKVACLTKHPSNIRELLNTGLLEGMPVRYI 404
Query: 331 --SVAREELRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIY 388
S + L+G+I G C C SCN +K + ++ FE+HAG KHP +HIY NG ++
Sbjct: 405 IPSSKKAVLKGVITGCNIRCFCLSCNGSKDVCSYFFEQHAGSNKKHPADHIYLGNGNSLR 464
Query: 389 QIVQELRSTPESSLFDTIQTIFGAPINQKAF 419
+++ S+P SL TI++ PI ++++
Sbjct: 465 DVLRACESSPLESLEKTIRSSID-PIAKRSY 494
>UniRef100_Q5Z411 PHD zinc finger protein-like [Oryza sativa]
Length = 779
Score = 93.6 bits (231), Expect = 1e-17
Identities = 51/154 (33%), Positives = 82/154 (53%), Gaps = 8/154 (5%)
Query: 269 LEASNSDVVASTPLVTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVK 328
L + S+ +S+P +T S +K ++K ++K S P N++ L++TG+L+G PVK
Sbjct: 122 LSSRTSNSSSSSPSAST------STSKMELKMSKKISFTRIPRNLKDLLATGLLEGHPVK 175
Query: 329 YV--SVAREELRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKT 386
Y+ R LRG+IK LC C SC ++ + FE HAG KHP+++I+ ENG
Sbjct: 176 YIMRKGKRAVLRGVIKRVGILCSCSSCKGRTVVSPYYFEVHAGSTKKHPSDYIFLENGNN 235
Query: 387 IYQIVQELRSTPESSLFDTIQTIFGAPINQKAFR 420
++ I++ L IQ G ++ FR
Sbjct: 236 LHDILRACSDATLDMLQSAIQNAIGPAPKKRTFR 269
>UniRef100_Q6JJ55 Putative PHD zinc finger protein [Ipomoea trifida]
Length = 1047
Score = 89.7 bits (221), Expect = 2e-16
Identities = 46/144 (31%), Positives = 80/144 (54%), Gaps = 6/144 (4%)
Query: 275 DVVASTPLVTTKKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAR 334
D A L + + + +K ++K ++K + PT ++ L++TG+L+G+PV+YV V +
Sbjct: 335 DTKAEDALESDEASAIGTTSKLEMKMSKKVALVKIPTKLKGLLATGLLEGLPVRYVRVTK 394
Query: 335 EE------LRGIIKGTTYLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIY 388
L+G+I+G+ LC CQ+C K + +FE HAG +K P +IY +NGKT+
Sbjct: 395 ARGRPEKGLQGVIQGSGILCFCQNCGGTKVVTPNQFEMHAGSSNKRPPEYIYLQNGKTLR 454
Query: 389 QIVQELRSTPESSLFDTIQTIFGA 412
++ + P +L I+ GA
Sbjct: 455 DVLVACKDAPADALEAAIRNATGA 478
Score = 43.5 bits (101), Expect = 0.014
Identities = 33/119 (27%), Positives = 52/119 (42%), Gaps = 19/119 (15%)
Query: 298 IKSTRKESPNTFPTN------------VRSLISTG--MLDGVPVKYVSVAREELRGIIKG 343
+KST + S T P + + L+ G + DG + Y ++ L G KG
Sbjct: 568 LKSTERMSSGTCPPSKVHGRLTRKDLRMHKLVFEGDVLPDGTALAYYVRGKKLLEGYKKG 627
Query: 344 TTYLCGCQSCNYAKGLNAFEFEKHAGCKSKH-PNNHIYFENGKTIYQIVQELRSTPESS 401
C C + +FE HAGC S+ P +HIY NG +++++ +L SS
Sbjct: 628 GAIFCYCCQSEVSPS----QFEAHAGCASRRKPYSHIYTSNGVSLHELSIKLSMERRSS 682
>UniRef100_Q9ZQA2 Putative PHD-type zinc finger protein [Arabidopsis thaliana]
Length = 958
Score = 75.5 bits (184), Expect = 3e-12
Identities = 42/120 (35%), Positives = 65/120 (54%), Gaps = 3/120 (2%)
Query: 310 PTNVRSLISTGMLDGVPVKYVSVAREE---LRGIIKGTTYLCGCQSCNYAKGLNAFEFEK 366
P VR L TG+LDG+ V Y+ + + LRGII+ LC C SC++A ++ +FE
Sbjct: 261 PETVRDLFETGLLDGLSVVYMGTVKSQAFPLRGIIRDGGILCSCSSCDWANVISTSKFEI 320
Query: 367 HAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAFRIWKGKE 426
HA + + + +I FENGK++ ++ R+TP +L TI +K F + KE
Sbjct: 321 HACKQYRRASQYICFENGKSLLDVLNISRNTPLHALEATILDAVDYASKEKRFTCKRCKE 380
>UniRef100_Q8RWG0 Putative PHD-type zinc finger protein [Arabidopsis thaliana]
Length = 1007
Score = 73.9 bits (180), Expect = 1e-11
Identities = 40/113 (35%), Positives = 62/113 (54%), Gaps = 3/113 (2%)
Query: 310 PTNVRSLISTGMLDGVPVKYVSVAREE---LRGIIKGTTYLCGCQSCNYAKGLNAFEFEK 366
P VR L TG+LDG+ V Y+ + + LRGII+ LC C SC++A ++ +FE
Sbjct: 261 PETVRDLFETGLLDGLSVVYMGTVKSQAFPLRGIIRDGGILCSCSSCDWANVISTSKFEI 320
Query: 367 HAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDTIQTIFGAPINQKAF 419
HA + + + +I FENGK++ ++ R+TP +L TI +K F
Sbjct: 321 HACKQYRRASQYICFENGKSLLDVLNISRNTPLHALEATILDAVDYASKEKRF 373
>UniRef100_Q8IJY4 Regulator of nonsense transcripts, putative [Plasmodium falciparum]
Length = 1554
Score = 45.4 bits (106), Expect = 0.004
Identities = 71/297 (23%), Positives = 123/297 (40%), Gaps = 38/297 (12%)
Query: 37 DFLPNKKQAIEDANERSSSGFSNVNFTPWENNHNFHSDPSHQNQLIDRLFGSETRPVNFT 96
+F NK + +D + ++ +N N T NN+N +++ ++ N+ D+LF N +
Sbjct: 1268 NFQKNKTYSYQDKFKSNTYKDNNTNTTNQYNNNNNNNNNNNNNKNDDQLF-------NSS 1320
Query: 97 EKNTYVSGDGSDVRSKMIANHYGD---GASFGLSISHSTEDSEPCMN---FGGI-KKVKV 149
N Y S S NH D + F L++S+ + S+ MN + I K K
Sbjct: 1321 HNNHYYS--SYYYYSHFNKNHLPDENTNSQFNLNLSNYSYCSDHFMNNDLYNYINKNKKK 1378
Query: 150 NQVNP-----------SDVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGLAYGSGDAR 198
N VN +D HN D + LH Y+ ++ S S
Sbjct: 1379 NIVNSYVNNLSQMSQFNDYPTETNHNNDDHNYLHLHDKYNDTLDDDKKSSNKCNISSSLN 1438
Query: 199 IRPFGTPYGKVDNTVLSIAESYNKDETNIISFGGFPDER-----GVISVGRAATDYEQLY 253
F P K+ + +SYN D TN+IS D + +++ Y
Sbjct: 1439 NSEFSYPNNKMSSN--HSIDSYNHD-TNLISMSKNADNLLPYLFYYKNNQNSSSQPYMYY 1495
Query: 254 NQSSVHVSTTAHEKELEAS--NSDVVASTPLVTTKKPESVSKNKQDIKSTRKESPNT 308
NQ+S ++++ +K L+ + N ++T +V+ K V+ K + + +K S NT
Sbjct: 1496 NQNSEQINSSKTQKGLKNTVLNKSSTSNT-MVSQKDSIIVNDQKLNCDNLKKVSENT 1551
>UniRef100_Q9SJJ4 Hypothetical protein At2g27980 [Arabidopsis thaliana]
Length = 1008
Score = 41.2 bits (95), Expect = 0.070
Identities = 32/133 (24%), Positives = 59/133 (44%), Gaps = 27/133 (20%)
Query: 286 KKPESVSKNKQDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKGTT 345
+ P + NK + K+ FP ++ + G+L+G+ V YV RG
Sbjct: 357 QSPSVTTPNK---RGRPKKFLRNFPAKLKDIFDCGILEGLIVYYV-------RG------ 400
Query: 346 YLCGCQSCNYAKGLNAFEFEKHAGCKSKHPNNHIYFENGKTIYQIVQELRSTPESSLFDT 405
AK ++ FE HA +K P +I E+G T+ ++ + P ++L +
Sbjct: 401 ----------AKVVSPAMFELHASSNNKRPPEYILLESGFTLRDVMNACKENPLATLEEK 450
Query: 406 IQTIFGAPINQKA 418
++ + G PI +K+
Sbjct: 451 LRVVVG-PILKKS 462
>UniRef100_Q8IJI4 10b antigen, putative [Plasmodium falciparum]
Length = 2290
Score = 40.4 bits (93), Expect = 0.12
Identities = 62/300 (20%), Positives = 129/300 (42%), Gaps = 53/300 (17%)
Query: 41 NKKQAIEDANERSSSGFSNVNFTPWENNHNFHSDPS---HQNQLIDRLFGSETRPVNFTE 97
NKK++ ++ N + S +N N NN+N ++ + N++I+ + +N T+
Sbjct: 583 NKKKSKKNKNNNNDSNNNNDN-----NNNNIMNENKKDINDNKIINNNNNNNNNNLNATK 637
Query: 98 KNTYVSGDGSDV-RSKMIANHYGDGASFGLSISHSTED---SEPCMNFGGIKKVKVNQVN 153
KN ++ + + + ++ I N++ + ++ H++ + MNF + K N
Sbjct: 638 KNNTININNTIIDKNNNIENNFAFVNNLNENLEHNSMKLIGNNYPMNFENNEIKKKNITA 697
Query: 154 PSDVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGLAYGSGDARIRPFGTPYGKVDNTV 213
+V P ++N+D + G+L+++Y+ L Y + + + + P+ +DN
Sbjct: 698 SKNV--PNKYNYDMINQGNLNYMYN----------PLNYYNMN---KYYDMPF--IDNIA 740
Query: 214 LSIAESY-------NKDETNIISFGGFPDERGVISVG---RAATDYEQLYNQSSV----- 258
++ Y N NII++ G + + + T+ +YN +S
Sbjct: 741 NNLNPMYFDNDMQRNHMMENIINYQGMYNMNNIYNTNNNMEQNTNNNNMYNHNSYANYKH 800
Query: 259 -HVSTTAHEKELEASNSDVVASTPLVTTKKPESVSKNKQDIKSTRK--ESPN--TFPTNV 313
H+S +E LE +N KK N+Q++ + +K PN FP N+
Sbjct: 801 EHLSLNVYEGPLEKNNE----WGKKKMNKKNNQSGMNQQNVMNQQKVMNQPNAMNFPNNM 856
>UniRef100_UPI00004671EF UPI00004671EF UniRef100 entry
Length = 931
Score = 39.7 bits (91), Expect = 0.20
Identities = 56/283 (19%), Positives = 108/283 (37%), Gaps = 33/283 (11%)
Query: 50 NERSSSGFSNVNFTPWENNHNFHSDPSHQNQLIDRLFGSETRPVNFTEKNTY-------- 101
N+ S + S V + EN N + S N + G TR NFT+KN++
Sbjct: 326 NQESQNYDSKVKYDKDENGKNIGNGKS--NLSYENKSGVNTRSGNFTKKNSFNNKNNNSW 383
Query: 102 -----VSGDGSDVRSKMIANHYGDGASFGLSISHSTEDSEPCMN-FGGIKKVKVNQVNPS 155
+ + SDV ++YG ++ E+ + +N G ++ +++ N +
Sbjct: 384 SEERHIDYNNSDVNMSEQRSYYG---TYNNGNGCEIEEGKNGVNKLYGYEENELDSKNDN 440
Query: 156 DVQAPEQHNFDRQSTGDLHHVYHGEVETRSGSIGLAYGSGDAR------------IRPFG 203
+ + ++ + + H+ + + G+ + Y + + I P G
Sbjct: 441 IINSGKRGYTKENNFNNNGHIKYHNNKYNDGNNTITYAKSNTKKGCDNNIIDNSVILPQG 500
Query: 204 TPYGKVDNTVLSIAESYNKDETNIISFGGFPDERGVISVGRAATDYEQLYNQSSVH-VST 262
Y + S K E S F +ER A +E LY +++ +
Sbjct: 501 VFYQDSKKAFCANWYSNGKQEKRYFSINKFGEERARSLAIAARKKFENLYKKNNKRDYVS 560
Query: 263 TAHEKELEASNSDVVASTPLVTTK-KPESVSKNKQDIKSTRKE 304
AH +++E NS++ S + S+ KN+ T+KE
Sbjct: 561 GAHTQQIENGNSEISVSKKNDNLNGENMSLEKNESGTDYTKKE 603
>UniRef100_UPI00002EA680 UPI00002EA680 UniRef100 entry
Length = 189
Score = 39.3 bits (90), Expect = 0.27
Identities = 27/92 (29%), Positives = 43/92 (46%), Gaps = 9/92 (9%)
Query: 110 RSKMIANHYGDGASFGLSISHSTEDSEPCMNFGGIKKV-----KVNQ--VNPSDVQAP-E 161
+ I H GDG+ +G+ + +S ED++P GGI K ++NQ V SD+
Sbjct: 26 KGDQIEKHIGDGSRYGIRVKYSREDAQPLETLGGILKALPILGEINQFVVINSDIWTDYP 85
Query: 162 QHNFDRQSTGDLHHVYHGEVETRSGSIGLAYG 193
HN R + H V + ++ +G L G
Sbjct: 86 MHNLQRAALNS-HIVLAPKSDSDNGDFDLRQG 116
>UniRef100_UPI00003C203D UPI00003C203D UniRef100 entry
Length = 779
Score = 38.1 bits (87), Expect = 0.59
Identities = 24/65 (36%), Positives = 35/65 (52%), Gaps = 3/65 (4%)
Query: 296 QDIKSTRKESPNTFPTNVRSLISTGMLDGVPVKYVSVAREELRGIIKG---TTYLCGCQS 352
Q S+ SP+T+ ++V +L+ G+L VP K VS +R+ +R KG T L C S
Sbjct: 677 QSAVSSSTASPSTWASDVGTLLWDGILRAVPKKKVSHSRKSMRAANKGLKDRTDLVHCSS 736
Query: 353 CNYAK 357
C K
Sbjct: 737 CGKPK 741
>UniRef100_Q7RNT0 Plant adhesion molecule 1-related [Plasmodium yoelii yoelii]
Length = 515
Score = 37.4 bits (85), Expect = 1.0
Identities = 57/272 (20%), Positives = 104/272 (37%), Gaps = 49/272 (18%)
Query: 267 KELEASNSDVVASTPLVTTKKP----------ESVSKNKQDIKSTRKESPNTFPTNVRSL 316
K E +N D+ S P P +++S+N D K R E+ +PT++ ++
Sbjct: 39 KSNEENNFDIFKSNPEKNEISPSVKNKICFNFDNLSENDCDFKKGRNENVPCYPTSIDNI 98
Query: 317 ISTGMLDGVPVKYVSVAREELRGIIKG-----TTYLCGCQSCN-----YAKGLNAFEFEK 366
I G + +K + + + G I Y+ + N K N + +K
Sbjct: 99 IGDGTKKSIQIKKDKI-NDNMIGNIDSDCNNEENYILKNEKKNIYYKYIQKKKNVLDKKK 157
Query: 367 HAGCKSKHPNNHIYFENGKTIYQIVQELRSTP----------ESSLFDTIQTIFGAPINQ 416
H K N+IY N K + + P + L +T ++Q
Sbjct: 158 H--MKISQNENNIYSNNSKNDEDLSSNICRIPLINNHMPTYISTDLILKKKTFMTMSLSQ 215
Query: 417 -KAFRIWKGKEIPNLFKHKQTNGCIGNVEFYCLGFVWYVINPLFV----TLA----NGLQ 467
K + K+I ++ K++ G + Y GFVW ++ +V T A N +
Sbjct: 216 KKKYFAEHSKKINSIIKNEIVKG----IPDYLRGFVWQILLQSYVYKDRTYADKDINNNE 271
Query: 468 N---HFKQQHASFNGFMEMKNVTSKSPPSLRK 496
N + Q + G+ ++T+K S++K
Sbjct: 272 NNSEYINQSNKCEKGYKYYLSITNKYESSIKK 303
>UniRef100_Q7PDN3 Erythrocyte membrane protein PFEMP3 [Plasmodium yoelii yoelii]
Length = 1781
Score = 37.0 bits (84), Expect = 1.3
Identities = 26/129 (20%), Positives = 55/129 (42%)
Query: 41 NKKQAIEDANERSSSGFSNVNFTPWENNHNFHSDPSHQNQLIDRLFGSETRPVNFTEKNT 100
N A++ N + +N N NN+N +SD + N + + + +N T+ N
Sbjct: 88 NSLNAVDQNNNAYNEEINNSNSLDSNNNNNTNSDVTSTNTIGKQNIAIKKSSLNTTDSND 147
Query: 101 YVSGDGSDVRSKMIANHYGDGASFGLSISHSTEDSEPCMNFGGIKKVKVNQVNPSDVQAP 160
S +G + + + +N+ + + + + + N G K +++ NP+ V
Sbjct: 148 NASSEGINNLNILESNNNNACCNIDKNATTISNTNGNSTNASGSSKHTIDKNNPNIVDIN 207
Query: 161 EQHNFDRQS 169
+ H D+ S
Sbjct: 208 DTHVDDQNS 216
>UniRef100_Q9LHN9 Emb|CAB68186.1 [Arabidopsis thaliana]
Length = 605
Score = 36.6 bits (83), Expect = 1.7
Identities = 31/107 (28%), Positives = 52/107 (47%), Gaps = 10/107 (9%)
Query: 220 YNKDETNIISFGGFPDERGV--ISVGRAATDYEQ--LYNQSSVHVSTTAHEKELEASNS- 274
Y E N + F F E V V + TD ++ +Y Q S H+ + E++LEA +
Sbjct: 346 YQPTEENFMDFLSFLRENDVDITDVKMSPTDEDEFSIYKQRSTHMRNHSLEEDLEAEKTI 405
Query: 275 ----DVVASTPLVTTKKPESVSKNKQDIKSTRK-ESPNTFPTNVRSL 316
V S T + ES+S+ + D+++ K ES +TF ++S+
Sbjct: 406 SFQDKVDPSGEEQTLMRNESISRKQSDLETPEKMESFSTFGDEIQSV 452
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.314 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 900,627,585
Number of Sequences: 2790947
Number of extensions: 40108228
Number of successful extensions: 73564
Number of sequences better than 10.0: 60
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 41
Number of HSP's that attempted gapping in prelim test: 73491
Number of HSP's gapped (non-prelim): 92
length of query: 500
length of database: 848,049,833
effective HSP length: 132
effective length of query: 368
effective length of database: 479,644,829
effective search space: 176509297072
effective search space used: 176509297072
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 77 (34.3 bits)
Medicago: description of AC139841.9


