

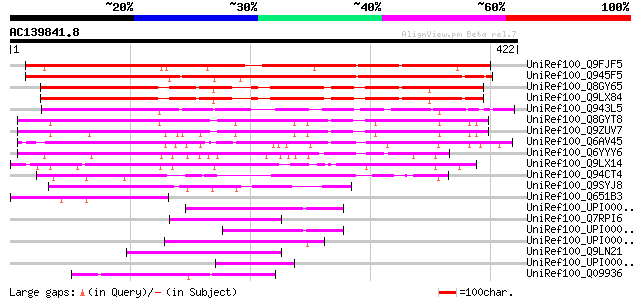
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.8 + phase: 0
(422 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FJF5 Gb|AAC98451.1 [Arabidopsis thaliana] 391 e-107
UniRef100_Q945F5 Auxin-regulated protein [Lycopersicon esculentum] 389 e-107
UniRef100_Q8GY65 Hypothetical protein At3g46110/F12M12_80 [Arabi... 298 2e-79
UniRef100_Q9LX84 Hypothetical protein F12M12_80 [Arabidopsis tha... 292 1e-77
UniRef100_Q943L5 P0031D11.20 protein [Oryza sativa] 258 2e-67
UniRef100_Q8GYT8 Hypothetical protein At2g28150 [Arabidopsis tha... 256 6e-67
UniRef100_Q9ZUV7 Hypothetical protein At2g28150 [Arabidopsis tha... 245 2e-63
UniRef100_Q6AV45 Expressed protein [Oryza sativa] 225 2e-57
UniRef100_Q6YYY6 Putative auxin-regulated protein [Oryza sativa] 176 1e-42
UniRef100_Q9LX14 Hypothetical protein T31P16_140 [Arabidopsis th... 160 5e-38
UniRef100_Q94CT4 P0459B04.11 protein [Oryza sativa] 123 9e-27
UniRef100_Q9SYJ8 F3F20.2 protein [Arabidopsis thaliana] 115 3e-24
UniRef100_Q651B3 Hypothetical protein B1331F11.4 [Oryza sativa] 109 1e-22
UniRef100_UPI000042E1DD UPI000042E1DD UniRef100 entry 54 6e-06
UniRef100_Q7RPI6 Hypothetical protein [Plasmodium yoelii yoelii] 52 4e-05
UniRef100_UPI00004532EC UPI00004532EC UniRef100 entry 51 5e-05
UniRef100_UPI000004DF1F UPI000004DF1F UniRef100 entry 49 2e-04
UniRef100_Q9LN21 F14O10.11 protein [Arabidopsis thaliana] 49 2e-04
UniRef100_UPI000024D6C3 UPI000024D6C3 UniRef100 entry 49 3e-04
UniRef100_Q09936 Hypothetical protein C53C9.2 in chromosome X [C... 49 3e-04
>UniRef100_Q9FJF5 Gb|AAC98451.1 [Arabidopsis thaliana]
Length = 423
Score = 391 bits (1004), Expect = e-107
Identities = 226/414 (54%), Positives = 274/414 (65%), Gaps = 43/414 (10%)
Query: 14 RETSPERTKVWTEP--KPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIR 71
++TSP+R ++W+EP KP RKV VVYYL RNG L+HPHF+EV LSS GLYLKDVI R
Sbjct: 24 QDTSPDRNRIWSEPRLKPVVNRKVPVVYYLCRNGQLDHPHFIEVTLSSHDGLYLKDVINR 83
Query: 72 LNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEH------ 125
LN LRGKGM ++YSWSSKRSYKNGFVWHDL+E DFI+P QG +Y+LKGSE+L+
Sbjct: 84 LNDLRGKGMASLYSWSSKRSYKNGFVWHDLSEDDFIFPVQGQEYVLKGSEVLDSCLISNP 143
Query: 126 ----ETTA-------RPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYK----SEPLGEN 170
ET++ P D P VI RRRNQSWSSIDL+EY+VYK S +
Sbjct: 144 RSLLETSSFRDPRSLNPDKNSGDDIPAVINRRRNQSWSSIDLSEYKVYKATESSAESTQR 203
Query: 171 IAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPP 230
+AADASTQT+++RRRRR +EE EE + +NQSTELSR+EISPP
Sbjct: 204 LAADASTQTDDRRRRRRPAKEEIEEV-------------KSPASYENQSTELSRDEISPP 250
Query: 231 PSDSSPETLGTLMKVDGRLGLR--SVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKE 288
PSDSSPETL L+K DGRL LR ++ TVES SGRMRAS+VL+QL+SCG +SFKE
Sbjct: 251 PSDSSPETLENLIKADGRLILRPSESSTDHRTVESLSSGRMRASAVLMQLISCGTMSFKE 310
Query: 289 GSGASSGKDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLIE 348
G KDQ +L G + RG ++ + V E+ RV+LEDKEYFSGSLIE
Sbjct: 311 -CGPVLLKDQGLALNGRSGCTITRGAEDNGEERVDK--ELKSFGRVQLEDKEYFSGSLIE 367
Query: 349 TKKVEAPAFKRSSSYNADSSSRL--QIMEHEGDMVRAKCIPRKSKTLLTKKEEG 400
TKK PA KRSSSYNAD SR+ + E + VRAKCIPRK K + + G
Sbjct: 368 TKKELVPALKRSSSYNADRCSRMGPTTEKDEEEAVRAKCIPRKPKPVAKRNNGG 421
>UniRef100_Q945F5 Auxin-regulated protein [Lycopersicon esculentum]
Length = 433
Score = 389 bits (999), Expect = e-107
Identities = 230/415 (55%), Positives = 280/415 (67%), Gaps = 30/415 (7%)
Query: 14 RETSPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLN 73
RETSPERTKVWTEP KV VVYYLSRNG LEHPHFMEVPLSS GLYL+DVI RLN
Sbjct: 16 RETSPERTKVWTEPPNHKLSKVPVVYYLSRNGQLEHPHFMEVPLSSRDGLYLRDVINRLN 75
Query: 74 LLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARP-- 131
LRGKGM +MYSWS+KRSY+NGFVWHDLTE+DFIYP G +Y+LKGSE+L+ ++P
Sbjct: 76 CLRGKGMASMYSWSAKRSYRNGFVWHDLTENDFIYPAHGQEYVLKGSELLDSALPSQPDE 135
Query: 132 --------------PLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGE---NIAAD 174
L E+ + P V RRRNQSWSS D +EY VYK+E GE I AD
Sbjct: 136 IACSNSRNTVPEKQKLSEDREFPAV-ARRRNQSWSSSDFHEYLVYKAESPGEILGRIGAD 194
Query: 175 ASTQTEEKRRRRRVLR--EEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPS 232
ASTQTE++RRRR +R EE+EE E ++D E + + NQSTEL+R EISPP S
Sbjct: 195 ASTQTEDRRRRRGGMRIVEEEEELSENRIIESDCKEPDEVEHSPNQSTELNRGEISPPLS 254
Query: 233 DSSPETLGTLMKVDGRLGLR-SVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSG 291
DSS ETL TLMK DG+L LR E+PT + SG+ +A+SVL+QLL CG++SFK+ G
Sbjct: 255 DSSSETLETLMKADGKLILRPDTISEDPTANTHSSGKSKAASVLMQLLYCGSMSFKQ-CG 313
Query: 292 ASSGKDQRFSLVGHYKSRLPRGG-GNHVGKEV-GTLVEI-PDLNRVRLEDKEYFSGSLIE 348
GK+ FSL+ YK+RL G N K+V +VE R++LEDKEYFSGSLIE
Sbjct: 314 PGYGKENGFSLISQYKNRLSHGRCTNQAVKDVESPIVEYHGSEERIKLEDKEYFSGSLIE 373
Query: 349 TKKVEA-PAFKRSSSYNADSSSRLQIMEHEGDMVRAKCIPRKSKTLLTKKEEGTS 402
KK E PA KRSSSYN + S++L++ E + + + KC RK K T +EEG S
Sbjct: 374 MKKKEKFPALKRSSSYNVERSTKLELTEKQ-EEGKTKCYLRKQKNQST-REEGDS 426
>UniRef100_Q8GY65 Hypothetical protein At3g46110/F12M12_80 [Arabidopsis thaliana]
Length = 343
Score = 298 bits (763), Expect = 2e-79
Identities = 187/379 (49%), Positives = 239/379 (62%), Gaps = 59/379 (15%)
Query: 26 EPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYS 85
+ KP R V VVYYLSRNG L+HPHF+EVPLSS +GLYLKDVI RLN LRG GM +YS
Sbjct: 11 DSKPSRERIVPVVYYLSRNGRLDHPHFIEVPLSSHNGLYLKDVINRLNDLRGNGMACLYS 70
Query: 86 WSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITR 145
WSSKR+YKNGFVW+DL++ DFI+P G +Y+LKGS+IL+ L+ S + +T
Sbjct: 71 WSSKRTYKNGFVWYDLSDEDFIFPVHGQEYVLKGSQILD--------LDNNSGNFSAVTH 122
Query: 146 RRNQSWSSIDLNEYRVYKSEPLG----ENIAADASTQTEEKRRRRRVLREEDEEQEEEEK 201
RRNQSWSS+D Y+VYK+ L ++ DASTQT+++RRR K
Sbjct: 123 RRNQSWSSVD--HYKVYKASELNAEATRKLSMDASTQTDDRRRR---------------K 165
Query: 202 EKNDEIENEGETENQNQSTELSREEI-SPPPSDSSPETLGTLMKVDGRLGLRSVPKE-NP 259
DE+ N+ TELSREEI SPP SDSSPETL +LM+ DGRL L +E N
Sbjct: 166 SPVDEV---------NEVTELSREEITSPPQSDSSPETLESLMRADGRLILLQEDQELNR 216
Query: 260 TVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSLVGHYKSRLPRGGGNHVG 319
TVE +MR S+VL+QL+SCGA+SFK K + G+ +S RG GN+
Sbjct: 217 TVE-----KMRPSAVLMQLISCGAMSFK--------KCGPTLMNGNTRSTAVRGTGNYRL 263
Query: 320 KEVGTLVEIPDLNRVRLEDKEYFSGSLIE--TKKVEAPAFKRSSSYNADSSSRLQI-MEH 376
+ E+ RV+LE+KEYFSGSLI+ +KK PA KRSSSYN D SSR+ + E
Sbjct: 264 ERAEK--ELKSFGRVKLEEKEYFSGSLIDESSKKELVPALKRSSSYNIDRSSRMGLTKEK 321
Query: 377 EG-DMVRAKCIPRKSKTLL 394
EG ++ RA IPR +++
Sbjct: 322 EGEELARANFIPRNPNSVV 340
>UniRef100_Q9LX84 Hypothetical protein F12M12_80 [Arabidopsis thaliana]
Length = 340
Score = 292 bits (747), Expect = 1e-77
Identities = 183/377 (48%), Positives = 234/377 (61%), Gaps = 58/377 (15%)
Query: 26 EPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYS 85
+ KP R V VVYYLSRNG L+HPHF+EVPLSS +GLYLKDVI RLN LRG GM +YS
Sbjct: 11 DSKPSRERIVPVVYYLSRNGRLDHPHFIEVPLSSHNGLYLKDVINRLNDLRGNGMACLYS 70
Query: 86 WSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITR 145
WSSKR+YKNGFVW+DL++ DFI+P G +Y+LKGS+IL+ L+ S + +T
Sbjct: 71 WSSKRTYKNGFVWYDLSDEDFIFPVHGQEYVLKGSQILD--------LDNNSGNFSAVTH 122
Query: 146 RRNQSWSSIDLNEYRVYKSEPLG----ENIAADASTQTEEKRRRRRVLREEDEEQEEEEK 201
RRNQSWSS+D Y+VYK+ L ++ DASTQT+++RRR K
Sbjct: 123 RRNQSWSSVD--HYKVYKASELNAEATRKLSMDASTQTDDRRRR---------------K 165
Query: 202 EKNDEIENEGETENQNQSTELSREEI-SPPPSDSSPETLGTLMKVDGRLGLRSVPKE-NP 259
DE+ N+ TELSREEI SPP SDSSPETL +LM+ DGRL L +E N
Sbjct: 166 SPVDEV---------NEVTELSREEITSPPQSDSSPETLESLMRADGRLILLQEDQELNR 216
Query: 260 TVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSLVGHYKSRLPRGGGNHVG 319
TVE +MR S+VL+QL+SCGA+SFK K + G+ +S RG GN+
Sbjct: 217 TVE-----KMRPSAVLMQLISCGAMSFK--------KCGPTLMNGNTRSTAVRGTGNYRL 263
Query: 320 KEVGTLVEIPDLNRVRLEDKEYFSGSLIE--TKKVEAPAFKRSSSYNADSSSRLQIMEHE 377
+ E+ RV+LE+KEYFSGSLI+ +KK PA KRSSSYN D + E E
Sbjct: 264 ERAEK--ELKSFGRVKLEEKEYFSGSLIDESSKKELVPALKRSSSYNIDRMGLTKEKEGE 321
Query: 378 GDMVRAKCIPRKSKTLL 394
++ RA IPR +++
Sbjct: 322 -ELARANFIPRNPNSVV 337
>UniRef100_Q943L5 P0031D11.20 protein [Oryza sativa]
Length = 411
Score = 258 bits (660), Expect = 2e-67
Identities = 172/423 (40%), Positives = 228/423 (53%), Gaps = 77/423 (18%)
Query: 27 PKPKT-PRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYS 85
P+P+ P + +VVYYLSRNGHLEHPHFMEV ++SP GLYL+DVI RL+ LRGKGM MYS
Sbjct: 37 PRPRPGPARAAVVYYLSRNGHLEHPHFMEVAVASPDGLYLRDVIDRLDALRGKGMARMYS 96
Query: 86 WSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEIL--------------------EH 125
W+SKRSY+NGFVWHDL + D+IYP G +Y+LKG+E L +
Sbjct: 97 WASKRSYRNGFVWHDLADDDYIYPVAGREYVLKGTERLHPIQLPLLDAAAASSCSSGSQE 156
Query: 126 ETTARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRR 185
T+ PP E +++ ++ +L EYRVYK+E AADA+TQTE+ R
Sbjct: 157 TATSSPPGWENGTGEA--RQKKGAGINTSELCEYRVYKAEDPAA-AAADAATQTEDGYRS 213
Query: 186 RRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPETLGTLMKV 245
R + ++ ELSREE SPP + +SPETL L+K
Sbjct: 214 SRGRHAQRAAAAAAQE-------------------ELSREETSPPTASTSPETLEALIKA 254
Query: 246 DGRLGLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSLVGH 305
DGR+ GR RASSVL+QL+SCG+VS K ++ H
Sbjct: 255 DGRV----------MAAVTGGGRTRASSVLMQLISCGSVSVK----STLASPVMARTAAH 300
Query: 306 YKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLIETKKVEAPA--------F 357
Y+ R PR T EIP+ + +EDKEYFSGSL+ETK+ +PA
Sbjct: 301 YRPRPPR-----PPTLASTTTEIPNYRQKIVEDKEYFSGSLVETKR-SSPADTSQDIAVL 354
Query: 358 KRSSSYNADSSSRLQIMEHEGDMVRAKCIPRKSKTLLTKKEEGTSSSMDSVSRSQHGSKR 417
+RSSSYNAD +++ DM +CIPR+ + KK+ G + + S Q+GSKR
Sbjct: 355 RRSSSYNADRVQKVEPSTEAVDM-HDRCIPRRPR---GKKDGG--AYLISGGNGQYGSKR 408
Query: 418 FEG 420
G
Sbjct: 409 HGG 411
>UniRef100_Q8GYT8 Hypothetical protein At2g28150 [Arabidopsis thaliana]
Length = 540
Score = 256 bits (655), Expect = 6e-67
Identities = 180/481 (37%), Positives = 250/481 (51%), Gaps = 105/481 (21%)
Query: 7 SRGRPGTRETSPERTKVWTEPKPKTP---RKVSVVYYLSRNGHLEHPHFMEVPLSSPHGL 63
+R + +RE SPER KVWTE PK +KV +VYYLS+N LEHPHFMEV +SSP+GL
Sbjct: 3 ARMKKYSREVSPERAKVWTEKSPKYHQKIKKVQIVYYLSKNRQLEHPHFMEVLISSPNGL 62
Query: 64 YLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEIL 123
YL+DVI RLN+LRG+GM +MYSWSSKRSY+NGFVWHDL+E D I P G++Y+LKGSE+
Sbjct: 63 YLRDVIERLNVLRGRGMASMYSWSSKRSYRNGFVWHDLSEDDLILPANGNEYVLKGSELF 122
Query: 124 ----------------------------------------------------EHETTARP 131
E + + P
Sbjct: 123 DESNSDHFSPIVNLATQNMKQIVVEPPSSRSMDDSSSSSSMNNGKGTNKHSHEDDELSPP 182
Query: 132 PLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRR----- 186
L S S V R ++ SS L EY+VYKSE L ADASTQT+E R
Sbjct: 183 ALRSVSSSGVSPDSRDAKNSSSWCLAEYKVYKSEGL-----ADASTQTDETVSGRSKTPI 237
Query: 187 ----RVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSS------P 236
R + +++ E E +N+ + + +S E+SR +SPP S+S+
Sbjct: 238 ETFSRGVSTDEDVSSEPETSENNLVSEASCAGKERESAEISRNSVSPPFSNSASSLGGKT 297
Query: 237 ETLGTLMKVD--GRLGLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASS 294
+TL +L++ D R + +E+ + + P R+RAS++L+QL+SCG++S
Sbjct: 298 DTLESLIRADVSKMNSFRILEQEDVRMPAIP--RLRASNMLMQLISCGSISV-------- 347
Query: 295 GKDQRFSLV-------GHYKSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLI 347
KD F LV GH K P + + ++ L E P L +R+E+KEYFSGSL+
Sbjct: 348 -KDNNFGLVPTYKPKFGHSKFPSPFFSSSFMMGDLDRLSETPSLMGLRMEEKEYFSGSLV 406
Query: 348 ETKKVEAPA------FKRSSSYNADSSSRLQIMEHEGDMV--RAKCIP--RKSKTLLTKK 397
ETK + A KRSSSYN D +S + GD +K P RK+ ++L K+
Sbjct: 407 ETKLQKKDAADSNASLKRSSSYNGDRASNQMGVAENGDSKPDSSKNNPSSRKASSILGKQ 466
Query: 398 E 398
+
Sbjct: 467 Q 467
>UniRef100_Q9ZUV7 Hypothetical protein At2g28150 [Arabidopsis thaliana]
Length = 541
Score = 245 bits (625), Expect = 2e-63
Identities = 182/500 (36%), Positives = 255/500 (50%), Gaps = 124/500 (24%)
Query: 7 SRGRPGTRETSPERTKVWTEPKPKTP---RKVSVVYYLSRNGHLEHPHFMEVPLSSPHGL 63
+R + +RE SPER KVWTE PK +KV +VYYLS+N LEHPHFMEV +SSP+GL
Sbjct: 3 ARMKKYSREVSPERAKVWTEKSPKYHQKIKKVQIVYYLSKNRQLEHPHFMEVLISSPNGL 62
Query: 64 YL--------KDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDY 115
YL KDVI RLN+LRG+GM +MYSWSSKRSY+NGFVWHDL+E D I P G++Y
Sbjct: 63 YLRGNVIIYFKDVIERLNVLRGRGMASMYSWSSKRSYRNGFVWHDLSEDDLILPANGNEY 122
Query: 116 ILKGSEILEHETT--ARPPLEEESD--SPVV---------------ITRRRNQSWSSIDL 156
+LKGSE+ + + R P+ +D SP+V +R + S SS +
Sbjct: 123 VLKGSELFDESNSEALRKPMLLCTDHFSPIVNLATQNMKQIVVEPPSSRSMDDSSSSSSM 182
Query: 157 N--------------------------------------------EYRVYKSEPLGENIA 172
N EY+VYKSE L
Sbjct: 183 NNGKGTNKHSHEDDELSPPALRSVSSSGVSPDSRDAKNSSSWCLAEYKVYKSEGL----- 237
Query: 173 ADASTQTEEKRRRR---------RVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELS 223
ADASTQT+E R R + +++ E E +N+ + + +S E+S
Sbjct: 238 ADASTQTDETVSGRSKTPIETFSRGVSTDEDVSSEPETSENNLVSEASCAGKERESAEIS 297
Query: 224 REEISPPPSDSS------PETLGTLMKVD--GRLGLRSVPKENPTVESCPSGRMRASSVL 275
R +SPP S+S+ +TL +L++ D R + +E+ + + P R+RAS++L
Sbjct: 298 RNSVSPPFSNSASSLGGKTDTLESLIRADVSKMNSFRILEQEDVRMPAIP--RLRASNML 355
Query: 276 LQLLSCGAVSFKEGSGASSGKDQRFSLV-------GHYKSRLPRGGGNHVGKEVGTLVEI 328
+QL+SCG++S KD F LV GH K P + + ++ L E
Sbjct: 356 MQLISCGSISV---------KDNNFGLVPTYKPKFGHSKFPSPFFSSSFMMGDLDRLSET 406
Query: 329 PDLNRVRLEDKEYFSGSLIETKKVEAPA------FKRSSSYNADSSSRLQIMEHEGDMV- 381
P L +R+E+KEYFSGSL+ETK + A KRSSSYN D +S + GD
Sbjct: 407 PSLMGLRMEEKEYFSGSLVETKLQKKDAADSNASLKRSSSYNGDRASNQMGVAENGDSKP 466
Query: 382 -RAKCIP--RKSKTLLTKKE 398
+K P RK+ ++L K++
Sbjct: 467 DSSKNNPSSRKASSILGKQQ 486
>UniRef100_Q6AV45 Expressed protein [Oryza sativa]
Length = 521
Score = 225 bits (574), Expect = 2e-57
Identities = 188/520 (36%), Positives = 256/520 (49%), Gaps = 135/520 (25%)
Query: 7 SRGRPGTRETSPERTKVWTEPKPKTPRKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLK 66
SRGR R SPER + RKV VVYYL+R+ HLEHPHF+EVP+SSP GLYL+
Sbjct: 3 SRGR---RPRSPERQR-------PAARKVPVVYYLTRSRHLEHPHFVEVPVSSPEGLYLR 52
Query: 67 DVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEHE 126
DVI LN++RGKGM MYSWS KRSYKNGFVWHDL E D ++P +Y+LKGSE+L+
Sbjct: 53 DVISHLNMVRGKGMAAMYSWSCKRSYKNGFVWHDLGEDDLVHPATDGEYVLKGSELLDQS 112
Query: 127 TT--------------------ARPPLEEE----SDSPVVITR----RRNQSWSSIDLN- 157
++ AR PL E S P VI R RR+ S S++ +
Sbjct: 113 SSGQFYQGTNGNQKQQSRLKEGARLPLPREASYSSSPPSVIVREAKPRRSPSVPSLEEDD 172
Query: 158 ---------------------------------EYRVYKSEPLGENIAADASTQTEEKRR 184
E+RVYK P G DA+TQT++ R
Sbjct: 173 SPVQCRVTSLENMSPESEPQRTLLSRAGSASPAEFRVYK--PTG---CVDAATQTDDLGR 227
Query: 185 RRRVLREEDEEQEEEEKEKNDEIENEGETENQNQ---STEL---SREEISP-------PP 231
R +R+ E ++ +D + E Q+Q S EL +RE +S P
Sbjct: 228 RS--VRKVPEMHKKSLSTDHDSVVREITEYRQSQPRRSAELQGIAREAMSQCHTPLSIPS 285
Query: 232 SDSSPETLGTLMKVDGRL--GLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEG 289
S E+L +L++ D R + +++ + +CP ++R ++VL+QL++CG++S K+
Sbjct: 286 SRGKSESLESLIRADNNALNSFRILEEDDIIMPTCP--KLRPANVLMQLITCGSLSVKD- 342
Query: 290 SGASSGKDQRFSLVGHYKSRLPRG-------GGNHVGKEVGTLVEIPDLNRVRLEDKEYF 342
+ LV YK R P + E+ L E P L +RLEDKEYF
Sbjct: 343 -------HENIRLVEGYKPRFPNMKFPSPLISRTMMMGELDYLSENPRLMGMRLEDKEYF 395
Query: 343 SGSLIETK-KVEAP-----AFKRSSSYNAD-SSSRLQIMEHEGDMV---RAKCIPRKS-- 390
SGSLIETK + + P A KRSSSYNA+ S+ L + D V RA+C+PR
Sbjct: 396 SGSLIETKMQRDVPADRYSALKRSSSYNAERSNETLDCARPDEDTVNTSRARCLPRTPIL 455
Query: 391 KTLLTKKEEGTSSSMD------------SVSRSQHGSKRF 418
+ L K E S + S++ S GSKRF
Sbjct: 456 SSFLHPKSEAMKSPISDCRRSSSAGPDCSLASSGDGSKRF 495
>UniRef100_Q6YYY6 Putative auxin-regulated protein [Oryza sativa]
Length = 502
Score = 176 bits (445), Expect = 1e-42
Identities = 154/505 (30%), Positives = 224/505 (43%), Gaps = 158/505 (31%)
Query: 7 SRGRPGTRETSPERTKVWTEP--------------KPKTPRKVSVVYYLSRNGHLEHPHF 52
+R R SP R KVW EP P + ++V+VVYYL RN HLEHPHF
Sbjct: 6 TRTRRRAATASPGRNKVWVEPPGKTHHLRSPPPPPSPSSSKRVAVVYYLCRNRHLEHPHF 65
Query: 53 MEVPLSSPHGLYLK-------------------------------DVIIRLNLLRGKGMP 81
+EVPL+SP GLYL+ DVI RLN+LRGKGM
Sbjct: 66 IEVPLASPDGLYLRGLYPSLFLVQASVFFFFPFSIPTNDAGGVDADVINRLNVLRGKGMA 125
Query: 82 TMYSWSSKRSYKNGFVWHDLTESDFIYPTQGHDYILKGSEILEH---------------E 126
MYSWS KRSYKNGFVWHDL+E D + P QG++YILKGSE+L+ E
Sbjct: 126 AMYSWSCKRSYKNGFVWHDLSEDDLVLPAQGNEYILKGSELLDRSPPDRQQNGVGEPKVE 185
Query: 127 TTARPPLE-----------EESDSPVVITRR----------RNQSWSSIDL--------- 156
T PP E S SP +T+ + Q+ S+ L
Sbjct: 186 TLKHPPEESPHSRGSQEGCSSSSSPSAVTKEASPPPPTPQPQQQAQSATLLPSSSASTNR 245
Query: 157 --NEYRVYKS--------EPLGENI--AADASTQTEEKRRRRRVLREEDEEQEEEEKEKN 204
++ R +S EP G ++AS+ + R + + +D + ++ E++
Sbjct: 246 EDDQCRTPRSGSSGNMSPEPAGRVAPPLSEASSPGPLEYRVCKPIGAQDASTQTDDSERD 305
Query: 205 DEIENEGE----TENQNQSTELSR---------------EEISPP--PSDSSP----ETL 239
N TEN +E+ SPP SD+SP ETL
Sbjct: 306 APERNSRMAGVCTENSTSDSEIQECHPRSTQPSPKGPGVVRESPPVCSSDASPGGRVETL 365
Query: 240 GTLMKVDG--RLGLRSVPKENPTVESCPSG-RMRASSVLLQLLSCGAVSFKEGSGASSGK 296
+L++ + R R++ +E+ P G + + +++L+QL++CG++S K+
Sbjct: 366 ESLIRAEASRRSSFRALEEEH---LFAPMGVKFKPANLLMQLITCGSISVKD-------- 414
Query: 297 DQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLNRV---------RLEDKEYFSGSLI 347
+ F L+ Y+ R + T + + L+ + R+ + EYFSGSL+
Sbjct: 415 HRSFGLIPTYRPRFTQ--VEFPSPMFSTPLALRHLDNIPCNARTIGMRIPESEYFSGSLV 472
Query: 348 ETKKVE------APAFKRSSSYNAD 366
ETKK + P KRSSS + D
Sbjct: 473 ETKKQDESGKGGTPTLKRSSSCSED 497
>UniRef100_Q9LX14 Hypothetical protein T31P16_140 [Arabidopsis thaliana]
Length = 414
Score = 160 bits (406), Expect = 5e-38
Identities = 130/425 (30%), Positives = 211/425 (49%), Gaps = 60/425 (14%)
Query: 1 MSVTSSSRGRPGTRETSPERT-----------KVWTEPKPKTP--RKVSVVYYLSRNGHL 47
M RGR + SPER ++ E K K P R+V VVYYL+RNGHL
Sbjct: 1 MEAVRCRRGRENNK--SPERIIRSLNHHQHDEELEEEVKTKKPIFRRVQVVYYLTRNGHL 58
Query: 48 EHPHFMEV--PLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESD 105
EHPHF+EV P++ P L L+DV+ RL +LRGK M + Y+WS KRSY+NGFVW+DL E+D
Sbjct: 59 EHPHFIEVISPVNQP--LRLRDVMNRLTILRGKCMTSQYAWSCKRSYRNGFVWNDLAEND 116
Query: 106 FIYPTQGHDYILKGSEILE--HETTARPPLE---EESDSPVVITRRRNQSWSSIDLNEYR 160
IYP+ +Y+LKGSEI + E PL +E+ ++ + + ++
Sbjct: 117 VIYPSDCAEYVLKGSEITDKFQEVHVNRPLSGSIQEAPKSRLLRSKLKPQNRTASFDDAE 176
Query: 161 VYKSEPLGE-----NIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETEN 215
+Y E E + + ++ T + R R E E ++K + + E + +
Sbjct: 177 LYVGEEEEEEDGEYELYEEKTSYTSSTTPQSRCSRGVSTETMESTEQKPNLTKTEQDLQV 236
Query: 216 QNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVESCPSGRMRASSVL 275
++ S++L+R S+P +V R+ ++ VE SGR S+
Sbjct: 237 RSDSSDLTR---------SNPVVKPRRHEVSTRV------EDGDPVEP-GSGR---GSMW 277
Query: 276 LQLLSCGAVSFKEGSGASSG---KDQRFSLVGHYKSRLPRGGGNHVGKEVGTLVEIPDLN 332
LQ++SCG ++ K + + K++ K+ + + + + + + E P
Sbjct: 278 LQMISCGHIATKYYAPSVMNPRQKEENLRKGVLCKNIVKKTVVDDEREMIRFMSENPRFG 337
Query: 333 RVRLEDKEYFSGSLIETKKVE----APAFKRSSSYNADSSSRLQI-----MEHEGDMVRA 383
+ E+KEYFSGS++E+ E P+ +RS+S+N + S +++ + E M +
Sbjct: 338 NPQAEEKEYFSGSIVESVSQERVTAEPSLRRSNSFNEERSKIVEMAKETKKKEERSMAKV 397
Query: 384 KCIPR 388
KCIPR
Sbjct: 398 KCIPR 402
>UniRef100_Q94CT4 P0459B04.11 protein [Oryza sativa]
Length = 392
Score = 123 bits (309), Expect = 9e-27
Identities = 112/362 (30%), Positives = 161/362 (43%), Gaps = 103/362 (28%)
Query: 23 VW-TEPKPKTPRKV--SVVYYLSR-NGHLEHPHFMEVPLSSPHG-------LYLKDVIIR 71
VW TE K K ++V VVYYL R +G L+HPHF+ V + S L+L+D I R
Sbjct: 2 VWRTELKKKQMKQVVVGVVYYLCRQDGQLDHPHFVHVHVPSDSDSDHPRPRLHLRDFIAR 61
Query: 72 LNLLRGKGMPTMYSWSSKRSYKN--GFVWHDLTESDFI-YPTQGH-DYILKGSEILEHET 127
L+ LRG MP YSWS+K +Y+ G+VW DLT D I P+ H +Y+LKGS +L H +
Sbjct: 62 LSDLRGAAMPAAYSWSAKTTYRRNAGYVWQDLTADDLIPAPSTNHEEYVLKGSPLLHHNS 121
Query: 128 TARPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRR 187
P + + +S DL +Y ++P+ AA
Sbjct: 122 NTPP--------------QHRRCMTSFDLADYH-RTTDPVPVPAAA-------------- 152
Query: 188 VLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSP-ETLGTLMKVD 246
Q + + +EISPPPS SSP +T L+ +
Sbjct: 153 ------------------------------QQSLIGIDEISPPPSSSSPDDTTTQLVTLK 182
Query: 247 GRLGLRSVPKENPTVESCPSGRMRASSVLLQLLSCGAVSFKEGSGASSGKDQRFSLVGHY 306
+ + P+GRMR S++L++L+SCGA S KE L G
Sbjct: 183 QKQQEEDGCTPQQQAATTPAGRMRTSAMLMKLISCGASSIKE-------------LQGQA 229
Query: 307 KSRLPRGGGNHVGKEVGTLVEIPDLNRVRLEDKEYFSGSLIETKKVEAP---AFKRSSSY 363
+S+ R H K PD+ ++ ++YFSGSL++ P +RSSS
Sbjct: 230 QSQRCRATAWHNNK--------PDI----MDHRDYFSGSLLDNNTTTHPIDLTLRRSSSC 277
Query: 364 NA 365
NA
Sbjct: 278 NA 279
>UniRef100_Q9SYJ8 F3F20.2 protein [Arabidopsis thaliana]
Length = 372
Score = 115 bits (287), Expect = 3e-24
Identities = 82/261 (31%), Positives = 120/261 (45%), Gaps = 44/261 (16%)
Query: 33 RKVSVVYYLSRNGHLEHPHFMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSY 92
R+V++VY+LSR+GH++HPH + V S +G++L+DV L RG MP +SWS KR Y
Sbjct: 11 RRVNLVYFLSRSGHVDHPHLLRVHHLSRNGVFLRDVKKWLADARGDAMPDAFSWSCKRRY 70
Query: 93 KNGFVWHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITRR----RN 148
KNG+VW DL + D I P ++Y+LKGSEIL P E+ +TR
Sbjct: 71 KNGYVWQDLLDDDLITPISDNEYVLKGSEILLSSPKEDYPNVEKK---AWVTRNGGIDAE 127
Query: 149 QSWSSIDLNEYRVYKSEPL--GENIAADASTQTEEKRRRRR--VLREEDEEQEEEEKEKN 204
+ + L ++ K P+ + A ST TEE VL+++D K
Sbjct: 128 EKLQKLKLTSEKIQKESPVFCSQRSTATTSTVTEESTTNEEGFVLKKQD--------PKT 179
Query: 205 DEIENEGETEN-QNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVES 263
+ +G TEN E R +S S SS +++
Sbjct: 180 VSGQRDGSTENGSGNDVESGRPSVSSTTSSSS------------------------YIKN 215
Query: 264 CPSGRMRASSVLLQLLSCGAV 284
+RAS VL L+ CG +
Sbjct: 216 KSYSSVRASHVLRNLMKCGGL 236
>UniRef100_Q651B3 Hypothetical protein B1331F11.4 [Oryza sativa]
Length = 519
Score = 109 bits (273), Expect = 1e-22
Identities = 63/140 (45%), Positives = 78/140 (55%), Gaps = 8/140 (5%)
Query: 1 MSVTSSSRGRPGTRETSPERTKVWTEPKPKTPRKVSVVYYL--SRNGHLEHPHFMEVPLS 58
+S + G + + E T + K RKV+VVYYL SR G LEHPH MEV +
Sbjct: 4 LSQAEAGGGGRSKKSAAGELTTTSEKKKKTRRRKVAVVYYLCRSRQGGLEHPHLMEVEVG 63
Query: 59 SPHG-----LYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTE-SDFIYPTQG 112
L L+DV RL+ LRGKGM MYSWS KRSY+ G+VWHDL+ D + PT
Sbjct: 64 DGEEQVHVQLRLRDVTRRLDALRGKGMAAMYSWSCKRSYRGGYVWHDLSHPDDLLLPTGP 123
Query: 113 HDYILKGSEILEHETTARPP 132
HDY+LK S + H PP
Sbjct: 124 HDYVLKASLLHLHHLIDPPP 143
>UniRef100_UPI000042E1DD UPI000042E1DD UniRef100 entry
Length = 363
Score = 54.3 bits (129), Expect = 6e-06
Identities = 35/132 (26%), Positives = 62/132 (46%), Gaps = 2/132 (1%)
Query: 147 RNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDE 206
RN + S + RV + + + + EE+ EE+EE+EEEE+E+ +E
Sbjct: 234 RNSNRSKTTTSNNRVKRDKYSDDECEDHEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 293
Query: 207 IENEGETENQNQSTELSREEISPPPSDSSPETLGTLMKVDGRLGLRSVPKENPTVESCPS 266
E E E E + + E EE + +P+ L T + GR ++ KE+ + + +
Sbjct: 294 EEEEEEEEEEEEEEEEEEEEEEEEEEEEAPKQLKTPQR--GRRAAENIAKEDDRLNNTYN 351
Query: 267 GRMRASSVLLQL 278
+R S+ L +L
Sbjct: 352 TSVRRSNRLKKL 363
>UniRef100_Q7RPI6 Hypothetical protein [Plasmodium yoelii yoelii]
Length = 426
Score = 51.6 bits (122), Expect = 4e-05
Identities = 29/93 (31%), Positives = 49/93 (52%)
Query: 134 EEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREED 193
E+ES + + + ++R + S+ DL E + GE IA + E EE+
Sbjct: 29 EDESTNDLPLEKKRKEHASNDDLPEPKKMPLIEGGEGIAKEEEEGEAENEEEEDEEEEEN 88
Query: 194 EEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
EE+E+EE+E+N+E E+E E E++ E +E
Sbjct: 89 EEEEDEEEEENEEEEDEDENEDEEDEDENEEDE 121
>UniRef100_UPI00004532EC UPI00004532EC UniRef100 entry
Length = 366
Score = 51.2 bits (121), Expect = 5e-05
Identities = 30/101 (29%), Positives = 52/101 (50%), Gaps = 2/101 (1%)
Query: 178 QTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPE 237
+ EE+ EE+EE+EEEE+E+ +E E E E E + + E EE + +P+
Sbjct: 268 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEAPK 327
Query: 238 TLGTLMKVDGRLGLRSVPKENPTVESCPSGRMRASSVLLQL 278
L T + GR ++ KE+ + + + +R S+ L +L
Sbjct: 328 QLKTPQR--GRRAAENIAKEDDRLNNTYNTSVRRSNRLKKL 366
>UniRef100_UPI000004DF1F UPI000004DF1F UniRef100 entry
Length = 849
Score = 49.3 bits (116), Expect = 2e-04
Identities = 28/135 (20%), Positives = 66/135 (48%), Gaps = 2/135 (1%)
Query: 130 RPPLEEESDSPVVITRRRNQSWSSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVL 189
+ P+ ++ D+P+V R+ Q + D N+ + I A EE++ +
Sbjct: 8 KSPVVKKKDTPIVRKRKPIQQEAEEDSNDEESEDELNVEGLIDASDDEDEEEEQEQEEQP 67
Query: 190 REEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSDSSPETLGTLMKVD--G 247
+EE+ ++++ + +D+ +N E +++ + +L EE PSD + + K D
Sbjct: 68 QEEENNSDDDDDDDDDDDDNNSEADSEEELNQLLGEEEEEDPSDYNSDEFSDEPKDDDLS 127
Query: 248 RLGLRSVPKENPTVE 262
R+ ++S+ +P+++
Sbjct: 128 RINIKSLSVSDPSIQ 142
>UniRef100_Q9LN21 F14O10.11 protein [Arabidopsis thaliana]
Length = 409
Score = 49.3 bits (116), Expect = 2e-04
Identities = 32/129 (24%), Positives = 54/129 (41%)
Query: 98 WHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLN 157
W + ++SDF T ++ +G+ + E EEE + + +
Sbjct: 249 WVNPSDSDFYKFTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 308
Query: 158 EYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQN 217
E + E E + + EE+ EE+EE+EEEE+E+ +E E E E E +
Sbjct: 309 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 368
Query: 218 QSTELSREE 226
+ E EE
Sbjct: 369 EEEEEEEEE 377
Score = 47.4 bits (111), Expect = 8e-04
Identities = 32/129 (24%), Positives = 51/129 (38%)
Query: 98 WHDLTESDFIYPTQGHDYILKGSEILEHETTARPPLEEESDSPVVITRRRNQSWSSIDLN 157
W L + ++ P+ Y S + E T EEE + + +
Sbjct: 241 WRVLCSTKWVNPSDSDFYKFTPSMVHVGEGTIVQAEEEEEEEEEEEEEEEEEEEEEEEEE 300
Query: 158 EYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQN 217
E + E E + + EE+ EE+EE+EEEE+E+ +E E E E E +
Sbjct: 301 EEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEE 360
Query: 218 QSTELSREE 226
+ E EE
Sbjct: 361 EEEEEEEEE 369
>UniRef100_UPI000024D6C3 UPI000024D6C3 UniRef100 entry
Length = 473
Score = 48.9 bits (115), Expect = 3e-04
Identities = 25/66 (37%), Positives = 38/66 (56%)
Query: 172 AADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPP 231
AA + TQ EE+ EE+EE+EEEE E+ +E E + E E ++Q E ++EE P
Sbjct: 359 AAASPTQEEEEEEEEAKEEEEEEEKEEEEAEEKEEEEEKEEEEEKDQEEEETKEEEEEPA 418
Query: 232 SDSSPE 237
+ + E
Sbjct: 419 KEKAKE 424
Score = 43.9 bits (102), Expect = 0.009
Identities = 19/64 (29%), Positives = 36/64 (55%)
Query: 174 DASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPPSD 233
+A + EE+ + E++EE+E+EE+E+ D+ E E + E + + E ++E+ P D
Sbjct: 373 EAKEEEEEEEKEEEEAEEKEEEEEKEEEEEKDQEEEETKEEEEEPAKEKAKEKPEPESKD 432
Query: 234 SSPE 237
E
Sbjct: 433 KGKE 436
Score = 36.2 bits (82), Expect = 1.8
Identities = 24/97 (24%), Positives = 43/97 (43%), Gaps = 8/97 (8%)
Query: 172 AADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREEISPPP 231
A +A T ++ + +EE+EE+EE ++E+ +E + E E E + + E EE
Sbjct: 347 ADEAITSSQAQDAAASPTQEEEEEEEEAKEEEEEEEKEEEEAEEKEEEEEKEEEEEKDQE 406
Query: 232 SDSSPETLGTLMKVDGRLGLRSVPKENPTVESCPSGR 268
+ + E + + KE P ES G+
Sbjct: 407 EEETKE--------EEEEPAKEKAKEKPEPESKDKGK 435
Score = 35.0 bits (79), Expect = 4.0
Identities = 17/60 (28%), Positives = 31/60 (51%)
Query: 167 LGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTELSREE 226
+ + +AD + + + + +E+EE+EEE KE+ +E E E E + + E EE
Sbjct: 341 VSSSFSADEAITSSQAQDAAASPTQEEEEEEEEAKEEEEEEEKEEEEAEEKEEEEEKEEE 400
>UniRef100_Q09936 Hypothetical protein C53C9.2 in chromosome X [Caenorhabditis
elegans]
Length = 397
Score = 48.9 bits (115), Expect = 3e-04
Identities = 40/173 (23%), Positives = 79/173 (45%), Gaps = 5/173 (2%)
Query: 52 FMEVPLSSPHGLYLKDVIIRLNLLRGKGMPTMYSWSSKRSYKNGFVWHDLTESDFIYPTQ 111
F++V PH + KD+ L + +G+ + S ++K + + G + +
Sbjct: 227 FLKVRDVLPHTVGGKDIEEELKQ-KSEGIVPLQSGTNKLASQRGMTGFGTPRNTQLRAGW 285
Query: 112 GHDYILKGSEILEHETTARPPLEEESDSPVVITRRR---NQSWSSIDLNEYRVYKSEPLG 168
++I L+ +PP S P +++ +S +++ V SEP+
Sbjct: 286 KKEWIEDYEAALKEWEETKPPGSASSVDPFGHYKKKFEERESSRQSEIDSQSVKASEPV- 344
Query: 169 ENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEGETENQNQSTE 221
E + + EE++ +EE+EE+EEEE+E+ +E+E E E E + + E
Sbjct: 345 EPEPEEEEEEEEEEKIEEPAAKEEEEEEEEEEEEEEEELEEEEEEEEEEEEDE 397
Score = 34.3 bits (77), Expect = 6.8
Identities = 20/86 (23%), Positives = 39/86 (45%)
Query: 152 SSIDLNEYRVYKSEPLGENIAADASTQTEEKRRRRRVLREEDEEQEEEEKEKNDEIENEG 211
S+ ++ + YK + + + ++ + V E +EE+EEEE+EK +E +
Sbjct: 308 SASSVDPFGHYKKKFEERESSRQSEIDSQSVKASEPVEPEPEEEEEEEEEEKIEEPAAKE 367
Query: 212 ETENQNQSTELSREEISPPPSDSSPE 237
E E + + E EE+ + E
Sbjct: 368 EEEEEEEEEEEEEEELEEEEEEEEEE 393
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.309 0.128 0.359
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 737,714,857
Number of Sequences: 2790947
Number of extensions: 33522111
Number of successful extensions: 400140
Number of sequences better than 10.0: 4013
Number of HSP's better than 10.0 without gapping: 1968
Number of HSP's successfully gapped in prelim test: 2210
Number of HSP's that attempted gapping in prelim test: 307544
Number of HSP's gapped (non-prelim): 30872
length of query: 422
length of database: 848,049,833
effective HSP length: 130
effective length of query: 292
effective length of database: 485,226,723
effective search space: 141686203116
effective search space used: 141686203116
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC139841.8


