

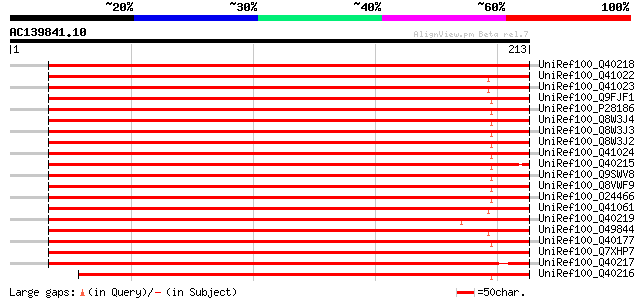
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139841.10 - phase: 0
(213 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q40218 RAB8D [Lotus japonicus] 370 e-101
UniRef100_Q41022 Small GTP-binding protein [Pisum sativum] 360 1e-98
UniRef100_Q41023 Small GTP-binding protein [Pisum sativum] 359 3e-98
UniRef100_Q9FJF1 Rab-type small GTP-binding protein-like [Arabid... 358 4e-98
UniRef100_P28186 Ras-related protein ARA-3 [Arabidopsis thaliana] 358 5e-98
UniRef100_Q8W3J4 Ras-related protein RAB8-1 [Nicotiana tabacum] 356 2e-97
UniRef100_Q8W3J3 Ras-related protein RAB8-3 [Nicotiana tabacum] 356 2e-97
UniRef100_Q8W3J2 Ras-related protein RAB8-5 [Nicotiana tabacum] 355 3e-97
UniRef100_Q41024 Small GTP-binding protein [Pisum sativum] 355 4e-97
UniRef100_Q40215 RAB8A [Lotus japonicus] 354 1e-96
UniRef100_Q9SWV8 Ethylene-responsive small GTP-binding protein [... 353 1e-96
UniRef100_Q8VWF9 Ras-related protein RAB8-4 [Nicotiana tabacum] 353 2e-96
UniRef100_O24466 GTPase AtRAB8 [Arabidopsis thaliana] 352 4e-96
UniRef100_Q41061 Small G protein [Pisum sativum] 350 1e-95
UniRef100_Q40219 RAB8E [Lotus japonicus] 350 1e-95
UniRef100_O49844 Small GTP-binding protein [Daucus carota] 350 2e-95
UniRef100_Q40177 GTP-binding protein [Lycopersicon esculentum] 348 5e-95
UniRef100_Q7XHP7 Putative ethylene-responsive small GTP-binding ... 347 2e-94
UniRef100_Q40217 RAB8C [Lotus japonicus] 345 5e-94
UniRef100_Q40216 RAB8B [Lotus japonicus] 343 2e-93
>UniRef100_Q40218 RAB8D [Lotus japonicus]
Length = 214
Score = 370 bits (949), Expect = e-101
Identities = 183/197 (92%), Positives = 193/197 (97%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKR+KLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRVKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDEASFNNIRNW+RNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWLRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD+KAEPTT+KINQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDHKAEPTTLKINQ 197
Query: 197 DSATGSGQAAQKSACCG 213
DSA G+G+AA KS+CCG
Sbjct: 198 DSAAGAGEAANKSSCCG 214
>UniRef100_Q41022 Small GTP-binding protein [Pisum sativum]
Length = 215
Score = 360 bits (924), Expect = 1e-98
Identities = 183/198 (92%), Positives = 189/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKIN- 195
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRLADTD+K+EP TIKIN
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLADTDSKSEPQTIKINQ 197
Query: 196 QDSATGSGQAAQKSACCG 213
QD A GQAA KSACCG
Sbjct: 198 QDPAANGGQAATKSACCG 215
>UniRef100_Q41023 Small GTP-binding protein [Pisum sativum]
Length = 215
Score = 359 bits (921), Expect = 3e-98
Identities = 182/198 (91%), Positives = 189/198 (94%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKIN- 195
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRLADTD+++EP TIKIN
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLADTDSRSEPQTIKINQ 197
Query: 196 QDSATGSGQAAQKSACCG 213
QD A GQAA KSACCG
Sbjct: 198 QDPAANGGQAATKSACCG 215
>UniRef100_Q9FJF1 Rab-type small GTP-binding protein-like [Arabidopsis thaliana]
Length = 216
Score = 358 bits (920), Expect = 4e-98
Identities = 181/198 (91%), Positives = 190/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVP SKGQALADEYGIKFFETSAKTNLNVEEVFFSIA+DIKQRLADTD++AEP TIKI+Q
Sbjct: 138 AVPKSKGQALADEYGIKFFETSAKTNLNVEEVFFSIAKDIKQRLADTDSRAEPATIKISQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A G+GQA QKSACCG
Sbjct: 198 TDQAAGAGQATQKSACCG 215
>UniRef100_P28186 Ras-related protein ARA-3 [Arabidopsis thaliana]
Length = 216
Score = 358 bits (919), Expect = 5e-98
Identities = 180/198 (90%), Positives = 190/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPT+KGQALADEYGIKFFETSAKTNLNVEEVFFSI RDIKQRL+DTD++AEP TIKI+Q
Sbjct: 138 AVPTAKGQALADEYGIKFFETSAKTNLNVEEVFFSIGRDIKQRLSDTDSRAEPATIKISQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A G+GQA QKSACCG
Sbjct: 198 TDQAAGAGQATQKSACCG 215
>UniRef100_Q8W3J4 Ras-related protein RAB8-1 [Nicotiana tabacum]
Length = 216
Score = 356 bits (914), Expect = 2e-97
Identities = 179/198 (90%), Positives = 188/198 (94%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRLA++DN+AEP TI+INQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLAESDNRAEPQTIRINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D G Q AQKSACCG
Sbjct: 198 PDQGAGGAQTAQKSACCG 215
>UniRef100_Q8W3J3 Ras-related protein RAB8-3 [Nicotiana tabacum]
Length = 216
Score = 356 bits (914), Expect = 2e-97
Identities = 179/198 (90%), Positives = 190/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRLA++D+KAEP TI+INQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLAESDSKAEPQTIRINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A G+ Q+ QKSACCG
Sbjct: 198 PDQAAGASQSVQKSACCG 215
>UniRef100_Q8W3J2 Ras-related protein RAB8-5 [Nicotiana tabacum]
Length = 216
Score = 355 bits (912), Expect = 3e-97
Identities = 179/198 (90%), Positives = 190/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELD KRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDSKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRLA++D+KAEP TI+INQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLAESDSKAEPQTIRINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A G+ Q+AQKSACCG
Sbjct: 198 PDQAAGAAQSAQKSACCG 215
>UniRef100_Q41024 Small GTP-binding protein [Pisum sativum]
Length = 216
Score = 355 bits (911), Expect = 4e-97
Identities = 178/198 (89%), Positives = 188/198 (94%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNI+NWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIKNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTN+NV+EVFFSIARDIKQRLA+TD+K EP T+KINQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVDEVFFSIARDIKQRLAETDSKTEPQTLKINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D GS QA QKSACCG
Sbjct: 198 PDQGAGSAQATQKSACCG 215
>UniRef100_Q40215 RAB8A [Lotus japonicus]
Length = 216
Score = 354 bits (908), Expect = 1e-96
Identities = 181/198 (91%), Positives = 190/198 (95%), Gaps = 2/198 (1%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 19 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 78
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDEASFNNI+NWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 79 RTITTAYYRGAMGILLVYDVTDEASFNNIKNWIRNIEQHASDNVNKILVGNKADMDESKR 138
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD++AEP TI+INQ
Sbjct: 139 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDSRAEPQTIQINQ 198
Query: 197 -DSATGSGQAAQKSACCG 213
D++ GQAAQKS CCG
Sbjct: 199 PDASASGGQAAQKS-CCG 215
>UniRef100_Q9SWV8 Ethylene-responsive small GTP-binding protein [Lycopersicon
esculentum]
Length = 216
Score = 353 bits (907), Expect = 1e-96
Identities = 177/198 (89%), Positives = 189/198 (95%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGS TTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSLTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPT+KGQALADEYGIKFFETSAKTNLNVE+VFFSIARDIKQRLADTD+KAEP+T+KINQ
Sbjct: 138 AVPTAKGQALADEYGIKFFETSAKTNLNVEQVFFSIARDIKQRLADTDSKAEPSTLKINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
++ G Q +QKSACCG
Sbjct: 198 PEAGAGGSQTSQKSACCG 215
>UniRef100_Q8VWF9 Ras-related protein RAB8-4 [Nicotiana tabacum]
Length = 216
Score = 353 bits (905), Expect = 2e-96
Identities = 178/198 (89%), Positives = 189/198 (94%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRL+++D+K EP I+INQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLSESDSKTEPQAIRINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A SGQAAQKS+CCG
Sbjct: 198 SDQAGTSGQAAQKSSCCG 215
>UniRef100_O24466 GTPase AtRAB8 [Arabidopsis thaliana]
Length = 216
Score = 352 bits (903), Expect = 4e-96
Identities = 178/198 (89%), Positives = 187/198 (93%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASD+VNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDSVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVP SKGQALADEYG+KFFETSAKTNLNVEEVFFSIA+DIKQRLADTD +AEP TIKINQ
Sbjct: 138 AVPKSKGQALADEYGMKFFETSAKTNLNVEEVFFSIAKDIKQRLADTDARAEPQTIKINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D G+ QA QKSACCG
Sbjct: 198 SDQGAGTSQATQKSACCG 215
>UniRef100_Q41061 Small G protein [Pisum sativum]
Length = 215
Score = 350 bits (899), Expect = 1e-95
Identities = 179/198 (90%), Positives = 185/198 (93%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ VGKSCLLLRFSDGSFTTSFI TIGIDFKIRTIELDGK IKLQIWDTAGQERF
Sbjct: 18 LLLIGDRRVGKSCLLLRFSDGSFTTSFIATIGIDFKIRTIELDGKPIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKIN- 195
AVPTSKGQALADEYGIKFFETSAKTN+NVEEVFFSIARDIKQRLADTD+++EP TIKIN
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNMNVEEVFFSIARDIKQRLADTDSRSEPQTIKINQ 197
Query: 196 QDSATGSGQAAQKSACCG 213
QD A GQAA KSACCG
Sbjct: 198 QDPAANGGQAATKSACCG 215
>UniRef100_Q40219 RAB8E [Lotus japonicus]
Length = 215
Score = 350 bits (899), Expect = 1e-95
Identities = 178/198 (89%), Positives = 186/198 (93%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNI+NWI NIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIKNWIHNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD-NKAEPTTIKIN 195
AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD +K EPT IKIN
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDSSKTEPTGIKIN 197
Query: 196 QDSATGSGQAAQKSACCG 213
+G+AAQKSACCG
Sbjct: 198 PQDKGSAGEAAQKSACCG 215
>UniRef100_O49844 Small GTP-binding protein [Daucus carota]
Length = 216
Score = 350 bits (897), Expect = 2e-95
Identities = 175/198 (88%), Positives = 187/198 (94%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIE+DGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIEMDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKIN- 195
AVPTSKGQALADEYGIKFFE SAKTN+NVEEVFFSIA+DIKQRLA+TD+K EP TIKIN
Sbjct: 138 AVPTSKGQALADEYGIKFFEASAKTNMNVEEVFFSIAKDIKQRLAETDSKTEPQTIKINQ 197
Query: 196 QDSATGSGQAAQKSACCG 213
Q+ G+ A+QKSACCG
Sbjct: 198 QEQGAGTSAASQKSACCG 215
>UniRef100_Q40177 GTP-binding protein [Lycopersicon esculentum]
Length = 216
Score = 348 bits (893), Expect = 5e-95
Identities = 175/198 (88%), Positives = 187/198 (94%), Gaps = 1/198 (0%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDTGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSI +DIKQRL+++D+K EP +I+INQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIGKDIKQRLSESDSKTEPQSIRINQ 197
Query: 197 -DSATGSGQAAQKSACCG 213
D A +GQ AQKS+CCG
Sbjct: 198 SDQAGTAGQGAQKSSCCG 215
>UniRef100_Q7XHP7 Putative ethylene-responsive small GTP-binding protein [Oryza
sativa]
Length = 215
Score = 347 bits (889), Expect = 2e-94
Identities = 174/197 (88%), Positives = 182/197 (92%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGAMGILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAMGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTNLNVE+VFFSIARDIKQRLA+TD+K E TIKIN+
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNLNVEQVFFSIARDIKQRLAETDSKTEDRTIKINK 197
Query: 197 DSATGSGQAAQKSACCG 213
QKSACCG
Sbjct: 198 PEGDAEATTLQKSACCG 214
>UniRef100_Q40217 RAB8C [Lotus japonicus]
Length = 212
Score = 345 bits (885), Expect = 5e-94
Identities = 177/197 (89%), Positives = 182/197 (91%), Gaps = 3/197 (1%)
Query: 17 ILLLFFAGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERF 76
+LL+ +GVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQER
Sbjct: 18 LLLIGDSGVGKSCLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERV 77
Query: 77 RTITTAYYRGAMGILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 136
RTITTAYYRGA GILLVYDVTDE+SFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR
Sbjct: 78 RTITTAYYRGAKGILLVYDVTDESSFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKR 137
Query: 137 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ 196
AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTD+KAEP TIKINQ
Sbjct: 138 AVPTSKGQALADEYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDSKAEPQTIKINQ 197
Query: 197 DSATGSGQAAQKSACCG 213
AAQ S CCG
Sbjct: 198 PDQPA---AAQNSTCCG 211
>UniRef100_Q40216 RAB8B [Lotus japonicus]
Length = 187
Score = 343 bits (879), Expect = 2e-93
Identities = 172/186 (92%), Positives = 178/186 (95%), Gaps = 1/186 (0%)
Query: 29 CLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTAYYRGAM 88
CLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTAYYRGAM
Sbjct: 1 CLLLRFSDGSFTTSFITTIGIDFKIRTIELDGKRIKLQIWDTAGQERFRTITTAYYRGAM 60
Query: 89 GILLVYDVTDEASFNNIRNWIRNIEQHASDNVNKILVGNKADMDESKRAVPTSKGQALAD 148
GILLVYDVTDE+SFNNI+NWIRNIEQHASDNVNKILVGNKADMDESKRAVPTSKGQALAD
Sbjct: 61 GILLVYDVTDESSFNNIKNWIRNIEQHASDNVNKILVGNKADMDESKRAVPTSKGQALAD 120
Query: 149 EYGIKFFETSAKTNLNVEEVFFSIARDIKQRLADTDNKAEPTTIKINQ-DSATGSGQAAQ 207
EYGIKFFETSAKTNLNV+EVFFSIARDIKQR+AD D+KAEP T+KINQ D GS QAA
Sbjct: 121 EYGIKFFETSAKTNLNVDEVFFSIARDIKQRVADNDSKAEPQTLKINQPDQGAGSAQAAP 180
Query: 208 KSACCG 213
KSACCG
Sbjct: 181 KSACCG 186
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 329,763,645
Number of Sequences: 2790947
Number of extensions: 12446414
Number of successful extensions: 44950
Number of sequences better than 10.0: 4089
Number of HSP's better than 10.0 without gapping: 3288
Number of HSP's successfully gapped in prelim test: 801
Number of HSP's that attempted gapping in prelim test: 37572
Number of HSP's gapped (non-prelim): 4203
length of query: 213
length of database: 848,049,833
effective HSP length: 122
effective length of query: 91
effective length of database: 507,554,299
effective search space: 46187441209
effective search space used: 46187441209
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC139841.10


