

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139745.7 + phase: 0
(240 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
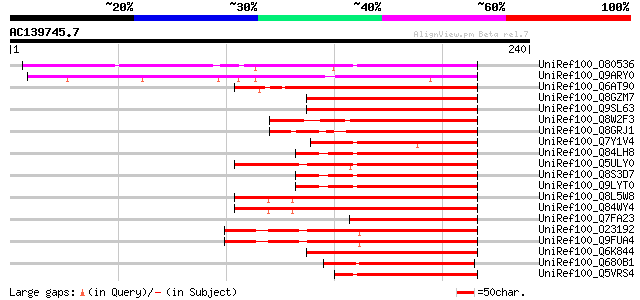
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O80536 Phytochrome-interacting factor 3 [Arabidopsis t... 111 1e-23
UniRef100_Q9ARY0 P0498A12.24 protein [Oryza sativa] 101 1e-20
UniRef100_Q6AT90 Hypothetical protein OSJNBa0069I13.9 [Oryza sat... 99 7e-20
UniRef100_Q8GZM7 Putative bHLH transcription factor [Arabidopsis... 93 5e-18
UniRef100_Q9SL63 Hypothetical protein At2g20180 [Arabidopsis tha... 93 5e-18
UniRef100_Q8W2F3 Phytochrome-interacting factor 4 [Arabidopsis t... 93 7e-18
UniRef100_Q8GRJ1 Transcription factor BHLH9-like protein [Oryza ... 92 9e-18
UniRef100_Q7Y1V4 BP-5 protein [Oryza sativa] 91 3e-17
UniRef100_Q84LH8 PIF3 like basic Helix Loop Helix protein [Arabi... 91 3e-17
UniRef100_Q5ULY0 Basic helix-loop-helix protein [Fragaria ananassa] 91 3e-17
UniRef100_Q8S3D7 Putative bHLH transcription factor [Arabidopsis... 91 3e-17
UniRef100_Q9LYT0 Hypothetical protein F17J16_110 [Arabidopsis th... 91 3e-17
UniRef100_Q8L5W8 PIF3 like basic Helix Loop Helix protein [Arabi... 89 7e-17
UniRef100_Q84WY4 Hypothetical protein At2g46970/F14M4.20 [Arabid... 89 7e-17
UniRef100_Q7FA23 OSJNBa0058K23.6 protein [Oryza sativa] 88 2e-16
UniRef100_O23192 Hypothetical protein C7A10.430 [Arabidopsis tha... 86 6e-16
UniRef100_Q9FUA4 SPATULA [Arabidopsis thaliana] 86 6e-16
UniRef100_Q6K844 Hypothetical protein OJ1548_F12.19 [Oryza sativa] 86 1e-15
UniRef100_Q680B1 Putative bHLH transcription factor [Arabidopsis... 86 1e-15
UniRef100_Q5VRS4 Basic helix-loop-helix protein SPATULA-like [Or... 86 1e-15
>UniRef100_O80536 Phytochrome-interacting factor 3 [Arabidopsis thaliana]
Length = 524
Score = 111 bits (278), Expect = 1e-23
Identities = 79/212 (37%), Positives = 116/212 (54%), Gaps = 9/212 (4%)
Query: 7 KNGKVNFSNFSIPAVFLKSNQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSGYERSRE 66
K VNFS+F PA F K+ + + + K+ Q + L A + S
Sbjct: 194 KPSLVNFSHFLRPATFAKTTNNNLHDTKEKSPQSPPNVFQTRV-LGAKDSEDKVLNESVA 252
Query: 67 LLPTVDEHSEAAASHTNAPRIRGKRKASAINLCNAQKPSSVCSLEA-SNDLNFGVRKSHE 125
D S + + + KA +C++ + SL+ S + +++ H
Sbjct: 253 SATPKDNQKACLISEDSCRKDQESEKAV---VCSSVGSGN--SLDGPSESPSLSLKRKHS 307
Query: 126 DTDDSPYLSDNDEETQENIVKEK-PVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRAL 184
+ D S++ EE + KE P R G KRS R+A+VHNLSER+RRD+INEK+RAL
Sbjct: 308 NIQDIDCHSEDVEEESGDGRKEAGPSRTGLGSKRS-RSAEVHNLSERRRRDRINEKMRAL 366
Query: 185 KELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ELIPNCNK+DKASMLD+AI+YLK+L+LQ+Q+
Sbjct: 367 QELIPNCNKVDKASMLDEAIEYLKSLQLQVQI 398
>UniRef100_Q9ARY0 P0498A12.24 protein [Oryza sativa]
Length = 895
Score = 101 bits (252), Expect = 1e-20
Identities = 79/239 (33%), Positives = 123/239 (51%), Gaps = 35/239 (14%)
Query: 9 GKVNFSNFSIPAVFLKS---NQQRVQEKESKAVACTKFQQQKHLTLNAATKSNSG----- 60
G +NFS FS PAV ++ + QR Q ++KA T + + + A+ S
Sbjct: 475 GVMNFSLFSRPAVLARATLESAQRTQGTDNKASNVTASNRVESTVVQTASGPRSAPAFAD 534
Query: 61 -----YERSRELLPTVDEHSEAAASHTNAPRIRGKRKASA---INLCNAQKP-------S 105
+ + +P + A N G+ +A ++ A+K S
Sbjct: 535 QRAAAWPPQPKEMPFASTAAAPMAPAVNLHHEMGRDRAGRTMPVHKTEARKAPEATVATS 594
Query: 106 SVCSLEA--SNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNA 163
SVCS S++L ++ + + D+D + + +++ + G R + R A
Sbjct: 595 SVCSGNGAGSDELWRQQKRKCQAQAECSASQDDDLDDEPGVLR----KSGTRSTKRSRTA 650
Query: 164 KVHNLSERKRRDKINEKIRALKELIPNCNK------MDKASMLDDAIDYLKTLKLQLQV 216
+VHNLSER+RRD+INEK+RAL+ELIPNCNK +DKASMLD+AI+YLKTL+LQ+Q+
Sbjct: 651 EVHNLSERRRRDRINEKMRALQELIPNCNKACYVLQIDKASMLDEAIEYLKTLQLQVQM 709
>UniRef100_Q6AT90 Hypothetical protein OSJNBa0069I13.9 [Oryza sativa]
Length = 505
Score = 99.4 bits (246), Expect = 7e-20
Identities = 57/113 (50%), Positives = 78/113 (68%), Gaps = 4/113 (3%)
Query: 105 SSVCSLEASN-DLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNA 163
SSVCS LN+ R SH + + + + DE ++ + R R + R A
Sbjct: 281 SSVCSGNGDRRQLNW--RDSHNN-QSAEWSASQDELDLDDELAGVHRRSAARSSKRSRTA 337
Query: 164 KVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+VHNLSER+RRD+INEK+RAL+ELIPNCNK+DKASML++AI+YLKTL+LQ+Q+
Sbjct: 338 EVHNLSERRRRDRINEKMRALQELIPNCNKIDKASMLEEAIEYLKTLQLQVQM 390
>UniRef100_Q8GZM7 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 478
Score = 93.2 bits (230), Expect = 5e-18
Identities = 44/79 (55%), Positives = 61/79 (76%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
+ET+ + K R + R A+VHNLSERKRRD+INE+++AL+ELIP CNK DKA
Sbjct: 261 DETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKA 320
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+Y+K+L+LQ+Q+
Sbjct: 321 SMLDEAIEYMKSLQLQIQM 339
>UniRef100_Q9SL63 Hypothetical protein At2g20180 [Arabidopsis thaliana]
Length = 490
Score = 93.2 bits (230), Expect = 5e-18
Identities = 44/79 (55%), Positives = 61/79 (76%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
+ET+ + K R + R A+VHNLSERKRRD+INE+++AL+ELIP CNK DKA
Sbjct: 261 DETESRSEETKQARVSTTSTKRSRAAEVHNLSERKRRDRINERMKALQELIPRCNKSDKA 320
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+Y+K+L+LQ+Q+
Sbjct: 321 SMLDEAIEYMKSLQLQIQM 339
>UniRef100_Q8W2F3 Phytochrome-interacting factor 4 [Arabidopsis thaliana]
Length = 430
Score = 92.8 bits (229), Expect = 7e-18
Identities = 53/96 (55%), Positives = 69/96 (71%), Gaps = 9/96 (9%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
RK TD+S LSD I + R G+ R R A+VHNLSER+RRD+INE+
Sbjct: 226 RKRINHTDESVSLSDA-------IGNKSNQRSGSN--RRSRAAEVHNLSERRRRDRINER 276
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
++AL+ELIP+C+K DKAS+LD+AIDYLK+L+LQLQV
Sbjct: 277 MKALQELIPHCSKTDKASILDEAIDYLKSLQLQLQV 312
>UniRef100_Q8GRJ1 Transcription factor BHLH9-like protein [Oryza sativa]
Length = 417
Score = 92.4 bits (228), Expect = 9e-18
Identities = 49/96 (51%), Positives = 73/96 (76%), Gaps = 10/96 (10%)
Query: 121 RKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEK 180
++ E+ DS LS+ +ET+ + ++P KR R A+VHNLSER+RRD+INEK
Sbjct: 198 KRGREELVDS--LSEVADETRPS---KRPA-----AKRRTRAAEVHNLSERRRRDRINEK 247
Query: 181 IRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+RAL+EL+P+CNK DKAS+LD+AI+YLK+L++Q+Q+
Sbjct: 248 LRALQELVPHCNKTDKASILDEAIEYLKSLQMQVQI 283
>UniRef100_Q7Y1V4 BP-5 protein [Oryza sativa]
Length = 335
Score = 90.9 bits (224), Expect = 3e-17
Identities = 48/79 (60%), Positives = 64/79 (80%), Gaps = 3/79 (3%)
Query: 140 TQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKEL--IPNCNKMDKA 197
++ +V KP + +RS R A+VHNLSER+RRD+INEK+RAL+EL IP+CNK DKA
Sbjct: 146 SRSRLVARKPPAKMTTARRS-RAAEVHNLSERRRRDRINEKMRALQELELIPHCNKTDKA 204
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLD+AI+YLK+L+LQL+V
Sbjct: 205 SMLDEAIEYLKSLQLQLRV 223
>UniRef100_Q84LH8 PIF3 like basic Helix Loop Helix protein [Arabidopsis thaliana]
Length = 444
Score = 90.5 bits (223), Expect = 3e-17
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>UniRef100_Q5ULY0 Basic helix-loop-helix protein [Fragaria ananassa]
Length = 298
Score = 90.5 bits (223), Expect = 3e-17
Identities = 55/114 (48%), Positives = 76/114 (66%), Gaps = 6/114 (5%)
Query: 105 SSVCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRV--KRSYRN 162
++V + N + V S + D+ S EE E +V+E V+ G R KRS R
Sbjct: 90 TAVVTASGPNVSSSSVGASENEADEYDCES---EEGLEALVEEAAVKSGGRSSSKRS-RA 145
Query: 163 AKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
A+VHNLSE++RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q+
Sbjct: 146 AEVHNLSEKRRRSRINEKMKALQNLIPNSNKTDKASMLDEAIEYLKQLQLQVQM 199
>UniRef100_Q8S3D7 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 442
Score = 90.5 bits (223), Expect = 3e-17
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>UniRef100_Q9LYT0 Hypothetical protein F17J16_110 [Arabidopsis thaliana]
Length = 442
Score = 90.5 bits (223), Expect = 3e-17
Identities = 46/84 (54%), Positives = 66/84 (77%), Gaps = 5/84 (5%)
Query: 133 LSDNDEETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCN 192
L+ D++T N K + +RS R A+VHNLSER+RRD+INE+++AL+ELIP+C+
Sbjct: 233 LTSTDDQTMGN----KSSQRSGSTRRS-RAAEVHNLSERRRRDRINERMKALQELIPHCS 287
Query: 193 KMDKASMLDDAIDYLKTLKLQLQV 216
+ DKAS+LD+AIDYLK+L++QLQV
Sbjct: 288 RTDKASILDEAIDYLKSLQMQLQV 311
>UniRef100_Q8L5W8 PIF3 like basic Helix Loop Helix protein [Arabidopsis thaliana]
Length = 416
Score = 89.4 bits (220), Expect = 7e-17
Identities = 48/115 (41%), Positives = 77/115 (66%), Gaps = 3/115 (2%)
Query: 105 SSVCSLEASNDLNF-GVRKSHEDTDD--SPYLSDNDEETQENIVKEKPVREGNRVKRSYR 161
SS S S DL+ +++ + D ++ S YLS+N ++ ++ + R V + R
Sbjct: 170 SSKFSRGTSRDLSCCSLKRKYGDIEEEESTYLSNNSDDESDDAKTQVHARTRKPVTKRKR 229
Query: 162 NAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ +VH L ERKRRD+ N+K+RAL++L+PNC K DKAS+LD+AI Y++TL+LQ+Q+
Sbjct: 230 STEVHKLYERKRRDEFNKKMRALQDLLPNCYKDDKASLLDEAIKYMRTLQLQVQM 284
>UniRef100_Q84WY4 Hypothetical protein At2g46970/F14M4.20 [Arabidopsis thaliana]
Length = 416
Score = 89.4 bits (220), Expect = 7e-17
Identities = 48/115 (41%), Positives = 77/115 (66%), Gaps = 3/115 (2%)
Query: 105 SSVCSLEASNDLNF-GVRKSHEDTDD--SPYLSDNDEETQENIVKEKPVREGNRVKRSYR 161
SS S S DL+ +++ + D ++ S YLS+N ++ ++ + R V + R
Sbjct: 170 SSKFSRGTSRDLSCCSLKRKYGDIEEEESTYLSNNSDDESDDAKTQVHARTRKPVTKRKR 229
Query: 162 NAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
+ +VH L ERKRRD+ N+K+RAL++L+PNC K DKAS+LD+AI Y++TL+LQ+Q+
Sbjct: 230 STEVHKLYERKRRDEFNKKMRALQDLLPNCYKDDKASLLDEAIKYMRTLQLQVQM 284
>UniRef100_Q7FA23 OSJNBa0058K23.6 protein [Oryza sativa]
Length = 181
Score = 88.2 bits (217), Expect = 2e-16
Identities = 40/59 (67%), Positives = 52/59 (87%)
Query: 158 RSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQV 216
R R+A+ HN SER+RRD+INEK++AL+EL+PNC K DK SMLD+AIDYLK+L+LQLQ+
Sbjct: 10 RRSRSAEFHNFSERRRRDRINEKLKALQELLPNCTKTDKVSMLDEAIDYLKSLQLQLQM 68
>UniRef100_O23192 Hypothetical protein C7A10.430 [Arabidopsis thaliana]
Length = 415
Score = 86.3 bits (212), Expect = 6e-16
Identities = 56/121 (46%), Positives = 75/121 (61%), Gaps = 12/121 (9%)
Query: 100 NAQKPSSVCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRS 159
N Q SS + +S+ V S +TD+ S EE E +V E P + RS
Sbjct: 140 NVQGNSSGTRVSSSS-----VGASGNETDEYDCES---EEGGEAVVDEAPSSKSGPSSRS 191
Query: 160 Y----RNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQ 215
R A+VHNLSE++RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q
Sbjct: 192 SSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSNKTDKASMLDEAIEYLKQLQLQVQ 251
Query: 216 V 216
+
Sbjct: 252 M 252
>UniRef100_Q9FUA4 SPATULA [Arabidopsis thaliana]
Length = 373
Score = 86.3 bits (212), Expect = 6e-16
Identities = 56/121 (46%), Positives = 75/121 (61%), Gaps = 12/121 (9%)
Query: 100 NAQKPSSVCSLEASNDLNFGVRKSHEDTDDSPYLSDNDEETQENIVKEKPVREGNRVKRS 159
N Q SS + +S+ V S +TD+ S EE E +V E P + RS
Sbjct: 140 NVQGNSSGTRVSSSS-----VGASGNETDEYDCES---EEGGEAVVDEAPSSKSGPSSRS 191
Query: 160 Y----RNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTLKLQLQ 215
R A+VHNLSE++RR +INEK++AL+ LIPN NK DKASMLD+AI+YLK L+LQ+Q
Sbjct: 192 SSKRCRAAEVHNLSEKRRRSRINEKMKALQSLIPNSNKTDKASMLDEAIEYLKQLQLQVQ 251
Query: 216 V 216
+
Sbjct: 252 M 252
>UniRef100_Q6K844 Hypothetical protein OJ1548_F12.19 [Oryza sativa]
Length = 110
Score = 85.5 bits (210), Expect = 1e-15
Identities = 43/79 (54%), Positives = 58/79 (72%)
Query: 138 EETQENIVKEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKA 197
EE + E+P R + R A+VHNLSE++RR +INEK++AL+ LIPN +K DKA
Sbjct: 9 EEALGSSESEQPTRPARPRGKRSRAAEVHNLSEKRRRSRINEKMKALQSLIPNSSKTDKA 68
Query: 198 SMLDDAIDYLKTLKLQLQV 216
SMLDDAI+YLK L+LQ+Q+
Sbjct: 69 SMLDDAIEYLKQLQLQVQM 87
>UniRef100_Q680B1 Putative bHLH transcription factor [Arabidopsis thaliana]
Length = 210
Score = 85.5 bits (210), Expect = 1e-15
Identities = 43/70 (61%), Positives = 57/70 (81%), Gaps = 1/70 (1%)
Query: 146 KEKPVREGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAID 205
K ++ N +KRS +A+ HNLSE+KRR KINEK++AL++LIPN NK DKASMLD+AI+
Sbjct: 79 KRSGAKQRNSLKRSI-DAQFHNLSEKKRRSKINEKMKALQKLIPNSNKTDKASMLDEAIE 137
Query: 206 YLKTLKLQLQ 215
YLK L+LQ+Q
Sbjct: 138 YLKQLQLQVQ 147
>UniRef100_Q5VRS4 Basic helix-loop-helix protein SPATULA-like [Oryza sativa]
Length = 315
Score = 85.5 bits (210), Expect = 1e-15
Identities = 44/66 (66%), Positives = 56/66 (84%), Gaps = 1/66 (1%)
Query: 151 REGNRVKRSYRNAKVHNLSERKRRDKINEKIRALKELIPNCNKMDKASMLDDAIDYLKTL 210
R G+ KRS R A+VHNLSE++RR KINEK++AL+ LIPN NK DKASMLD+AI+YLK L
Sbjct: 94 RGGSGSKRS-RAAEVHNLSEKRRRSKINEKMKALQSLIPNSNKTDKASMLDEAIEYLKQL 152
Query: 211 KLQLQV 216
+LQ+Q+
Sbjct: 153 QLQVQM 158
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.130 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 367,985,729
Number of Sequences: 2790947
Number of extensions: 14465809
Number of successful extensions: 67040
Number of sequences better than 10.0: 1522
Number of HSP's better than 10.0 without gapping: 707
Number of HSP's successfully gapped in prelim test: 817
Number of HSP's that attempted gapping in prelim test: 65671
Number of HSP's gapped (non-prelim): 1760
length of query: 240
length of database: 848,049,833
effective HSP length: 124
effective length of query: 116
effective length of database: 501,972,405
effective search space: 58228798980
effective search space used: 58228798980
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 73 (32.7 bits)
Medicago: description of AC139745.7


