

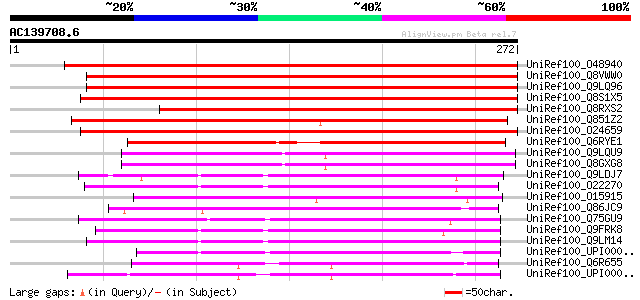
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139708.6 - phase: 0
(272 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O48940 Polyphosphoinositide binding protein Ssh2p [Gly... 372 e-102
UniRef100_Q8VWW0 Polyphosphoinositide binding protein [Gossypium... 350 2e-95
UniRef100_Q9LQ96 T1N6.1 protein [Arabidopsis thaliana] 322 9e-87
UniRef100_Q8S1X5 Sec14 like protein [Oryza sativa] 269 5e-71
UniRef100_Q8RXS2 Putative polyphosphoinositide binding protein [... 269 5e-71
UniRef100_Q851Z2 Putative phosphatidylinositol/phosphatidylcholi... 266 4e-70
UniRef100_O24659 Sec14 like protein [Oryza sativa] 263 3e-69
UniRef100_Q6RYE1 Phosphatidylinositol phosphatidylcholine transf... 213 4e-54
UniRef100_Q9LQU9 F10B6.22 [Arabidopsis thaliana] 159 1e-37
UniRef100_Q8GXG8 Putative phosphatidylinositol/ phosphatidylchol... 159 1e-37
UniRef100_Q9LDJ7 ESTs AU081555 [Oryza sativa] 118 1e-25
UniRef100_O22270 Putative phosphoglyceride transfer protein [Ara... 115 1e-24
UniRef100_O15915 Random slug cDNA5 protein [Dictyostelium discoi... 113 6e-24
UniRef100_Q86JC9 Hypothetical protein [Dictyostelium discoideum] 112 1e-23
UniRef100_Q75GU9 Putative cellular retinaldehyde-binding protein... 109 7e-23
UniRef100_Q9FRK8 Hypothetical protein F22H5.20 [Arabidopsis thal... 108 2e-22
UniRef100_Q9LM14 F16L1.9 protein [Arabidopsis thaliana] 106 7e-22
UniRef100_UPI00003C22FE UPI00003C22FE UniRef100 entry 102 8e-21
UniRef100_Q6R655 Sec14-like [Melampsora lini] 102 1e-20
UniRef100_UPI000023CF54 UPI000023CF54 UniRef100 entry 101 2e-20
>UniRef100_O48940 Polyphosphoinositide binding protein Ssh2p [Glycine max]
Length = 256
Score = 372 bits (956), Expect = e-102
Identities = 174/243 (71%), Positives = 214/243 (87%)
Query: 30 LVQDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFL 89
L Q DG DSTE ELTKI L+R +VE+RDPSSKE DD MIRRFLRARDLDV+KASAM L
Sbjct: 14 LGQGDGVAKDSTETELTKIRLLRAIVETRDPSSKEEDDFMIRRFLRARDLDVEKASAMLL 73
Query: 90 KYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFK 149
KY+KWR SFVP+GSVS S++ ++LAQ+K+++QG DK GRPI++ F +HFQNK+GLD FK
Sbjct: 74 KYLKWRNSFVPNGSVSVSDVPNELAQDKVFMQGHDKIGRPILMVFGGRHFQNKDGLDEFK 133
Query: 150 RYVVFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFI 209
R+VV+ L+K+ + MPPG+EKFV IA++KGWGY+NSD+RGYL AL+ILQDYYPERLGKLFI
Sbjct: 134 RFVVYVLDKVCASMPPGQEKFVGIAELKGWGYSNSDVRGYLSALSILQDYYPERLGKLFI 193
Query: 210 VHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPI 269
V+APY+FMKVW+I+YPFID+ TKKKIVFVE K+K+TLLEE++ESQ+PEI+GG LPLVPI
Sbjct: 194 VNAPYIFMKVWQIVYPFIDNKTKKKIVFVEKNKVKSTLLEEMEESQVPEIFGGSLPLVPI 253
Query: 270 QDS 272
QDS
Sbjct: 254 QDS 256
>UniRef100_Q8VWW0 Polyphosphoinositide binding protein [Gossypium hirsutum]
Length = 247
Score = 350 bits (898), Expect = 2e-95
Identities = 159/231 (68%), Positives = 202/231 (86%)
Query: 42 EAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPS 101
E E+TKI+LMR+LVE++DPSSKEVDD+ IRRFLRAR+LDV+KAS+MFLKY+KWR+SFVP+
Sbjct: 17 EEEITKISLMRSLVETQDPSSKEVDDMTIRRFLRARELDVEKASSMFLKYLKWRRSFVPN 76
Query: 102 GSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLIS 161
G +SPSE+ ++ Q K+++QG D KGRPI V AA+HFQ+ GLD FKR++++ +K+++
Sbjct: 77 GFISPSELTHEIQQNKMFLQGSDNKGRPISVLLAARHFQHNGGLDEFKRFILYIFDKILA 136
Query: 162 RMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWK 221
RMPPG++KF+ I D+ GWGYAN DIR YL AL++LQDYYPERLGK+FIVHAPY+FM WK
Sbjct: 137 RMPPGQDKFIVIGDLDGWGYANCDIRAYLAALSLLQDYYPERLGKMFIVHAPYVFMAAWK 196
Query: 222 IIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQDS 272
I++PFID T+KKIVFVENK LK+TLLEEIDESQLPE+YGG LPL+PIQDS
Sbjct: 197 IVHPFIDVKTRKKIVFVENKSLKSTLLEEIDESQLPEMYGGTLPLIPIQDS 247
>UniRef100_Q9LQ96 T1N6.1 protein [Arabidopsis thaliana]
Length = 255
Score = 322 bits (824), Expect = 9e-87
Identities = 151/231 (65%), Positives = 188/231 (81%)
Query: 42 EAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPS 101
E E +K+ +MR L + +DP +KEVDDLMIRRFLRARDLD++KAS MFL Y+ W++S +P
Sbjct: 25 EIERSKVGIMRALCDRQDPETKEVDDLMIRRFLRARDLDIEKASTMFLNYLTWKRSMLPK 84
Query: 102 GSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLIS 161
G + +EIA+DL+ K+ +QG DK GRPI VA +H +K D FKR+VV+ LEK+ +
Sbjct: 85 GHIPEAEIANDLSHNKMCMQGHDKMGRPIAVAIGNRHNPSKGNPDEFKRFVVYTLEKICA 144
Query: 162 RMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWK 221
RMP G+EKFV+I D++GWGY+N DIRGYL AL+ LQD YPERLGKL+IVHAPY+FM WK
Sbjct: 145 RMPRGQEKFVAIGDLQGWGYSNCDIRGYLAALSTLQDCYPERLGKLYIVHAPYIFMTAWK 204
Query: 222 IIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQDS 272
+IYPFID NTKKKIVFVENKKL TLLE+IDESQLP+IYGGKLPLVPIQ++
Sbjct: 205 VIYPFIDANTKKKIVFVENKKLTPTLLEDIDESQLPDIYGGKLPLVPIQET 255
>UniRef100_Q8S1X5 Sec14 like protein [Oryza sativa]
Length = 247
Score = 269 bits (688), Expect = 5e-71
Identities = 123/234 (52%), Positives = 173/234 (73%)
Query: 39 DSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSF 98
D+ E E K+ +R VE++DP +KEVD+L +RRFLRARD +V+KASAM LK ++WR+
Sbjct: 8 DAGEGEWLKVAELRATVEAQDPHAKEVDNLTLRRFLRARDHNVEKASAMLLKALRWRREA 67
Query: 99 VPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEK 158
VP GSV ++ DL +K+Y+ G D+ GRPI++AF AKHF K + FK Y V+ L+
Sbjct: 68 VPGGSVPEEKVQSDLDDDKVYMGGADRTGRPILLAFPAKHFSAKRDMPKFKSYCVYLLDS 127
Query: 159 LISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMK 218
+ +R+P G+EKFV I D+KGWGY+N DIR Y+ A+ I+Q+YYPERLGK ++H PYMFMK
Sbjct: 128 ICARIPRGQEKFVCIVDLKGWGYSNCDIRAYIAAIEIMQNYYPERLGKALMIHVPYMFMK 187
Query: 219 VWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQDS 272
WK+IYPFID+ T+ K VFV++K L+ L +EID+SQ+P+ GGKL V ++++
Sbjct: 188 AWKMIYPFIDNVTRDKFVFVDDKSLQEVLHQEIDDSQIPDTLGGKLAPVSLKNN 241
>UniRef100_Q8RXS2 Putative polyphosphoinositide binding protein [Arabidopsis
thaliana]
Length = 192
Score = 269 bits (688), Expect = 5e-71
Identities = 125/192 (65%), Positives = 156/192 (81%)
Query: 81 VDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQ 140
++KAS MFL Y+ W++S +P G + +EIA+DL+ K+ +QG DK GRPI VA +H
Sbjct: 1 IEKASTMFLNYLTWKRSMLPKGHIPEAEIANDLSHNKMCMQGHDKMGRPIAVAIGNRHNP 60
Query: 141 NKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYY 200
+K D FKR+VV+ LEK+ +RMP G+EKFV+I D++GWGY+N DIRGYL AL+ LQD Y
Sbjct: 61 SKGNPDEFKRFVVYTLEKICARMPRGQEKFVAIGDLQGWGYSNCDIRGYLAALSTLQDCY 120
Query: 201 PERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIY 260
PERLGKL+IVHAPY+FM WK+IYPFID NTKKKIVFVENKKL TLLE+IDESQLP+IY
Sbjct: 121 PERLGKLYIVHAPYIFMTAWKVIYPFIDANTKKKIVFVENKKLTPTLLEDIDESQLPDIY 180
Query: 261 GGKLPLVPIQDS 272
GGKLPLVPIQ++
Sbjct: 181 GGKLPLVPIQET 192
>UniRef100_Q851Z2 Putative phosphatidylinositol/phosphatidylcholine transfer protein
[Oryza sativa]
Length = 261
Score = 266 bits (680), Expect = 4e-70
Identities = 126/238 (52%), Positives = 175/238 (72%), Gaps = 4/238 (1%)
Query: 34 DGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMK 93
DG +D E K+ +R +VE++DP+ KE DD +RRFLRARD ++ KASAM +KY++
Sbjct: 10 DGHRDDGDVTEWKKVAELRAVVEAQDPACKEEDDYQLRRFLRARDHNIGKASAMLVKYLQ 69
Query: 94 WRKSFVPSG-SVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYV 152
W++ P G +++ E+ +LAQEK+Y+QG D++GRP++ F A+HF + LD FKRYV
Sbjct: 70 WKREVKPGGRAIADEEVRGELAQEKLYMQGYDRQGRPLVYGFGARHFPARRDLDEFKRYV 129
Query: 153 VFALEKLISRMPP--GEEKFVSIADIKGWGY-ANSDIRGYLGALTILQDYYPERLGKLFI 209
V+ L++ +R+ G+EKF ++AD++GWGY N DIR Y+ AL I+Q+YYPERLG++F+
Sbjct: 130 VYVLDRTCARLGGNGGQEKFAAVADLQGWGYYGNCDIRAYVAALEIMQNYYPERLGRVFL 189
Query: 210 VHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLV 267
+H PY+FM WKIIYPFIDDNTKKK VFV +K L ATL + ID+S L E YGGKL LV
Sbjct: 190 IHVPYVFMAAWKIIYPFIDDNTKKKFVFVADKDLHATLRDAIDDSNLAEDYGGKLKLV 247
>UniRef100_O24659 Sec14 like protein [Oryza sativa]
Length = 247
Score = 263 bits (673), Expect = 3e-69
Identities = 120/234 (51%), Positives = 171/234 (72%)
Query: 39 DSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSF 98
D+ E E K+ +R VE++DP +KEVD+L +RRFLRARD +V+KASAM LK ++WR+
Sbjct: 8 DAGEGEWLKVAELRATVEAQDPHAKEVDNLTLRRFLRARDHNVEKASAMLLKALRWRREA 67
Query: 99 VPSGSVSPSEIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEK 158
VP GSV ++ DL +K+Y+ G D+ GRPI++ F K+F K + FK Y V+ L+
Sbjct: 68 VPGGSVPEEKVQSDLDDDKVYMGGADRTGRPILLGFPVKNFSAKRDMPKFKSYCVYLLDS 127
Query: 159 LISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMK 218
+ +R+P G+EKFV I D+KGWGY+N DIR Y+ A+ I+Q+YYPERLGK ++H PYMFMK
Sbjct: 128 ICARIPRGQEKFVCIVDLKGWGYSNCDIRAYIAAIEIMQNYYPERLGKALMIHVPYMFMK 187
Query: 219 VWKIIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQDS 272
WK+IYPFID+ T+ K VFV++K L+ L +EID+SQ+P+ GGKL V ++++
Sbjct: 188 AWKMIYPFIDNVTRDKFVFVDDKSLQEVLHQEIDDSQIPDTLGGKLAPVSLKNN 241
>UniRef100_Q6RYE1 Phosphatidylinositol phosphatidylcholine transfer protein sec14
cytosolic-like protein [Triticum monococcum]
Length = 240
Score = 213 bits (542), Expect = 4e-54
Identities = 97/203 (47%), Positives = 141/203 (68%), Gaps = 13/203 (6%)
Query: 64 EVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGL 123
E D+L +RRFLR R ++ KASAM LKY+ W+++ P G +S E+ + LAQEK+Y QG
Sbjct: 37 EEDNLTLRRFLRTRGHNIGKASAMLLKYLAWKRAVKPRGFISDDEVHNQLAQEKVYTQGF 96
Query: 124 DKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADIKGWGYAN 183
DK GRP++ FAA+HF ++ D KRYVV+ +EKF ++ D+KGWGY N
Sbjct: 97 DKMGRPMVYLFAARHFPRRD-FDELKRYVVY------------QEKFAAVVDLKGWGYVN 143
Query: 184 SDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKL 243
DI+ + L I+++YYPE+LG++F+VH P++FM WK+ F+D+NTKKK VF++++ L
Sbjct: 144 CDIKASVAGLDIIKNYYPEQLGQVFLVHVPFVFMAAWKLGCTFVDNNTKKKFVFIDDRDL 203
Query: 244 KATLLEEIDESQLPEIYGGKLPL 266
TL + +DESQLP++YGGK L
Sbjct: 204 SGTLRDVVDESQLPDVYGGKFKL 226
>UniRef100_Q9LQU9 F10B6.22 [Arabidopsis thaliana]
Length = 730
Score = 159 bits (401), Expect = 1e-37
Identities = 87/216 (40%), Positives = 127/216 (58%), Gaps = 6/216 (2%)
Query: 61 SSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVP-SGSVSPSEIADDLAQEKIY 119
S K D + RFL AR +D KA+ MF+ + KWR S VP +G + SE+ D+L K+
Sbjct: 74 SEKGYDKPTLMRFLVARSMDPVKAAKMFVDWQKWRASMVPPTGFIPESEVQDELEFRKVC 133
Query: 120 VQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEE----KFVSIAD 175
+QG K G P+++ +KHF +K+ + FK++VV+AL+K I+ G+E K V++ D
Sbjct: 134 LQGPTKSGHPLVLVITSKHFASKDPAN-FKKFVVYALDKTIASGNNGKEVGGEKLVAVID 192
Query: 176 IKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKI 235
+ Y N D RG + LQ YYPERL K +I+H P F+ VWK + F++ T++KI
Sbjct: 193 LANITYKNLDARGLITGFQFLQSYYPERLAKCYILHMPGFFVTVWKFVCRFLEKATQEKI 252
Query: 236 VFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQD 271
V V + + + EEI LPE YGG+ L IQD
Sbjct: 253 VIVTDGEEQRKFEEEIGADALPEEYGGRAKLTAIQD 288
>UniRef100_Q8GXG8 Putative phosphatidylinositol/ phosphatidylcholine transfer protein
[Arabidopsis thaliana]
Length = 239
Score = 159 bits (401), Expect = 1e-37
Identities = 87/216 (40%), Positives = 127/216 (58%), Gaps = 6/216 (2%)
Query: 61 SSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVP-SGSVSPSEIADDLAQEKIY 119
S K D + RFL AR +D KA+ MF+ + KWR S VP +G + SE+ D+L K+
Sbjct: 9 SEKGYDKPTLMRFLVARSMDPVKAAKMFVDWQKWRASMVPPTGFIPESEVQDELEFRKVC 68
Query: 120 VQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEE----KFVSIAD 175
+QG K G P+++ +KHF +K+ + FK++VV+AL+K I+ G+E K V++ D
Sbjct: 69 LQGPTKSGHPLVLVITSKHFASKDPAN-FKKFVVYALDKTIASGNNGKEVGGEKLVAVID 127
Query: 176 IKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKI 235
+ Y N D RG + LQ YYPERL K +I+H P F+ VWK + F++ T++KI
Sbjct: 128 LANITYKNLDARGLITGFQFLQSYYPERLAKCYILHMPGFFVTVWKFVCRFLEKATQEKI 187
Query: 236 VFVENKKLKATLLEEIDESQLPEIYGGKLPLVPIQD 271
V V + + + EEI LPE YGG+ L IQD
Sbjct: 188 VIVTDGEEQRKFEEEIGADALPEEYGGRAKLTAIQD 223
>UniRef100_Q9LDJ7 ESTs AU081555 [Oryza sativa]
Length = 318
Score = 118 bits (296), Expect = 1e-25
Identities = 76/235 (32%), Positives = 123/235 (52%), Gaps = 12/235 (5%)
Query: 38 NDSTEAELTKINLMRTLVESRDPSSKEVDDLM----IRRFLRARDLDVDKASAMFLKYMK 93
N + E + KIN +R E + SS+E+ D + RFLRAR+ +V KAS M +K
Sbjct: 16 NLTLEEQQEKINDLRK--ELGEHSSEEIQDFLSDASCLRFLRARNWNVQKASKMMKSAVK 73
Query: 94 WRKSFVPSGSVSPSEIADDLAQEKIY-VQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYV 152
WR S++P ++ ++A + KIY DK GR ++V +N +Y+
Sbjct: 74 WRVSYMPQ-KINWDDVAHEAETGKIYRADYKDKHGRTVLVLRPG--LENTTSGKGQIKYL 130
Query: 153 VFALEKLISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHA 212
V+ LEK I + +EK V + D + W ++ ++ + +LQD YPERLG + +
Sbjct: 131 VYCLEKAIMSLTEDQEKMVWLTDFQSWTLGSTPLKVTRETVNVLQDCYPERLGLAILYNP 190
Query: 213 PYMFMKVWKIIYPFIDDNTKKKIVFV--ENKKLKATLLEEIDESQLPEIYGGKLP 265
P +F WKI+ PF+D T KK+ FV +K+ + + + D +L +GG+ P
Sbjct: 191 PRIFESFWKIVKPFLDHETYKKVKFVYSSDKESQKIMADVFDLDKLDSAFGGRNP 245
>UniRef100_O22270 Putative phosphoglyceride transfer protein [Arabidopsis thaliana]
Length = 301
Score = 115 bits (288), Expect = 1e-24
Identities = 70/226 (30%), Positives = 115/226 (49%), Gaps = 7/226 (3%)
Query: 41 TEAELTKINLMRTLVESR-DPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFV 99
TE E KI +R L+ + S D + R+LRAR+ V KA+ M + +KWR +
Sbjct: 16 TEEEQAKIEEVRKLLGPLPEKLSSFCSDDAVLRYLRARNWHVKKATKMLKETLKWRVQYK 75
Query: 100 PSGSVSPSEIADDLAQEKIYVQG-LDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEK 158
P + E+A + KIY +DK GRP+++ + +N + RY+V+ +E
Sbjct: 76 PE-EICWEEVAGEAETGKIYRSSCVDKLGRPVLIMRPS--VENSKSVKGQIRYLVYCMEN 132
Query: 159 LISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMK 218
+ +PPGEE+ V + D G+ AN +R +LQ++YPERL + + P F
Sbjct: 133 AVQNLPPGEEQMVWMIDFHGYSLANVSLRTTKETAHVLQEHYPERLAFAVLYNPPKFFEP 192
Query: 219 VWKIIYPFIDDNTKKKIVFV--ENKKLKATLLEEIDESQLPEIYGG 262
WK+ PF++ T+ K+ FV ++ K + E D ++ +GG
Sbjct: 193 FWKVARPFLEPKTRNKVKFVYSDDPNTKVIMEENFDMEKMELAFGG 238
>UniRef100_O15915 Random slug cDNA5 protein [Dictyostelium discoideum]
Length = 324
Score = 113 bits (282), Expect = 6e-24
Identities = 72/205 (35%), Positives = 106/205 (51%), Gaps = 7/205 (3%)
Query: 67 DLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKK 126
D+ R+LRAR+ V K+ M ++WRK F P +I + + +YV DKK
Sbjct: 73 DMCFLRYLRARNYIVSKSEKMLRDTLEWRKKFRPQDIQLGGDIREIGSAGCVYVNKRDKK 132
Query: 127 GRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRM--PPGEEKFVSIADIKGWGYANS 184
GRPII A + + +V+ LE+ SRM P G E+F I D K +G N
Sbjct: 133 GRPIIFAVPRNDTLKNVPSELKFKNLVYWLEQGFSRMDEPKGIEQFCFIVDYKDFGSGNM 192
Query: 185 DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLK 244
D++ L A+ L D+ PER+G+ + P +F WKII PF+++ T K+ F+ +KK+
Sbjct: 193 DMKTNLEAMHFLLDHCPERMGQSLFLDPPALFWFAWKIISPFLNEVTLSKVRFINSKKVD 252
Query: 245 -----ATLLEEIDESQLPEIYGGKL 264
A LLE +D L + GG L
Sbjct: 253 GKRTFAELLEYVDIENLEQNLGGNL 277
>UniRef100_Q86JC9 Hypothetical protein [Dictyostelium discoideum]
Length = 247
Score = 112 bits (279), Expect = 1e-23
Identities = 68/215 (31%), Positives = 111/215 (51%), Gaps = 10/215 (4%)
Query: 54 LVESRDP----SSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSG--SVSPS 107
+ E DP +++DD M RFLRAR ++ + M + +K+R +F G ++ S
Sbjct: 23 IAEINDPVVEKEIQQLDDSMTLRFLRARKWNLKDSFDMLYEALKFRATFQDVGVEGITES 82
Query: 108 EIADDLAQEKIYVQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGE 167
+ ++L K Y G+DK GRP+ + ++H L+ RY V+ +E S + G
Sbjct: 83 MVVNELKSGKSYFHGVDKGGRPVCIVKTSRHDSYNRDLNESMRYCVYVMENGKSMLKDGI 142
Query: 168 EKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFI 227
E I D+ + N D + + Q +YPE L K I++AP++FM +W II ++
Sbjct: 143 ETCTLIFDMSDFSSKNMDYPLVKFMVELFQKFYPESLQKCLILNAPWIFMGIWHIIKHWL 202
Query: 228 DDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
D NT K+ FV+ K+ L++ I + QL YGG
Sbjct: 203 DPNTASKVSFVKTKQ----LVDYIPKDQLESSYGG 233
>UniRef100_Q75GU9 Putative cellular retinaldehyde-binding protein [Oryza sativa]
Length = 299
Score = 109 bits (273), Expect = 7e-23
Identities = 72/231 (31%), Positives = 124/231 (53%), Gaps = 8/231 (3%)
Query: 38 NDSTEAELTKINLMRTLVESRDPSSKEV-DDLMIRRFLRARDLDVDKASAMFLKYMKWRK 96
+D E + KI +R + S K+ + +RR+L AR+ +VDK+ M + +KWR
Sbjct: 14 SDDIEQQEAKIQELRAALGPLSSSGKKYCTEACLRRYLEARNWNVDKSRKMLEESLKWRT 73
Query: 97 SFVPSGSVSPSEIADDLAQEKIY-VQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFA 155
++ P P EI+ + K+Y +D++GR +++ AK QN + + R++V+
Sbjct: 74 AYRPEDIRWP-EISVESETGKMYRASFVDREGRTVVIMRPAK--QNTSSHEGQVRFLVYT 130
Query: 156 LEKLISRMPPGEEKFVSIADIKGWGYANSD-IRGYLGALTILQDYYPERLGKLFIVHAPY 214
LE I +P +EK V + D GW AN+ I+ ILQ++YPERL + + P
Sbjct: 131 LENAILSLPEDQEKMVWLIDFTGWTLANATPIKTARECANILQNHYPERLAIGILFNPPK 190
Query: 215 MFMKVWKIIYPFIDDNTKKKI--VFVENKKLKATLLEEIDESQLPEIYGGK 263
+F WK++ F+D + +K+ V+++N++ L + ID LP +GGK
Sbjct: 191 VFEAFWKVVKHFLDPKSIQKVNFVYLKNEESMKILHKYIDPEVLPVEFGGK 241
>UniRef100_Q9FRK8 Hypothetical protein F22H5.20 [Arabidopsis thaliana]
Length = 296
Score = 108 bits (269), Expect = 2e-22
Identities = 68/222 (30%), Positives = 115/222 (51%), Gaps = 8/222 (3%)
Query: 47 KINLMRTLV-ESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVS 105
K+ ++TL+ + +S D ++R+L AR+ +V KA M + +KWR SF P +
Sbjct: 23 KMKELKTLIGQLSGRNSLYCSDACLKRYLEARNWNVGKAKKMLEETLKWRSSFKPE-EIR 81
Query: 106 PSEIADDLAQEKIYVQGL-DKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMP 164
+E++ + K+Y G D+ GR +++ QN L+ +++V+ +E I +P
Sbjct: 82 WNEVSGEGETGKVYKAGFHDRHGRTVLILRPG--LQNTKSLENQMKHLVYLIENAILNLP 139
Query: 165 PGEEKFVSIADIKGWGYANS-DIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKII 223
+E+ + D GW + S I+ + ILQ++YPERL F+ + P +F WKI+
Sbjct: 140 EDQEQMSWLIDFTGWSMSTSVPIKSARETINILQNHYPERLAVAFLYNPPRLFEAFWKIV 199
Query: 224 YPFIDDNT--KKKIVFVENKKLKATLLEEIDESQLPEIYGGK 263
FID T K K V+ +N + + DE LP +GGK
Sbjct: 200 KYFIDAKTFVKVKFVYPKNSESVELMSTFFDEENLPTEFGGK 241
>UniRef100_Q9LM14 F16L1.9 protein [Arabidopsis thaliana]
Length = 314
Score = 106 bits (264), Expect = 7e-22
Identities = 73/226 (32%), Positives = 116/226 (51%), Gaps = 7/226 (3%)
Query: 42 EAELTKINLMRTLVES-RDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVP 100
E L KIN +RTL+ + SS+ D I R+L AR+ V KA+ M + +KWR + P
Sbjct: 20 EEYLNKINEVRTLLGPLTEKSSEFCSDAAITRYLAARNGHVKKATKMLKETLKWRAQYKP 79
Query: 101 SGSVSPSEIADDLAQEKIY-VQGLDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKL 159
+ EIA + KIY DK GR ++V + QN R +V+ +E
Sbjct: 80 E-EIRWEEIAREAETGKIYRANCTDKYGRTVLVMRPS--CQNTKSYKGQIRILVYCMENA 136
Query: 160 ISRMPPGEEKFVSIADIKGWGYANSDIRGYLGALTILQDYYPERLGKLFIVHAPYMFMKV 219
I +P +E+ V + D G+ ++ ++ +LQ++YPERLG + + P +F
Sbjct: 137 ILNLPDNQEQMVWLIDFHGFNMSHISLKVSRETAHVLQEHYPERLGLAIVYNPPKIFESF 196
Query: 220 WKIIYPFIDDNTKKKIVFV-ENKKLKATLLEEI-DESQLPEIYGGK 263
+K++ PF++ T K+ FV + L LLE++ D QL +GGK
Sbjct: 197 YKMVKPFLEPKTSNKVKFVYSDDNLSNKLLEDLFDMEQLEVAFGGK 242
>UniRef100_UPI00003C22FE UPI00003C22FE UniRef100 entry
Length = 697
Score = 102 bits (255), Expect = 8e-21
Identities = 68/197 (34%), Positives = 98/197 (49%), Gaps = 11/197 (5%)
Query: 69 MIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQGLDKKGR 128
+ +R+LRA D+D A + WR+ F P ++P +A + K V G D GR
Sbjct: 91 LYQRYLRAAKGDLDNAKKRIKSTLDWRRDFRPE-IIAPGSVAKEAETGKQIVSGFDNDGR 149
Query: 129 PIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFVSIADI-KGWGYANSDIR 187
P+I A+ +N DA RY+V+ LE+ I MPPG E + I D K +N +
Sbjct: 150 PLIYLRPAR--ENTTPSDAQVRYLVWTLERAIDLMPPGVENYAIIIDYHKATTQSNPSLS 207
Query: 188 GYLGALTILQDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATL 247
ILQ++Y ERLG+ FIV+ P+ + I PF+D TK KI F A L
Sbjct: 208 TARAVANILQNHYVERLGRAFIVNVPWFINAFFSAIVPFLDPVTKDKIRF------NANL 261
Query: 248 LEEIDESQL-PEIYGGK 263
++ + QL E GG+
Sbjct: 262 VDFVPADQLDAEFTGGR 278
>UniRef100_Q6R655 Sec14-like [Melampsora lini]
Length = 285
Score = 102 bits (253), Expect = 1e-20
Identities = 72/221 (32%), Positives = 108/221 (48%), Gaps = 32/221 (14%)
Query: 66 DDLMIRRFLRARDLDVDKASAMFLKYMKWRKSFVPSGSVSPSEIADDLAQEKIYVQ---G 122
DD + RFLRAR D++K+ MF KWRK F + E + + IY Q
Sbjct: 1 DDATLLRFLRARKFDLEKSKLMFTDCEKWRKEFKVDELYATFEYPEKKEVDAIYPQFYHK 60
Query: 123 LDKKGRPIIVAFAAKHFQNKNGLDAFKRYVVFALEKLISRMPPGEEKFV----------- 171
+K GRPI + K LD K Y V E+ + R+ EKF+
Sbjct: 61 TEKDGRPIYIEQLGK-------LDLTKLYKVTTPERQLQRLVVEYEKFLRDRLPVCSVQQ 113
Query: 172 --------SIADIKGWGYANS-DIRGYLGALTIL-QDYYPERLGKLFIVHAPYMFMKVWK 221
+I D+ G G + ++ Y+ + L Q+YYPE +GK +I++APY+F VW
Sbjct: 114 GKLVETSCTIMDLSGVGLSQFWKVKNYVQQASHLSQNYYPETMGKFYIINAPYLFSTVWS 173
Query: 222 IIYPFIDDNTKKKIVFVENKKLKATLLEEIDESQLPEIYGG 262
++ P++D+ T KKI +++ K TLLE+I LP+ G
Sbjct: 174 LVKPWLDEVTVKKISILDSSYHK-TLLEQIPAESLPKSLKG 213
>UniRef100_UPI000023CF54 UPI000023CF54 UniRef100 entry
Length = 337
Score = 101 bits (252), Expect = 2e-20
Identities = 73/256 (28%), Positives = 120/256 (46%), Gaps = 33/256 (12%)
Query: 32 QDDGALNDSTEAELTKINLMRTLVESRDPSSKEVDDLMIRRFLRARDLDVDKASAMFLKY 91
+ DG TE + K+ R ++ES + + +D L + RFLRAR DV+ + AMFL
Sbjct: 19 KQDGHAGYLTEDQTAKLEQFRMMLESEGCTDR-LDTLTLLRFLRARKFDVEASKAMFLDT 77
Query: 92 MKWRKSFVPSGSVSPSEIADDLAQEKIYVQ---GLDKKGRPIIVAFAAKHFQNKNGLDAF 148
KWRK +V + + K Y Q DK GRPI + + G+D
Sbjct: 78 EKWRKETKLDETVPVWDYPEKAEINKYYTQFYHKTDKDGRPIYI-------ETLGGIDLN 130
Query: 149 KRYVVFALEKLISRMPPGEEKFV-------------------SIADIKGWGYAN-SDIRG 188
Y + E++++ + E+ ++ D+KG +
Sbjct: 131 AMYKITTAERMLTNLAVEYERVADPRLPACSRKAGHLLETCCTVMDLKGVSIGKVPQVYS 190
Query: 189 YLGALTIL-QDYYPERLGKLFIVHAPYMFMKVWKIIYPFIDDNTKKKIVFVENKKLKATL 247
Y+ +++ Q+YYPERLGKL++++AP+ F VW I+ ++D T KI + K L
Sbjct: 191 YVKQASVISQNYYPERLGKLYMINAPWGFSTVWSIVKGWLDPVTVSKINIL-GSGYKGEL 249
Query: 248 LEEIDESQLPEIYGGK 263
L++I LP+ +GG+
Sbjct: 250 LKQIPAENLPKAFGGE 265
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.137 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 464,413,688
Number of Sequences: 2790947
Number of extensions: 19416396
Number of successful extensions: 51746
Number of sequences better than 10.0: 482
Number of HSP's better than 10.0 without gapping: 355
Number of HSP's successfully gapped in prelim test: 127
Number of HSP's that attempted gapping in prelim test: 50877
Number of HSP's gapped (non-prelim): 544
length of query: 272
length of database: 848,049,833
effective HSP length: 125
effective length of query: 147
effective length of database: 499,181,458
effective search space: 73379674326
effective search space used: 73379674326
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC139708.6


