

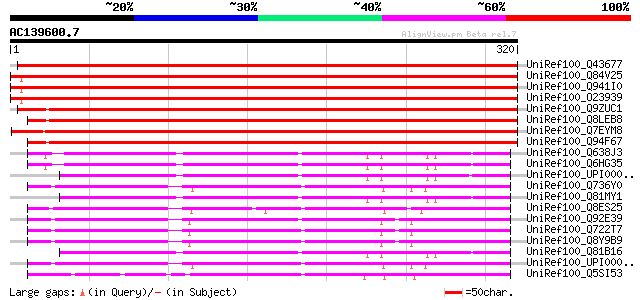
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.7 - phase: 0
(320 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q43677 Auxin-induced protein [Vigna radiata] 518 e-146
UniRef100_Q84V25 Quinone oxidoreductase [Fragaria ananassa] 471 e-131
UniRef100_Q941I0 Putative quinone oxidoreductase [Fragaria anana... 464 e-129
UniRef100_O23939 Ripening-induced protein [Fragaria vesca] 464 e-129
UniRef100_Q9ZUC1 Quinone oxidoreductase-like protein At1g23740, ... 407 e-112
UniRef100_Q8LEB8 Quinone oxidoreductase-like protein [Arabidopsi... 401 e-110
UniRef100_Q7EYM8 Putative oxidoreductase, zinc-binding [Oryza sa... 392 e-108
UniRef100_Q94F67 Quinone oxidoreductase-like protein [Helianthus... 389 e-107
UniRef100_Q638J3 Quinone oxidoreductase [Bacillus cereus] 184 3e-45
UniRef100_Q6HG35 Quinone oxidoreductase [Bacillus thuringiensis] 184 4e-45
UniRef100_UPI00003CB31E UPI00003CB31E UniRef100 entry 182 1e-44
UniRef100_Q736Y0 Oxidoreductase, zinc-binding [Bacillus cereus] 179 9e-44
UniRef100_Q81MY1 Alcohol dehydrogenase, zinc-containing [Bacillu... 179 1e-43
UniRef100_Q8ES25 Zinc-binding oxidoreductase [Oceanobacillus ihe... 178 2e-43
UniRef100_Q92E39 Lin0622 protein [Listeria innocua] 177 4e-43
UniRef100_Q722T7 Alcohol dehydrogenase, zinc-dependent [Listeria... 176 6e-43
UniRef100_Q8Y9B9 Lmo0613 protein [Listeria monocytogenes] 176 7e-43
UniRef100_Q81B16 Quinone oxidoreductase [Bacillus cereus] 174 4e-42
UniRef100_UPI00003CC0B8 UPI00003CC0B8 UniRef100 entry 173 5e-42
UniRef100_Q5SI53 NADPH-quinone reductase [Thermus thermophilus HB8] 169 7e-41
>UniRef100_Q43677 Auxin-induced protein [Vigna radiata]
Length = 317
Score = 518 bits (1335), Expect = e-146
Identities = 255/315 (80%), Positives = 282/315 (88%)
Query: 6 SIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD 65
SIPSH +AW+YS+YGNIEE+LKFD NVPTPH E+QVLIKVVAAA+NPVD KRALGHFKD
Sbjct: 3 SIPSHMRAWIYSEYGNIEEVLKFDPNVPTPHLMENQVLIKVVAAAINPVDYKRALGHFKD 62
Query: 66 IDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEK 125
IDSPLPT PGYDVAGVVV VG QVKKF+VGDEVYGDINE+TL +PKTIG+L+EYT EEK
Sbjct: 63 IDSPLPTAPGYDVAGVVVKVGNQVKKFQVGDEVYGDINEVTLDHPKTIGSLAEYTAVEEK 122
Query: 126 VLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
VLAHKPSNLSFVEAASLPLAIITAYQGLE +FS GKS+LVLGGAGGVGSLVIQ+AKH+F
Sbjct: 123 VLAHKPSNLSFVEAASLPLAIITAYQGLETAQFSVGKSILVLGGAGGVGSLVIQIAKHIF 182
Query: 186 EASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEG 245
AS+IAAT ST KL+ LG DL IDYTKEN+EEL EKFDVVYDAVG SERA+K+ EG
Sbjct: 183 GASKIAATGSTGKLELFENLGTDLPIDYTKENFEELAEKFDVVYDAVGQSERALKSVKEG 242
Query: 246 GKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYL 305
GKVVTI+ P TPPAIPFLLTSDG +LEKL+PYLENG+VKPILDPKSPFPF QT+EAFSYL
Sbjct: 243 GKVVTIVAPATPPAIPFLLTSDGDLLEKLRPYLENGQVKPILDPKSPFPFSQTVEAFSYL 302
Query: 306 NTNRAIGKIVIHPIP 320
TNRA GK+VI+PIP
Sbjct: 303 KTNRATGKVVIYPIP 317
>UniRef100_Q84V25 Quinone oxidoreductase [Fragaria ananassa]
Length = 322
Score = 471 bits (1212), Expect = e-131
Identities = 234/322 (72%), Positives = 274/322 (84%), Gaps = 2/322 (0%)
Query: 1 MASTPS--IPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKR 58
MA+ PS IPS KAWVYS+YG ++LKFD +V P KEDQVLIKVVAA+LNPVD KR
Sbjct: 1 MAAAPSESIPSVNKAWVYSEYGKTSDVLKFDPSVAVPEVKEDQVLIKVVAASLNPVDFKR 60
Query: 59 ALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSE 118
ALG+FKD DSPLPTVPGYDVAGVVV VG QV KFKVGDEVYGD+NE L NP G+L+E
Sbjct: 61 ALGYFKDTDSPLPTVPGYDVAGVVVKVGSQVTKFKVGDEVYGDLNEAALVNPTRFGSLAE 120
Query: 119 YTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVI 178
YT A+E+VLAHKP +LSF+EAASLPLAI TAY+GLER E S+GKS+LVLGGAGGVG+ +I
Sbjct: 121 YTAADERVLAHKPKDLSFIEAASLPLAIETAYEGLERAELSAGKSILVLGGAGGVGTHII 180
Query: 179 QLAKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERA 238
QLAKHVF AS++AATAST KLDFLR LG DLAIDYTKEN E+L EKFDVVYDAVG++++A
Sbjct: 181 QLAKHVFGASKVAATASTKKLDFLRTLGVDLAIDYTKENIEDLPEKFDVVYDAVGETDKA 240
Query: 239 VKAANEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQT 298
VKA EGGKVVTI+ P TPPAI F+LTS G+VLEKL+PYLE+GKVKP+LDP SP+PF +
Sbjct: 241 VKAVKEGGKVVTIVGPATPPAIHFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKL 300
Query: 299 LEAFSYLNTNRAIGKIVIHPIP 320
+EAF YL ++RA GK+V++PIP
Sbjct: 301 VEAFGYLESSRATGKVVVYPIP 322
>UniRef100_Q941I0 Putative quinone oxidoreductase [Fragaria ananassa]
Length = 339
Score = 464 bits (1195), Expect = e-129
Identities = 233/323 (72%), Positives = 274/323 (84%), Gaps = 3/323 (0%)
Query: 1 MASTPS--IPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKR 58
MA+ PS IPS KAWVYS+YG ++LKFD +V P KEDQVLIKVVAA+LNPVD KR
Sbjct: 17 MAAAPSESIPSVNKAWVYSEYGKTSDVLKFDPSVAVPEIKEDQVLIKVVAASLNPVDFKR 76
Query: 59 ALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSE 118
ALG+FKD DSPLPT+PGYDVAGVVV VG QV KFKVGDEVYGD+NE L NP G+L+E
Sbjct: 77 ALGYFKDTDSPLPTIPGYDVAGVVVKVGSQVTKFKVGDEVYGDLNETALVNPTRFGSLAE 136
Query: 119 YTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVI 178
YT A E+VLAHKP NLSF+EAASLPLAI TA++GLER E S+GKS+LVLGGAGGVG+ +I
Sbjct: 137 YTAAYERVLAHKPKNLSFIEAASLPLAIETAHEGLERAELSAGKSVLVLGGAGGVGTHII 196
Query: 179 QLAKHVFEASRIAATASTTKLDFLRKLG-ADLAIDYTKENYEELTEKFDVVYDAVGDSER 237
QLAKHVF AS++AATAST KLD LR LG ADLAIDYTKEN+E+L EKFDVVYDAVG++++
Sbjct: 197 QLAKHVFGASKVAATASTKKLDLLRTLGAADLAIDYTKENFEDLPEKFDVVYDAVGETDK 256
Query: 238 AVKAANEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQ 297
AVKA EGGKVVTI+ P TPPAI F+LTS G+VLEKL+PYLE+GKVKP+LDP SP+PF +
Sbjct: 257 AVKAVKEGGKVVTIVGPATPPAILFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTK 316
Query: 298 TLEAFSYLNTNRAIGKIVIHPIP 320
+EAF YL ++RA GK+V++PIP
Sbjct: 317 VVEAFGYLESSRATGKVVVYPIP 339
>UniRef100_O23939 Ripening-induced protein [Fragaria vesca]
Length = 337
Score = 464 bits (1194), Expect = e-129
Identities = 231/322 (71%), Positives = 273/322 (84%), Gaps = 2/322 (0%)
Query: 1 MASTPS--IPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKR 58
MA+ PS IPS KAWV S+YG ++LKFD +V P KEDQVLIKVVAA+LNPVD KR
Sbjct: 16 MAAAPSESIPSVNKAWVXSEYGKTSDVLKFDPSVAVPEIKEDQVLIKVVAASLNPVDFKR 75
Query: 59 ALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSE 118
ALG+FKD DSPLPT+PGY VAGVVV VG QV KFKVGDEVYGD+NE L NP G+L+E
Sbjct: 76 ALGYFKDTDSPLPTIPGYYVAGVVVKVGSQVTKFKVGDEVYGDLNETALVNPTRFGSLAE 135
Query: 119 YTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVI 178
YT A+E+VLAHKP NLSF+EAASLPLAI TA++GLER E S+GKS+LVLGGAGGVG+ +I
Sbjct: 136 YTAADERVLAHKPKNLSFIEAASLPLAIETAHEGLERAELSAGKSVLVLGGAGGVGTHII 195
Query: 179 QLAKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERA 238
QLAKHVF AS++AATAST KLD LR LGADLAIDYTKEN+E+L EKFDVVYDAVG++++A
Sbjct: 196 QLAKHVFGASKVAATASTKKLDLLRTLGADLAIDYTKENFEDLPEKFDVVYDAVGETDKA 255
Query: 239 VKAANEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQT 298
VKA EGGKVVTI+ P TPPAI F+LTS G+VLEKL+PYLE+GKVKP+LDP SP+PF +
Sbjct: 256 VKAVKEGGKVVTIVGPATPPAILFVLTSKGSVLEKLKPYLESGKVKPVLDPTSPYPFTKV 315
Query: 299 LEAFSYLNTNRAIGKIVIHPIP 320
+EAF YL ++RA GK+V++PIP
Sbjct: 316 VEAFGYLESSRATGKVVVYPIP 337
>UniRef100_Q9ZUC1 Quinone oxidoreductase-like protein At1g23740, chloroplast
precursor [Arabidopsis thaliana]
Length = 386
Score = 407 bits (1045), Expect = e-112
Identities = 203/315 (64%), Positives = 245/315 (77%), Gaps = 1/315 (0%)
Query: 6 SIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD 65
SIP KAWVYS YG ++ +LK +SN+ P KEDQVLIKVVAAALNPVD KR G FK
Sbjct: 73 SIPKEMKAWVYSDYGGVD-VLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKA 131
Query: 66 IDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEK 125
DSPLPTVPGYDVAGVVV VG VK K GDEVY +++E L PK G+L+EYT EEK
Sbjct: 132 TDSPLPTVPGYDVAGVVVKVGSAVKDLKEGDEVYANVSEKALEGPKQFGSLAEYTAVEEK 191
Query: 126 VLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
+LA KP N+ F +AA LPLAI TA +GL R EFS+GKS+LVL GAGGVGSLVIQLAKHV+
Sbjct: 192 LLALKPKNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLAKHVY 251
Query: 186 EASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEG 245
AS++AATAST KL+ +R LGADLAIDYTKEN E+L +K+DVV+DA+G ++AVK EG
Sbjct: 252 GASKVAATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEG 311
Query: 246 GKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYL 305
GKVV + TPP F++TS+G VL+KL PY+E+GKVKP++DPK PFPF + +AFSYL
Sbjct: 312 GKVVALTGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYL 371
Query: 306 NTNRAIGKIVIHPIP 320
TN A GK+V++PIP
Sbjct: 372 ETNHATGKVVVYPIP 386
>UniRef100_Q8LEB8 Quinone oxidoreductase-like protein [Arabidopsis thaliana]
Length = 309
Score = 401 bits (1030), Expect = e-110
Identities = 199/309 (64%), Positives = 242/309 (77%), Gaps = 1/309 (0%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KAWVYS YG ++ +LK +SN+ P KEDQVLIKVVAA LNPVD KR G FK DSPLP
Sbjct: 2 KAWVYSDYGGVD-VLKLESNIAVPEIKEDQVLIKVVAAGLNPVDAKRRQGKFKATDSPLP 60
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKP 131
TVPGYDVAGVVV VG VK FK GDEVY +++E L PK G+L+EYT EEK+LA KP
Sbjct: 61 TVPGYDVAGVVVKVGSAVKDFKEGDEVYANVSEKALEGPKQFGSLAEYTAVEEKLLALKP 120
Query: 132 SNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIA 191
N+ F +AA LPLAI TA +GL R EFS+GKS+LVL GAGGVGSL+IQLAKHV+ AS++A
Sbjct: 121 KNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLMIQLAKHVYGASKVA 180
Query: 192 ATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEGGKVVTI 251
ATAST KL+ +R LGADLAIDYTKEN E+L +K+DVV+DA+G ++AVK EGGKVV +
Sbjct: 181 ATASTGKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEGGKVVAL 240
Query: 252 LPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAI 311
TPP F++TS+G VL+KL PY+E+GKVKP++DPK PFPF + +AFSYL TN A
Sbjct: 241 TGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYLETNHAT 300
Query: 312 GKIVIHPIP 320
GK+V++PIP
Sbjct: 301 GKVVVYPIP 309
>UniRef100_Q7EYM8 Putative oxidoreductase, zinc-binding [Oryza sativa]
Length = 390
Score = 392 bits (1007), Expect = e-108
Identities = 197/319 (61%), Positives = 238/319 (73%), Gaps = 1/319 (0%)
Query: 2 ASTPSIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALG 61
A +P+ KAW Y YG+ +LK + P +DQVL++V AAALNPVD KR G
Sbjct: 73 AEAGEVPATMKAWAYDDYGD-GSVLKLNDAAAVPDIADDQVLVRVAAAALNPVDAKRRAG 131
Query: 62 HFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTV 121
FK DSPLPTVPGYDVAGVVV G +VK K GDEVYG+I+E L PK G+L+EYT
Sbjct: 132 KFKATDSPLPTVPGYDVAGVVVKAGRKVKGLKEGDEVYGNISEKALEGPKQSGSLAEYTA 191
Query: 122 AEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLA 181
EEK+LA KP +L F +AA LPLAI TA++GLER FS+GKS+L+LGGAGGVGSL IQLA
Sbjct: 192 VEEKLLALKPKSLGFAQAAGLPLAIETAHEGLERAGFSAGKSILILGGAGGVGSLAIQLA 251
Query: 182 KHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKA 241
KHV+ AS++AATAST KL+ L+ LGAD+AIDYTKEN+E+L +K+DVV DAVG E+AVK
Sbjct: 252 KHVYGASKVAATASTPKLELLKSLGADVAIDYTKENFEDLPDKYDVVLDAVGQGEKAVKV 311
Query: 242 ANEGGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEA 301
EGG VV + PP F++TSDG+VLEKL PYLE+GKVKP++DPK PF F Q +EA
Sbjct: 312 VKEGGSVVVLTGAVVPPGFRFVVTSDGSVLEKLNPYLESGKVKPLVDPKGPFAFSQVVEA 371
Query: 302 FSYLNTNRAIGKIVIHPIP 320
FSYL T RA GK+VI PIP
Sbjct: 372 FSYLETGRATGKVVISPIP 390
>UniRef100_Q94F67 Quinone oxidoreductase-like protein [Helianthus annuus]
Length = 309
Score = 389 bits (999), Expect = e-107
Identities = 190/309 (61%), Positives = 242/309 (77%), Gaps = 1/309 (0%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KAW Y +YG+++ +LK ++V P K+DQVL+KVVAAA+NPVD KR LG+FK IDSPLP
Sbjct: 2 KAWKYDEYGSVD-VLKLATDVAVPEIKDDQVLVKVVAAAVNPVDYKRRLGYFKAIDSPLP 60
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKP 131
+PG+DV GVV+ VG QVK K GDEVYGDIN+ L P GTL+EYT EE++LA KP
Sbjct: 61 IIPGFDVLGVVLKVGSQVKDLKEGDEVYGDINDKGLEGPTQFGTLAEYTAVEERLLALKP 120
Query: 132 SNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIA 191
NL F++AA+LPLAI TAY+GLER +FS GK++LVL GAGGVGS +IQLAKHV+ AS++A
Sbjct: 121 KNLDFIQAAALPLAIETAYEGLERAKFSEGKTILVLNGAGGVGSFIIQLAKHVYGASKVA 180
Query: 192 ATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEGGKVVTI 251
AT+ST KL+ L+ LGAD+AIDYTKEN+E+L +K+DVVYDA+G E+A+KA NE G V+I
Sbjct: 181 ATSSTGKLELLKSLGADVAIDYTKENFEDLPDKYDVVYDAIGQPEKALKAVNETGVAVSI 240
Query: 252 LPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAI 311
P PP F+LTSDG++L KL PYLE+GK+K DPKSPFPF + EA++YL +
Sbjct: 241 TGPIPPPGFSFILTSDGSILTKLNPYLESGKIKSDNDPKSPFPFDKLNEAYAYLESTGPT 300
Query: 312 GKIVIHPIP 320
GK+VI+PIP
Sbjct: 301 GKVVIYPIP 309
>UniRef100_Q638J3 Quinone oxidoreductase [Bacillus cereus]
Length = 331
Score = 184 bits (467), Expect = 3e-45
Identities = 123/340 (36%), Positives = 187/340 (54%), Gaps = 49/340 (14%)
Query: 12 KAWVYSQYGN-----IEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI 66
KA + +G+ +EE+ K P VLI V A ++NP+D K G +
Sbjct: 2 KAQIIHSFGDSSVFQLEEVAK-------PKLLPGHVLIHVKATSVNPIDTKMRSGAVSSV 54
Query: 67 DSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKV 126
P + DVAG+V+ VGE V KFK GDEVYG +T G L+E+ +A+ ++
Sbjct: 55 APEFPAILHGDVAGIVIEVGEGVSKFKTGDEVYGCAGGF----KETGGALAEFMLADARL 110
Query: 127 LAHKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
+AHKP N++ EAA+LPL ITA++ L +R SG+++L+ G GGVG + IQLAK +
Sbjct: 111 IAHKPKNVTMEEAAALPLVAITAWESLFDRANMKSGQNVLIHGATGGVGHVAIQLAK--W 168
Query: 186 EASRIAATAS-TTKLDFLRKLGADLAIDYTKENYEELTEK------FDVVYDAVG--DSE 236
+++ TAS K++ +LGAD+AI+Y +E+ +E +K F+V++D VG + +
Sbjct: 169 AGAKVFTTASQQNKMEIAHRLGADIAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLD 228
Query: 237 RAVKAANEGGKVVTILPPGTPPAIPF---------------LLTSD-----GAVLEKLQP 276
+ +AA G VVTI T P +L +D G +L K+
Sbjct: 229 NSFEAAAVNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITQ 288
Query: 277 YLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
+E GK++P+LD KS F F + +A YL +N+AIGKIV+
Sbjct: 289 IVEEGKLRPLLDSKS-FTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_Q6HG35 Quinone oxidoreductase [Bacillus thuringiensis]
Length = 331
Score = 184 bits (466), Expect = 4e-45
Identities = 123/340 (36%), Positives = 187/340 (54%), Gaps = 49/340 (14%)
Query: 12 KAWVYSQYGN-----IEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI 66
KA + +G+ +EE+ K P VLI V A ++NP+D K G +
Sbjct: 2 KAQIIHSFGDSSVFQLEEVAK-------PKLLPGHVLIHVKATSVNPIDTKMRSGAVSSV 54
Query: 67 DSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKV 126
P + DVAG+V+ VGE V KFK GDEVYG +T G L+E+ +A+ ++
Sbjct: 55 APEFPAILHGDVAGIVIEVGEGVSKFKTGDEVYGCAGGF----KETGGALAEFMLADARL 110
Query: 127 LAHKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
+AHKP N++ EAA+LPL ITA++ L +R SG+++L+ G GGVG + IQLAK +
Sbjct: 111 IAHKPKNITMEEAAALPLVAITAWESLFDRANIKSGQNVLIHGATGGVGHVAIQLAK--W 168
Query: 186 EASRIAATAS-TTKLDFLRKLGADLAIDYTKENYEELTEK------FDVVYDAVG--DSE 236
+++ TAS K++ +LGAD+AI+Y +E+ +E +K F+V++D VG + +
Sbjct: 169 AGAKVFTTASQQNKMEISHRLGADVAINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLD 228
Query: 237 RAVKAANEGGKVVTILPPGTPPAIPF---------------LLTSD-----GAVLEKLQP 276
+ +AA G VVTI T P +L +D G +L K+
Sbjct: 229 NSFEAAAVNGTVVTIAARSTHDLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITQ 288
Query: 277 YLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
+E GK++P+LD KS F F + +A YL +N+AIGKIV+
Sbjct: 289 IVEEGKLRPLLDSKS-FTFDEVAQAHEYLESNKAIGKIVL 327
>UniRef100_UPI00003CB31E UPI00003CB31E UniRef100 entry
Length = 331
Score = 182 bits (462), Expect = 1e-44
Identities = 118/315 (37%), Positives = 177/315 (55%), Gaps = 37/315 (11%)
Query: 32 VPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKK 91
V P VLI V A ++NP+D K G + P + DVAG+V+ VGE V K
Sbjct: 20 VSKPKLLPGHVLIHVKATSVNPIDTKMRSGAVSAVAPEFPAILHGDVAGIVIEVGEGVSK 79
Query: 92 FKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQ 151
FK GDEVYG +T G L+E+ +A+ +++AHKP+NL+ EAA+LPL ITA++
Sbjct: 80 FKSGDEVYGCAGGF----KETGGALAEFMLADARLIAHKPNNLTMEEAAALPLVAITAWE 135
Query: 152 GL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATAS-TTKLDFLRKLGADL 209
L +R G+++L+ G GGVG + IQLAK + +++ TAS +K++ LGAD+
Sbjct: 136 SLFDRANIKPGQNVLIHGATGGVGHVAIQLAK--WAGAKVFTTASQQSKMEISHHLGADI 193
Query: 210 AIDYTKENYEELTEK------FDVVYDAVG--DSERAVKAANEGGKVVTILPPGTPPAIP 261
AI+Y +E+ +E +K F+V++D VG + + + +AA G VVTI T P
Sbjct: 194 AINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLDHSFEAAAVNGTVVTIAARSTHDLSP 253
Query: 262 F---------------LLTSD-----GAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEA 301
+L +D G +L K+ +E GK++P+LD K PF F + +A
Sbjct: 254 LHTKGLTLHVTFMALKILHTDKRNHCGEILTKITQIIEEGKLRPLLDSK-PFTFDEVAQA 312
Query: 302 FSYLNTNRAIGKIVI 316
YL +N+AIGKIV+
Sbjct: 313 HEYLESNKAIGKIVL 327
>UniRef100_Q736Y0 Oxidoreductase, zinc-binding [Bacillus cereus]
Length = 308
Score = 179 bits (454), Expect = 9e-44
Identities = 122/316 (38%), Positives = 180/316 (56%), Gaps = 23/316 (7%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD-IDSPL 70
KA V QYG++EE+ + V P K ++VLI+++A ++NPVD K G ++ +
Sbjct: 2 KAIVIDQYGSVEELK--ERQVLKPVVKNNEVLIRILATSINPVDWKIRKGDLQEQLRFSF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIG--TLSEYTVAEEKVLA 128
P + G DVAGV+ VGE+++ FK+GD+VY P+ IG + +E+ + +++
Sbjct: 60 PIILGLDVAGVIEEVGEEIECFKIGDKVYT--------KPENIGQGSYAEFITVKSDLVS 111
Query: 129 HKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEA 187
+ P+NLSF EAAS+PLA +TA+Q L + + +L+ GAGGVGSL IQ+AK
Sbjct: 112 YMPTNLSFEEAASIPLAGLTAWQSLVDYAKIKEHDRVLIHAGAGGVGSLAIQIAKSF--G 169
Query: 188 SRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGD--SERAVKAANEG 245
+ +A TAS+ +FL+ LGADL IDY K+ +EEL FD+V D +G E++ + G
Sbjct: 170 AFVATTASSKNEEFLKSLGADLVIDYKKDKFEELLSDFDIVLDTIGGEVQEKSFQILKPG 229
Query: 246 GKVVTIL--PPGTPPAIP--FL-LTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLE 300
G +V+I+ P I FL L +G LE L + +GKVKPI+ PF E
Sbjct: 230 GVLVSIVHEPIQKVKEIKSGFLWLKPNGKQLEALSDLIVHGKVKPIISKVMPFSEEGIRE 289
Query: 301 AFSYLNTNRAIGKIVI 316
A GKIVI
Sbjct: 290 AHILSEGQHVRGKIVI 305
>UniRef100_Q81MY1 Alcohol dehydrogenase, zinc-containing [Bacillus anthracis]
Length = 335
Score = 179 bits (453), Expect = 1e-43
Identities = 117/319 (36%), Positives = 177/319 (54%), Gaps = 41/319 (12%)
Query: 32 VPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKK 91
V P VLI V A ++NP+D K G + P + DVAG+V+ VGE V K
Sbjct: 20 VSKPKLLPGHVLIDVKATSVNPIDTKMRSGAVSAVAPEFPAILHGDVAGIVIEVGEGVSK 79
Query: 92 FKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQ 151
FK GDEVYG +T G L+E+ +A+ +++AHKP+N++ EAA+LPL ITA++
Sbjct: 80 FKCGDEVYGCAGGF----KETGGALAEFMLADARLIAHKPNNITMEEAAALPLVAITAWE 135
Query: 152 GL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATAS-TTKLDFLRKLGADL 209
L +R SG+++L+ G GGVG + IQLAK + + + TAS K++ +LGAD+
Sbjct: 136 SLFDRANIKSGQNVLIHGATGGVGHVAIQLAK--WAGANVFTTASQQNKMEIAHRLGADV 193
Query: 210 AIDYTKENYEELTEK----------FDVVYDAVG--DSERAVKAANEGGKVVTILPPGTP 257
AI+Y +E+ +E ++ F+V++D VG + + + +AA G VVTI T
Sbjct: 194 AINYKEESVQESVQEYVQKHTNGNGFEVIFDTVGGKNLDNSFEAAAVNGTVVTIAARSTH 253
Query: 258 PAIPF---------------LLTSD-----GAVLEKLQPYLENGKVKPILDPKSPFPFYQ 297
P +L +D G +L K+ +E GK++P+LD KS F F +
Sbjct: 254 DLSPLHAKGLSLHVTFMALKILHTDKRNDCGEILTKITKIVEEGKLRPLLDSKS-FTFDE 312
Query: 298 TLEAFSYLNTNRAIGKIVI 316
+A YL +N+AIGKIV+
Sbjct: 313 VAQAHEYLESNKAIGKIVL 331
>UniRef100_Q8ES25 Zinc-binding oxidoreductase [Oceanobacillus iheyensis]
Length = 311
Score = 178 bits (452), Expect = 2e-43
Identities = 124/323 (38%), Positives = 178/323 (54%), Gaps = 34/323 (10%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD-IDSPL 70
KA V YG+ +E+ + +VP P ++Q+LI+ A ++NP+D K G+ KD +
Sbjct: 2 KAIVIENYGHADEL--HEQDVPKPTINDNQILIEQYATSINPIDWKLREGYLKDGFNFEF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTI--GTLSEYTVAEEKVLA 128
P + G+D AGVV VG+ V +FKVGD V+ P T GT +E+ EE +A
Sbjct: 60 PIILGWDSAGVVAEVGKNVSQFKVGDRVFS--------RPATTAQGTYAEFVAVEESRVA 111
Query: 129 HKPSNLSFVEAASLPLAIITAYQGLERVEFSS---GKSLLVLGGAGGVGSLVIQLAKHVF 185
P N+SF EAAS+PLA +TA+Q L V+FS+ G +L+ G+GGVGS IQ AKH
Sbjct: 112 KIPENVSFEEAASVPLAGLTAWQCL--VDFSAIKKGDKVLIHAGSGGVGSFAIQFAKHF- 168
Query: 186 EASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDS--ERAVKAAN 243
+ +A TAS +++LGAD I+Y +EN+ E+ E +D+V D +G E++ +
Sbjct: 169 -GAYVATTASGKNESLVKELGADRFINYKEENFNEVLEDYDIVVDTLGGDILEQSFEVLK 227
Query: 244 EGGKVVTILPPGTPP---------AIPFL-LTSDGAVLEKLQPYLENGKVKPILDPKSPF 293
EGGK+V+I G P FL L +G L ++ LENGKVK ++ P
Sbjct: 228 EGGKLVSI--AGNPDEEKAKEKGIKAGFLWLEENGKQLSEVADLLENGKVKSVIGHTFPL 285
Query: 294 PFYQTLEAFSYLNTNRAIGKIVI 316
EA T+ A GKIVI
Sbjct: 286 TEQGLREAHQLSETHHARGKIVI 308
>UniRef100_Q92E39 Lin0622 protein [Listeria innocua]
Length = 313
Score = 177 bits (448), Expect = 4e-43
Identities = 120/322 (37%), Positives = 175/322 (54%), Gaps = 31/322 (9%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI-DSPL 70
KA V YG EE+ + V P ++QV++K VA ++NP+D K G+ K + D
Sbjct: 2 KAVVIENYGGKEELK--EKEVAMPKAGKNQVIVKEVATSINPIDWKLREGYLKQMMDWEF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKT--IGTLSEYTVAEEKVLA 128
P + G+DVAGV+ VGE V +KVGDEV+ P+T GT +EYT ++ +LA
Sbjct: 60 PIILGWDVAGVISEVGEGVTDWKVGDEVFA--------RPETTRFGTYAEYTAVDDHLLA 111
Query: 129 HKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEA 187
P +SF EAAS+PLA +TA+Q L + + G+ +L+ GAGGVG+L IQLAK +
Sbjct: 112 PLPEGISFEEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAK--YAG 169
Query: 188 SRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVG-----DSERAVKAA 242
+ + TAS + L+ LGAD IDY + N++++ DVV+D +G DS +K
Sbjct: 170 AEVITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETDSYDVLKEG 229
Query: 243 NEGGKVVTIL--------PPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFP 294
G++V+I+ A L +G L+KL L N VKPI+ PF
Sbjct: 230 T--GRLVSIVGISNEDRAKEKNVTANGIWLQPNGEQLKKLGELLANKTVKPIVGATFPFS 287
Query: 295 FYQTLEAFSYLNTNRAIGKIVI 316
+A + T+ A+GKIVI
Sbjct: 288 EKGVFDAHALSETHHAVGKIVI 309
>UniRef100_Q722T7 Alcohol dehydrogenase, zinc-dependent [Listeria monocytogenes]
Length = 313
Score = 176 bits (447), Expect = 6e-43
Identities = 119/322 (36%), Positives = 174/322 (53%), Gaps = 31/322 (9%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI-DSPL 70
KA V YG EE+ + V P ++QV++K A ++NP+D K G+ K + D
Sbjct: 2 KAVVIENYGGKEELK--EKEVAMPKAGKNQVIVKEAATSINPIDWKLREGYLKQMMDWEF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKT--IGTLSEYTVAEEKVLA 128
P + G+DVAGV+ VGE V +KVGDEV+ P+T GT +EYT ++ +LA
Sbjct: 60 PIILGWDVAGVISEVGEGVTDWKVGDEVFA--------RPETTRFGTYAEYTAVDDHLLA 111
Query: 129 HKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEA 187
P +SF EAAS+PLA +TA+Q L + + G+ +L+ GAGGVG+L IQLAKH
Sbjct: 112 PLPEGISFDEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAKHA--G 169
Query: 188 SRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVG-----DSERAVKAA 242
+ + TAS + L+ LGAD IDY + N++++ DVV+D +G DS +K
Sbjct: 170 AEVITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETDSYDVLKEG 229
Query: 243 NEGGKVVTIL--------PPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFP 294
G++V+I+ A L +G L++L L N VKPI+ PF
Sbjct: 230 T--GRLVSIVGISNEERAKEKNVTATGIWLQPNGEQLKELGKLLANKTVKPIVGATFPFS 287
Query: 295 FYQTLEAFSYLNTNRAIGKIVI 316
+A + T+ A+GKIVI
Sbjct: 288 EKGVFDAHALSETHHAVGKIVI 309
>UniRef100_Q8Y9B9 Lmo0613 protein [Listeria monocytogenes]
Length = 313
Score = 176 bits (446), Expect = 7e-43
Identities = 118/322 (36%), Positives = 174/322 (53%), Gaps = 31/322 (9%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDI-DSPL 70
KA V YG EE+ + V P ++QV++K A ++NP+D K G+ K + D
Sbjct: 2 KAVVIENYGGKEELK--EKEVAMPKAGKNQVIVKEAATSINPIDWKLREGYLKQMMDWEF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKT--IGTLSEYTVAEEKVLA 128
P + G+DVAGV+ VGE V +KVGDEV+ P+T GT +EYT ++ +LA
Sbjct: 60 PIILGWDVAGVISEVGEGVTDWKVGDEVFA--------RPETTRFGTYAEYTAVDDHLLA 111
Query: 129 HKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEA 187
P +SF EAAS+PLA +TA+Q L + + G+ +L+ GAGGVG+L IQLAKH
Sbjct: 112 PLPEGISFDEAASIPLAGLTAWQALFDHAKLQKGEKVLIHAGAGGVGTLAIQLAKHA--G 169
Query: 188 SRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVG-----DSERAVKAA 242
+ + TAS + L+ LGAD IDY + N++++ DVV+D +G DS +K
Sbjct: 170 AEVITTASAKNHELLKSLGADQVIDYKEVNFKDVLSDIDVVFDTMGGQIETDSYDVLKEG 229
Query: 243 NEGGKVVTIL--------PPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFP 294
G++V+I+ A L +G L++L L N +KPI+ PF
Sbjct: 230 T--GRLVSIVGISNEDRAKEKNVTATGIWLQPNGEQLKELGKLLANKTIKPIVGATFPFS 287
Query: 295 FYQTLEAFSYLNTNRAIGKIVI 316
+A + T+ A+GKIVI
Sbjct: 288 EKGVFDAHALSETHHAVGKIVI 309
>UniRef100_Q81B16 Quinone oxidoreductase [Bacillus cereus]
Length = 331
Score = 174 bits (440), Expect = 4e-42
Identities = 115/315 (36%), Positives = 174/315 (54%), Gaps = 37/315 (11%)
Query: 32 VPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKK 91
V P VLI V A ++NP+D K G + P + DVAG+V+ VGE V K
Sbjct: 20 VSKPKLLPGHVLIDVKATSVNPIDTKMRSGAVSAVAPEFPAILHGDVAGIVIEVGEGVSK 79
Query: 92 FKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQ 151
FK GDEVYG +T G L+E+ +A+ +++AHKP +L+ AA+LPL ITA++
Sbjct: 80 FKCGDEVYGCAGGF----KETGGALAEFMLADARLIAHKPKSLTMEAAAALPLVAITAWE 135
Query: 152 GL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATAS-TTKLDFLRKLGADL 209
L +R SG+++L+ G GGVG + IQLAK + +++ TAS K++ +LGAD+
Sbjct: 136 SLFDRANIKSGQNVLIHGATGGVGHVAIQLAK--WAGAKVFTTASQQNKMEIAHRLGADI 193
Query: 210 AIDYTKENYEELTEK------FDVVYDAVG--DSERAVKAANEGGKVVTILPPGTPPAIP 261
AI+Y +E+ +E +K F+V++D VG + + + +AA G VVTI T P
Sbjct: 194 AINYKEESVQEYVQKHTNGNGFEVIFDTVGGKNLDNSFEAAAVNGTVVTIAARSTHDLSP 253
Query: 262 F---------------LLTSD-----GAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEA 301
+L +D +L KL +E G ++P+LD K+ F F + +A
Sbjct: 254 LHAKGLSLHVTFMALKILHTDKRDACREILTKLTQIVEEGHLRPLLDSKT-FTFDEVAQA 312
Query: 302 FSYLNTNRAIGKIVI 316
YL +N+AIGKIV+
Sbjct: 313 HEYLESNKAIGKIVL 327
>UniRef100_UPI00003CC0B8 UPI00003CC0B8 UniRef100 entry
Length = 308
Score = 173 bits (439), Expect = 5e-42
Identities = 120/316 (37%), Positives = 177/316 (55%), Gaps = 23/316 (7%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD-IDSPL 70
KA V QYG++EE+ + V P K ++VLI+++A ++NP D K G ++ +
Sbjct: 2 KAIVIDQYGSVEELK--ERRVLKPVVKNNEVLIRILATSINPGDWKIRKGDLQEQLGFSF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIG--TLSEYTVAEEKVLA 128
P + G DVAGV+ +GE+V+ FK+GD+V+ P+ IG + +E+ + +++
Sbjct: 60 PIILGLDVAGVIEEIGEEVECFKIGDKVFT--------KPENIGWGSYAEFITVKTDLVS 111
Query: 129 HKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEA 187
+ P+NLSFVEAAS+PLA +TA+Q L + + +L+ GAGGVGSL IQ+AK
Sbjct: 112 YMPTNLSFVEAASIPLAGLTAWQSLVDYAKIKEHDRVLIHAGAGGVGSLAIQIAKSF--G 169
Query: 188 SRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGD--SERAVKAANEG 245
+ +A TAS+ +FL+ LGADL IDY K+ +EEL FD+V +G E++ + G
Sbjct: 170 AFVATTASSKNEEFLKSLGADLVIDYKKDKFEELLSDFDIVLGTIGGEVQEKSFQILKPG 229
Query: 246 GKVVTIL--PPGTPPAIP--FL-LTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLE 300
G V+I+ P I FL L G LE L + +GKVKPI+ PF E
Sbjct: 230 GVFVSIVHEPIQKVKEIKSGFLWLKPSGKQLEALSDLIVHGKVKPIISKVMPFSEEGIRE 289
Query: 301 AFSYLNTNRAIGKIVI 316
A GKIVI
Sbjct: 290 AHILSEGQHVRGKIVI 305
>UniRef100_Q5SI53 NADPH-quinone reductase [Thermus thermophilus HB8]
Length = 318
Score = 169 bits (429), Expect = 7e-41
Identities = 119/323 (36%), Positives = 171/323 (52%), Gaps = 28/323 (8%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIK-RALGHFKDIDSPL 70
+A V +G +E + + D VP P P E VL++ +A A+NPVD K RA G + ++ P
Sbjct: 2 RAVVLKGFGGLEMLEERDLPVPEPGPGE--VLVRNLAVAVNPVDAKIRAAGRWAGVEPPF 59
Query: 71 PTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHK 130
V GYD AGVV VG VK K GDEVY NP GT +EYT ++A K
Sbjct: 60 --VLGYDAAGVVAKVGPGVKDLKEGDEVY--YTPEIFGNPH--GTYAEYTPVPAGIVAKK 113
Query: 131 PSNLSFVEAASLPLAIITAYQG-LERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASR 189
P NLSF EAA++PLA TA++ + R+ G+++LV+GGAGGVGS +Q AK +
Sbjct: 114 PKNLSFAEAAAIPLAGGTAWEAVVRRLAVRPGETVLVMGGAGGVGSFAVQFAKAA--GAY 171
Query: 190 IAATASTTKLDFLRKLGADLAIDYTKENYEELT-----EKFDVVYDAVGDS--ERAVKAA 242
+ ATAS L L++LGADLA+DY EE+ + D ++ G+S ER +
Sbjct: 172 VIATASAENLPVLKELGADLALDYRGPWAEEVLKATEGQGVDAAFETAGESLVERVIPVV 231
Query: 243 NEGGKVVTILPP---------GTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPF 293
G++ TILPP LT + L +++P E G +P++ PF
Sbjct: 232 RPFGRIATILPPQGNLSGLYTKNQTLYGVFLTRERKRLLEMRPLFERGLARPLIAEVLPF 291
Query: 294 PFYQTLEAFSYLNTNRAIGKIVI 316
+A + +++ GKIV+
Sbjct: 292 SLENLRKAHARMDSGHGRGKIVL 314
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 561,889,331
Number of Sequences: 2790947
Number of extensions: 24719276
Number of successful extensions: 71597
Number of sequences better than 10.0: 4143
Number of HSP's better than 10.0 without gapping: 1609
Number of HSP's successfully gapped in prelim test: 2534
Number of HSP's that attempted gapping in prelim test: 64417
Number of HSP's gapped (non-prelim): 5211
length of query: 320
length of database: 848,049,833
effective HSP length: 127
effective length of query: 193
effective length of database: 493,599,564
effective search space: 95264715852
effective search space used: 95264715852
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC139600.7


