

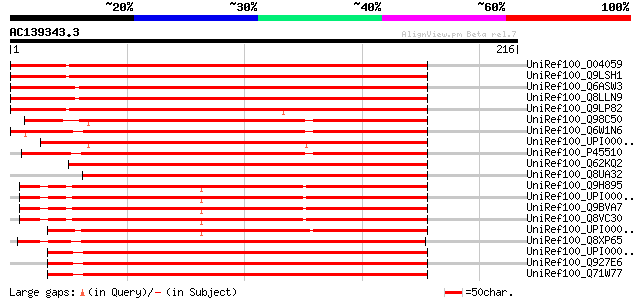
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139343.3 + phase: 0 /pseudo
(216 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O04059 Putative 3,4-dihydroxy-2-butanone kinase [Lycop... 294 9e-79
UniRef100_Q9LSH1 Dihydroxyacetone/glycerone kinase-like protein ... 285 7e-76
UniRef100_Q6ASW3 Putative DAK2 domain containing protein [Oryza ... 283 2e-75
UniRef100_Q8LLN9 3,4-dihydroxy-2-butanone kinase [Oryza sativa] 283 2e-75
UniRef100_Q9LP82 T1N15.4 [Arabidopsis thaliana] 275 6e-73
UniRef100_Q98C50 Dihydroxyacetone kinase [Rhizobium loti] 209 4e-53
UniRef100_Q6W1N6 Dihydroxyacetone kinase [Rhizobium sp.] 204 1e-51
UniRef100_UPI00002B0033 UPI00002B0033 UniRef100 entry 197 2e-49
UniRef100_P45510 Dihydroxyacetone kinase [Citrobacter freundii] 196 3e-49
UniRef100_Q62KQ2 Dihydroxyacetone kinase [Burkholderia mallei] 194 1e-48
UniRef100_Q8UA32 3,4-dihydroxy-2-butanone kinase [Agrobacterium ... 188 7e-47
UniRef100_Q9H895 Hypothetical protein FLJ13853 [Homo sapiens] 180 2e-44
UniRef100_UPI00001CEB5D UPI00001CEB5D UniRef100 entry 179 4e-44
UniRef100_Q9BVA7 DKFZP586B1621 protein [Homo sapiens] 179 6e-44
UniRef100_Q8VC30 CDNA sequence BC021917 [Mus musculus] 178 8e-44
UniRef100_UPI0000438A06 UPI0000438A06 UniRef100 entry 174 1e-42
UniRef100_Q8XP65 Dihydroxyacetone kinase [Clostridium perfringens] 170 3e-41
UniRef100_UPI00003CA2C5 UPI00003CA2C5 UniRef100 entry 169 4e-41
UniRef100_Q927E6 Lin2843 protein [Listeria innocua] 169 4e-41
UniRef100_Q71W77 Dihydroxyacetone kinase, Dak1 subunit, putative... 169 5e-41
>UniRef100_O04059 Putative 3,4-dihydroxy-2-butanone kinase [Lycopersicon esculentum]
Length = 594
Score = 294 bits (753), Expect = 9e-79
Identities = 150/178 (84%), Positives = 158/178 (88%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+E YP LQYLDG+P VKVV R DV G YDKVAIISGGGSGHEPAHAGFVGEGMLTAAI
Sbjct: 24 IENYPGLQYLDGFPEVKVVLRADV-SGAKYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
CGDVFASP V++ILAGIRAVTGPMGCLLIVKNYTGDRLNFG AAEQAKSEGYKVE VIV
Sbjct: 83 CGDVFASPNVDSILAGIRAVTGPMGCLLIVKNYTGDRLNFGLAAEQAKSEGYKVEMVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGT+LVHKVAGAAAA GL LADVAAEA+ ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGIAGRRGLAGTLLVHKVAGAAAACGLPLADVAAEAKRASEMVGTMGVAL 200
>UniRef100_Q9LSH1 Dihydroxyacetone/glycerone kinase-like protein [Arabidopsis
thaliana]
Length = 618
Score = 285 bits (728), Expect = 7e-76
Identities = 145/178 (81%), Positives = 157/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P VKVV R DV YDKVA+ISGGGSGHEPAHAG+VGEGMLTAAI
Sbjct: 24 VETYPGLQYLDGFPEVKVVLRADV-SAAKYDKVAVISGGGSGHEPAHAGYVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
CGDVFASP V++ILAGIRAVTG GCLLIVKNYTGDRLNFG AAEQAKSEGYKVE+VIV
Sbjct: 83 CGDVFASPPVDSILAGIRAVTGTEGCLLIVKNYTGDRLNFGLAAEQAKSEGYKVETVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+DCA+PPP + GRRGLAGT+LVHKVAGAAAAAGLSL VAAEA+ ASE VGTMGVAL
Sbjct: 143 EDCALPPPRGIAGRRGLAGTVLVHKVAGAAAAAGLSLEKVAAEAKCASEMVGTMGVAL 200
>UniRef100_Q6ASW3 Putative DAK2 domain containing protein [Oryza sativa]
Length = 594
Score = 283 bits (725), Expect = 2e-75
Identities = 143/178 (80%), Positives = 156/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P +KVV R DV G YDKVA+ISGGGSGHEP HAGFVG GMLTAA+
Sbjct: 24 VETYPGLQYLDGFPQIKVVLRADVVQGA-YDKVAVISGGGSGHEPTHAGFVGPGMLTAAV 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
GDVF SP V++ILA IRAVTGPMGCLLIVKNYTGDRLNFG AAEQAKSEGYK+E VIV
Sbjct: 83 SGDVFTSPPVDSILAAIRAVTGPMGCLLIVKNYTGDRLNFGLAAEQAKSEGYKMEMVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGTILVHKVAGAAA AGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGIAGRRGLAGTILVHKVAGAAADAGLSLAEVAAEAKHASEVVGTMGVAL 200
>UniRef100_Q8LLN9 3,4-dihydroxy-2-butanone kinase [Oryza sativa]
Length = 575
Score = 283 bits (725), Expect = 2e-75
Identities = 143/178 (80%), Positives = 156/178 (87%), Gaps = 1/178 (0%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG+P +KVV R DV G YDKVA+ISGGGSGHEP HAGFVG GMLTAA+
Sbjct: 24 VETYPGLQYLDGFPQIKVVLRADVVQGA-YDKVAVISGGGSGHEPTHAGFVGPGMLTAAV 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVA 120
GDVF SP V++ILA IRAVTGPMGCLLIVKNYTGDRLNFG AAEQAKSEGYK+E VIV
Sbjct: 83 SGDVFTSPPVDSILAAIRAVTGPMGCLLIVKNYTGDRLNFGLAAEQAKSEGYKMEMVIVG 142
Query: 121 DDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
DDCA+PPP + GRRGLAGTILVHKVAGAAA AGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 DDCALPPPRGIAGRRGLAGTILVHKVAGAAADAGLSLAEVAAEAKHASEVVGTMGVAL 200
>UniRef100_Q9LP82 T1N15.4 [Arabidopsis thaliana]
Length = 625
Score = 275 bits (703), Expect = 6e-73
Identities = 140/180 (77%), Positives = 157/180 (86%), Gaps = 3/180 (1%)
Query: 1 METYPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAI 60
+ETYP LQYLDG P +KVV R DV YDKVA+ISGGGSGHEPA AG+VGEGMLTAAI
Sbjct: 24 VETYPGLQYLDGLPEIKVVLRADV-SAANYDKVAVISGGGSGHEPAQAGYVGEGMLTAAI 82
Query: 61 CGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVE--SVI 118
CGDVFASPTV++ILA IRAVTGP GCLL+V NYTGDRLNFG AAEQAK+EG+ +E +VI
Sbjct: 83 CGDVFASPTVDSILARIRAVTGPKGCLLVVTNYTGDRLNFGLAAEQAKTEGFDIEVTTVI 142
Query: 119 VADDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
V DDCA+PPP + GRRGLAGT+LVHKVAGAAAAAGLSLA+VAAEA++ASE VGTMGVAL
Sbjct: 143 VGDDCALPPPRGVAGRRGLAGTVLVHKVAGAAAAAGLSLAEVAAEAKHASEMVGTMGVAL 202
>UniRef100_Q98C50 Dihydroxyacetone kinase [Rhizobium loti]
Length = 547
Score = 209 bits (532), Expect = 4e-53
Identities = 105/174 (60%), Positives = 141/174 (80%), Gaps = 11/174 (6%)
Query: 7 LQYLDGYPNVKVVFRKDVYPGPPYDK--VAIISGGGSGHEPAHAGFVGEGMLTAAICGDV 64
L LDGYP +KVV R D +DK V+++SGGG+GHEP+HAGFVG+GMLTAA+ G++
Sbjct: 28 LARLDGYPEIKVVLRAD------WDKSRVSVVSGGGAGHEPSHAGFVGKGMLTAAVSGEI 81
Query: 65 FASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCA 124
FASP+VEA+LA IRAVTGP GCLLIVKNYTGDRLNFG AAE+A++EG++VE VIVADD A
Sbjct: 82 FASPSVEAVLAAIRAVTGPAGCLLIVKNYTGDRLNFGLAAEKARAEGFRVEMVIVADDIA 141
Query: 125 IPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+P ++ RG+AGT+ VHK++G + AG +LAD+AA AR A++++ ++G++L
Sbjct: 142 LP---DIAQPRGVAGTLFVHKISGHLSEAGHNLADIAAAARAAAKDIVSLGMSL 192
>UniRef100_Q6W1N6 Dihydroxyacetone kinase [Rhizobium sp.]
Length = 547
Score = 204 bits (519), Expect = 1e-51
Identities = 108/180 (60%), Positives = 136/180 (75%), Gaps = 9/180 (5%)
Query: 1 METYP--SLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTA 58
++T P SL LD YP++KVV R D KVA+ISGGG+GHEP+HAGFVG GMLTA
Sbjct: 23 LQTAPAGSLARLDTYPDIKVVLRGDWDKA----KVAVISGGGAGHEPSHAGFVGRGMLTA 78
Query: 59 AICGDVFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVI 118
A+ G++FASP+VEA+L IRA TGP GCLLIVKNYTGDRLNFG AAE+A+ EG++VE VI
Sbjct: 79 AVSGEIFASPSVEAVLTAIRAATGPKGCLLIVKNYTGDRLNFGLAAEKARVEGFRVEMVI 138
Query: 119 VADDCAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
VADD A+P ++V RG+AGT+ VHK+AG A G L VAA AR A+ ++ ++GV+L
Sbjct: 139 VADDIALP---DIVQPRGVAGTLFVHKIAGHLAECGAGLETVAAAARSAARDIVSLGVSL 195
>UniRef100_UPI00002B0033 UPI00002B0033 UniRef100 entry
Length = 218
Score = 197 bits (500), Expect = 2e-49
Identities = 103/169 (60%), Positives = 129/169 (75%), Gaps = 4/169 (2%)
Query: 14 PNVKVVFRKDVYPGPPYDK---VAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTV 70
P++ + ++V P D+ VA+ISGGGSGHEPA AG++GEGMLTAA+CG+V+ASP+V
Sbjct: 26 PSIARIADQNVLVYQPRDRDETVAVISGGGSGHEPAMAGYLGEGMLTAAVCGNVYASPSV 85
Query: 71 EAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAI-PPPL 129
EA+L+ IRA G G +L+V NYTGDRLNFG AAE+AK EG+KVE VIV DDCA+ +
Sbjct: 86 EAVLSAIRATAGKAGAVLVVMNYTGDRLNFGLAAERAKLEGHKVEMVIVNDDCALASDKV 145
Query: 130 EMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+ GRRGLAGT+ HKVAGAAAAAG SL +V E + E+VGTMGVAL
Sbjct: 146 GIAGRRGLAGTLFAHKVAGAAAAAGKSLQEVVEETKACVESVGTMGVAL 194
>UniRef100_P45510 Dihydroxyacetone kinase [Citrobacter freundii]
Length = 552
Score = 196 bits (499), Expect = 3e-49
Identities = 101/173 (58%), Positives = 130/173 (74%), Gaps = 7/173 (4%)
Query: 6 SLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVF 65
+L L+ P +++V R+D+ + VA+ISGGGSGHEPAH GF+G+GMLTAA+CGDVF
Sbjct: 28 NLARLESDPAIRIVVRRDLNK----NNVAVISGGGSGHEPAHVGFIGKGMLTAAVCGDVF 83
Query: 66 ASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAI 125
ASP+V+A+L I+AVTG GCLLIVKNYTGDRLNFG AAE+A+ GY VE +IV DD ++
Sbjct: 84 ASPSVDAVLTAIQAVTGEAGCLLIVKNYTGDRLNFGLAAEKARRLGYNVEMLIVGDDISL 143
Query: 126 PPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
P + RG+AGTILVHK+AG A G +LA V EA+YA+ N ++GVAL
Sbjct: 144 P---DNKHPRGIAGTILVHKIAGYFAERGYNLATVLREAQYAASNTFSLGVAL 193
>UniRef100_Q62KQ2 Dihydroxyacetone kinase [Burkholderia mallei]
Length = 570
Score = 194 bits (493), Expect = 1e-48
Identities = 98/153 (64%), Positives = 118/153 (77%)
Query: 26 PGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMG 85
P P VA+ISGGGSGHEPAH G+VG GML+AA+CG+VF SP+ +A+LA IRA GP G
Sbjct: 41 PEPAKRAVAVISGGGSGHEPAHGGYVGAGMLSAAVCGEVFTSPSADAVLAAIRATAGPHG 100
Query: 86 CLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHK 145
LLIVKNYTGDRLNFG AAE A+++G VE V+VADD ++ E RRG+AGT+LVHK
Sbjct: 101 ALLIVKNYTGDRLNFGLAAELARADGIPVEIVVVADDVSLRALTERGRRRGIAGTVLVHK 160
Query: 146 VAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGAAA GL+LA VAA A A+ +GTMGVAL
Sbjct: 161 LAGAAAERGLTLAQVAAAANAAAARLGTMGVAL 193
>UniRef100_Q8UA32 3,4-dihydroxy-2-butanone kinase [Agrobacterium tumefaciens]
Length = 564
Score = 188 bits (478), Expect = 7e-47
Identities = 94/147 (63%), Positives = 115/147 (77%)
Query: 32 KVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAGIRAVTGPMGCLLIVK 91
KVA++SGGGSGHEPAHAG+VG GMLT A+ GDVF SP+ +A+LAGIRA GP G L+IVK
Sbjct: 47 KVAVLSGGGSGHEPAHAGYVGTGMLTVAVAGDVFTSPSTDAVLAGIRAAAGPAGALVIVK 106
Query: 92 NYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRGLAGTILVHKVAGAAA 151
NYTGDRLNFG AAE A++EG VE V+VADD A+ + RRG+AGT+LVHK+AGAAA
Sbjct: 107 NYTGDRLNFGLAAELARAEGIPVEIVVVADDVALKDTVPAERRRGIAGTVLVHKLAGAAA 166
Query: 152 AAGLSLADVAAEARYASENVGTMGVAL 178
GL L +VA AR A+ + +MGV+L
Sbjct: 167 EKGLPLQEVARIARDAAAKLSSMGVSL 193
>UniRef100_Q9H895 Hypothetical protein FLJ13853 [Homo sapiens]
Length = 575
Score = 180 bits (457), Expect = 2e-44
Identities = 98/176 (55%), Positives = 125/176 (70%), Gaps = 8/176 (4%)
Query: 5 PSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDV 64
P+LQ L G+ +V R D+ +VA++SGGGSGHEPAHAGF+G+GMLT AI G V
Sbjct: 26 PNLQLLQGH---RVALRSDL--DSLKGRVALLSGGGSGHEPAHAGFIGKGMLTGAIAGAV 80
Query: 65 FASPTVEAILAGIRAV--TGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADD 122
F SP V +ILA IRAV G +G LLIVKNYTGDRLNFG A EQA++EG VE V++ DD
Sbjct: 81 FTSPAVGSILAAIRAVAQAGTVGTLLIVKNYTGDRLNFGLAREQARAEGIPVEMVVIGDD 140
Query: 123 CAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
A L+ GRRGL GT+L+HKVAGA A AG+ L ++A + ++ +GT+GV+L
Sbjct: 141 SAF-TVLKKAGRRGLCGTVLIHKVAGALAEAGVGLEEIAKQVNVVTKAMGTLGVSL 195
>UniRef100_UPI00001CEB5D UPI00001CEB5D UniRef100 entry
Length = 578
Score = 179 bits (454), Expect = 4e-44
Identities = 99/176 (56%), Positives = 121/176 (68%), Gaps = 8/176 (4%)
Query: 5 PSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDV 64
P LQ L GY +V R D+ +VA++SGGGSGHEPAHAGF+G+GMLT I G V
Sbjct: 26 PDLQLLQGY---RVALRSDL--DSLKGRVALLSGGGSGHEPAHAGFIGKGMLTGVIAGAV 80
Query: 65 FASPTVEAILAGIRAV--TGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADD 122
FASP V +ILA IRAV G G LLIVKNYTGDRLNFG A EQAK+EG VE V++ DD
Sbjct: 81 FASPAVGSILAAIRAVAQAGTAGTLLIVKNYTGDRLNFGLAMEQAKAEGISVEMVVIEDD 140
Query: 123 CAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
A L+ GRRGL GTIL+HKVAGA A G+ L ++ + ++ +GT+GV+L
Sbjct: 141 SAF-TVLKKAGRRGLCGTILIHKVAGALAEEGMGLEEITKKVSVIAKAIGTLGVSL 195
>UniRef100_Q9BVA7 DKFZP586B1621 protein [Homo sapiens]
Length = 575
Score = 179 bits (453), Expect = 6e-44
Identities = 97/176 (55%), Positives = 124/176 (70%), Gaps = 8/176 (4%)
Query: 5 PSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDV 64
P+LQ L G+ +V R D+ +VA++SGGGSGHEPAHAGF+G+GMLT I G V
Sbjct: 26 PNLQLLQGH---RVALRSDL--DSLKGRVALLSGGGSGHEPAHAGFIGKGMLTGVIAGAV 80
Query: 65 FASPTVEAILAGIRAV--TGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADD 122
F SP V +ILA IRAV G +G LLIVKNYTGDRLNFG A EQA++EG VE V++ DD
Sbjct: 81 FTSPAVGSILAAIRAVAQAGTVGTLLIVKNYTGDRLNFGLAREQARAEGIPVEMVVIGDD 140
Query: 123 CAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
A L+ GRRGL GT+L+HKVAGA A AG+ L ++A + ++ +GT+GV+L
Sbjct: 141 SAF-TVLKKAGRRGLCGTVLIHKVAGALAEAGVGLEEIAKQVNVVAKAMGTLGVSL 195
>UniRef100_Q8VC30 CDNA sequence BC021917 [Mus musculus]
Length = 578
Score = 178 bits (452), Expect = 8e-44
Identities = 99/176 (56%), Positives = 121/176 (68%), Gaps = 8/176 (4%)
Query: 5 PSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDV 64
P LQ L G+ +V R D+ +VA++SGGGSGHEPAHAGF+G+GMLT I G V
Sbjct: 26 PDLQLLQGH---RVALRSDL--DTLKGRVALLSGGGSGHEPAHAGFIGKGMLTGVIAGSV 80
Query: 65 FASPTVEAILAGIRAV--TGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADD 122
FASP V +ILA IRAV G +G LLIVKNYTGDRLNFG A EQAK+EG VE VIV DD
Sbjct: 81 FASPPVGSILAAIRAVAQAGTVGTLLIVKNYTGDRLNFGLAMEQAKAEGISVEMVIVEDD 140
Query: 123 CAIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
A L+ GRRGL GT+L+HKVAGA A G+ L ++ ++ +GT+GV+L
Sbjct: 141 SAF-TVLKKAGRRGLCGTVLIHKVAGALAEEGMGLEEITKRVSVIAKTMGTLGVSL 195
>UniRef100_UPI0000438A06 UPI0000438A06 UniRef100 entry
Length = 572
Score = 174 bits (442), Expect = 1e-42
Identities = 91/164 (55%), Positives = 115/164 (69%), Gaps = 5/164 (3%)
Query: 17 KVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAG 76
+VV R D+ KVA++SGGGSGHEPAHAGF+GEGML+ A+ G VFASP ++LA
Sbjct: 32 RVVLRSDLQSIK--GKVALLSGGGSGHEPAHAGFIGEGMLSGAVAGSVFASPPPGSVLAA 89
Query: 77 IRAV--TGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGR 134
I + G G LLIVKNYTGDRLNFG A EQA+S+G V+ VIVADDCA P GR
Sbjct: 90 ILTLWKGGASGVLLIVKNYTGDRLNFGIAVEQARSQGVPVDMVIVADDCAFAQP-SKAGR 148
Query: 135 RGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
RGL GT+ +HK+AGA A G +L D+ A A++ +GT+G++L
Sbjct: 149 RGLCGTVFIHKLAGALAEKGCALGDIVARLNEATKRIGTLGISL 192
>UniRef100_Q8XP65 Dihydroxyacetone kinase [Clostridium perfringens]
Length = 582
Score = 170 bits (430), Expect = 3e-41
Identities = 91/174 (52%), Positives = 120/174 (68%), Gaps = 7/174 (4%)
Query: 4 YPSLQYLDGYPNVKVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGD 63
+P L++L+ Y KVV +KD+ DKV +ISGGGSGHEPAHAGF+G+GML A+CGD
Sbjct: 23 HPELEFLNKY---KVVKKKDINE----DKVTLISGGGSGHEPAHAGFIGKGMLDVAVCGD 75
Query: 64 VFASPTVEAILAGIRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDC 123
VFASP+ I I+A G LLI+KNY+GD +NF +AA A +G KV+ + V DD
Sbjct: 76 VFASPSQIQIYQAIKASKSNKGTLLIIKNYSGDMMNFKNAAHLATEDGIKVDYIKVDDDI 135
Query: 124 AIPPPLEMVGRRGLAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVA 177
A+ L VGRRG+AGT+LVHK+AGAAA G SL +V A+ A+ +V ++G A
Sbjct: 136 AVEDSLYTVGRRGVAGTVLVHKIAGAAAERGNSLEEVKRIAKKAASSVKSIGFA 189
>UniRef100_UPI00003CA2C5 UPI00003CA2C5 UniRef100 entry
Length = 329
Score = 169 bits (429), Expect = 4e-41
Identities = 88/162 (54%), Positives = 111/162 (68%), Gaps = 4/162 (2%)
Query: 17 KVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAG 76
+V+ R D PG KV ++SGGGSGHEPAHAG+VG GML+AA+CGDVF SPT + I G
Sbjct: 34 RVIARNDKRPG----KVGLVSGGGSGHEPAHAGYVGRGMLSAAVCGDVFTSPTPDQIYEG 89
Query: 77 IRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRG 136
I+A G LLIVKNYTGD +NF AA+ A ++ KVE ++V DD A+ GRRG
Sbjct: 90 IKAADQGAGVLLIVKNYTGDVMNFEMAADLADADDIKVEQIVVDDDIAVEDSTFTTGRRG 149
Query: 137 LAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGT+LVHK+ GAAA AG SL ++ A +V T+GVAL
Sbjct: 150 VAGTVLVHKIIGAAAEAGASLEELKALGEKVIASVKTLGVAL 191
>UniRef100_Q927E6 Lin2843 protein [Listeria innocua]
Length = 329
Score = 169 bits (429), Expect = 4e-41
Identities = 88/162 (54%), Positives = 111/162 (68%), Gaps = 4/162 (2%)
Query: 17 KVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAG 76
+V+ R D PG KV ++SGGGSGHEPAHAG+VG GML+AA+CGDVF SPT + I G
Sbjct: 34 RVIARNDKRPG----KVGLVSGGGSGHEPAHAGYVGRGMLSAAVCGDVFTSPTPDQIYEG 89
Query: 77 IRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRG 136
I+A G LLIVKNYTGD +NF AA+ A ++ KVE ++V DD A+ GRRG
Sbjct: 90 IKAADQGAGVLLIVKNYTGDVMNFEMAADLADADDIKVEQIVVDDDIAVEDSTFTTGRRG 149
Query: 137 LAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGT+LVHK+ GAAA AG SL ++ A +V T+GVAL
Sbjct: 150 VAGTVLVHKIIGAAAEAGASLEELKALGEKVIASVKTLGVAL 191
>UniRef100_Q71W77 Dihydroxyacetone kinase, Dak1 subunit, putative [Listeria
monocytogenes]
Length = 329
Score = 169 bits (428), Expect = 5e-41
Identities = 88/162 (54%), Positives = 111/162 (68%), Gaps = 4/162 (2%)
Query: 17 KVVFRKDVYPGPPYDKVAIISGGGSGHEPAHAGFVGEGMLTAAICGDVFASPTVEAILAG 76
+V+ R D PG KV ++SGGGSGHEPAHAG+VG GML+AA+CGDVF SPT + I G
Sbjct: 34 RVIARNDKRPG----KVGLVSGGGSGHEPAHAGYVGRGMLSAAVCGDVFTSPTPDQIYEG 89
Query: 77 IRAVTGPMGCLLIVKNYTGDRLNFGSAAEQAKSEGYKVESVIVADDCAIPPPLEMVGRRG 136
I+A G LLIVKNYTGD +NF AA+ A ++ KVE ++V DD A+ GRRG
Sbjct: 90 IKAADQGAGVLLIVKNYTGDVMNFEMAADLADADDIKVEQIVVDDDIAVEDSTFTTGRRG 149
Query: 137 LAGTILVHKVAGAAAAAGLSLADVAAEARYASENVGTMGVAL 178
+AGT+LVHK+ GAAA AG SL ++ A +V T+GVAL
Sbjct: 150 VAGTVLVHKIIGAAAEAGASLDELKALGEKVIASVKTLGVAL 191
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.137 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 305,788,963
Number of Sequences: 2790947
Number of extensions: 12096481
Number of successful extensions: 34627
Number of sequences better than 10.0: 266
Number of HSP's better than 10.0 without gapping: 239
Number of HSP's successfully gapped in prelim test: 27
Number of HSP's that attempted gapping in prelim test: 34269
Number of HSP's gapped (non-prelim): 268
length of query: 216
length of database: 848,049,833
effective HSP length: 122
effective length of query: 94
effective length of database: 507,554,299
effective search space: 47710104106
effective search space used: 47710104106
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC139343.3


