

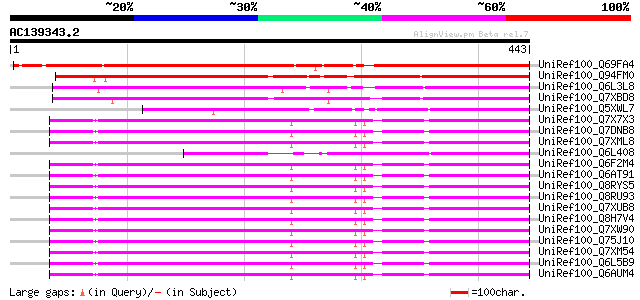
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139343.2 + phase: 0 /pseudo
(443 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q69FA4 Gag-pol polyprotein [Phaseolus vulgaris] 534 e-150
UniRef100_Q94FM0 Pol polyprotein [Citrus paradisi] 278 2e-73
UniRef100_Q6L3L8 Putative polyprotein [Solanum demissum] 240 7e-62
UniRef100_Q7XBD8 Putative retrotransposon RIRE1 poly protein [Ze... 228 2e-58
UniRef100_Q5XWL7 Pol polyprotein-like [Solanum tuberosum] 217 6e-55
UniRef100_Q7X7X3 OSJNBb0112E13.7 protein [Oryza sativa] 205 2e-51
UniRef100_Q7DNB8 OSJNBb0046K02.5 protein [Oryza sativa] 205 2e-51
UniRef100_Q7XML8 OSJNBa0040D17.9 protein [Oryza sativa] 204 4e-51
UniRef100_Q6L408 Putative polyprotein [Solanum demissum] 204 4e-51
UniRef100_Q6F2M4 Putative polyprotein [Oryza sativa] 204 5e-51
UniRef100_Q6AT91 Putative polyprotein [Oryza sativa] 204 5e-51
UniRef100_Q8RYS5 Putative polyprotein [Oryza sativa] 204 5e-51
UniRef100_Q8RU93 Putative polyprotein from transposon TNT 1-94 [... 204 5e-51
UniRef100_Q7XUB8 OSJNBb0078D11.12 protein [Oryza sativa] 204 5e-51
UniRef100_Q8H7V4 Putative polyprotein from transposon TNT [Oryza... 204 5e-51
UniRef100_Q7XW90 OSJNBb0043H09.7 protein [Oryza sativa] 204 5e-51
UniRef100_Q75J10 Putative gag and pol protein [Oryza sativa] 204 5e-51
UniRef100_Q7XM54 OSJNBb0018J12.5 protein [Oryza sativa] 203 7e-51
UniRef100_Q6L5B9 Putative polyprotein [Oryza sativa] 203 7e-51
UniRef100_Q6AUM4 Putative polyprotein [Oryza sativa] 203 7e-51
>UniRef100_Q69FA4 Gag-pol polyprotein [Phaseolus vulgaris]
Length = 1290
Score = 534 bits (1376), Expect = e-150
Identities = 278/449 (61%), Positives = 339/449 (74%), Gaps = 25/449 (5%)
Query: 4 ENSDNDKNKNVTDSTFSVATNPPAVVTYAKQFPDVSRIEVFKGQNFRRWNERVFTLLDVH 63
EN +N N N T +V + + +AK FPDVS+IEVF GQNFRRW ERV TLLD++
Sbjct: 3 ENPNN--NDNTAPETSNVVSATQTI--FAKLFPDVSKIEVFTGQNFRRWQERVSTLLDMY 58
Query: 64 GVASVLTDVKPDEAKSDAKQIEKWNNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNM 123
GVA LT KPD + AKQ++ W +ANKVCRHT+LS +S+ LFDVY SYK AKDIWD++
Sbjct: 59 GVAHALTTAKPDSTTA-AKQVDDWIHANKVCRHTLLSVLSNDLFDVYASYKNAKDIWDSL 117
Query: 124 NVKYTAEEATKQKFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAAL 183
+KYTAE+ +Q+FV+ Y W+M + K+IK+QINEYH+LIE++K E+I LPD FV+ L
Sbjct: 118 ILKYTAEDIVRQRFVIAKYYRWEMIKGKDIKIQINEYHKLIEDIKTESIKLPDEFVSELL 177
Query: 184 VEKLRSSWDDYKQQLKHKHKQMSLNDLIRHIIIEDTSRKECGAARAKALESRANLIQNNA 243
+EKL SW DYKQQLKH+ KQMSL+DLI HIIIEDT+RKEC AA+AKAL ++AN+I++
Sbjct: 178 IEKLPQSWTDYKQQLKHRQKQMSLSDLITHIIIEDTNRKECAAAKAKALSAKANVIEDKP 237
Query: 244 RKQQRYENKSDHALKV---------TNPNFKANLGECYVCGKKGHKAYHCRYRKVNGQPP 294
+RYE K DH K TNP FK G C+VCGK GH A CR+R N P
Sbjct: 238 -APKRYEKKFDHKKKPNNKFSRPSGTNPTFKKK-GNCFVCGKPGHHAPQCRHRAKNDYP- 294
Query: 295 KPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSYISVG 354
PKANL +G +D I AV+S+V+LV +V KWVVDSGATRHICA R FTSY SVG
Sbjct: 295 -PKANLAEG-------EDTIVAVVSQVNLVTNVSKWVVDSGATRHICANRNVFTSYTSVG 346
Query: 355 DDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKV 414
D E+QVYLGDSRTT V GKGKV+LKLTSGKTLAL+DVL+VP+IR NL+SVALL+KVGVKV
Sbjct: 347 DGEEQVYLGDSRTTPVLGKGKVLLKLTSGKTLALNDVLHVPSIRVNLVSVALLSKVGVKV 406
Query: 415 SFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
SFESDK+V++KNNVFVG GYCDQGLFVL+
Sbjct: 407 SFESDKIVITKNNVFVGKGYCDQGLFVLN 435
>UniRef100_Q94FM0 Pol polyprotein [Citrus paradisi]
Length = 701
Score = 278 bits (711), Expect = 2e-73
Identities = 149/410 (36%), Positives = 255/410 (61%), Gaps = 19/410 (4%)
Query: 40 RIEVFKGQNFRRWNERVFTLLDVHGVASVLTD--VKPDEAKSD---AKQIEKWNNANKVC 94
R E F GQNF+RW +++F L +A LT KP E ++D A I+ W++++ +C
Sbjct: 41 RPEKFNGQNFKRWQQKMFFYLTTLNLARFLTKDAPKPKEGETDIQVASAIDAWHHSDFLC 100
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
++ +++ +SD+L++VY K AK++W++++ KY E+A +KFVVG +L ++M + K +
Sbjct: 101 KNYVMNGLSDSLYNVYIGKKTAKELWESLDRKYKTEDAGTKKFVVGRFLDFKMVDSKTVI 160
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q+ E ++ E+ AE + L + F AA++EKL +W D+K LKHK K+M++ L+ +
Sbjct: 161 SQVQELQIILSEILAEGMHLSETFQVAAIIEKLPPAWKDFKSYLKHKQKEMNIEGLVVKL 220
Query: 215 IIEDTSRKECGAARAKALE-SRANLIQNNARKQQRYENKSDHALKVTNPNFKANLGECYV 273
IE+ +R A R A+ ++AN +++ +K+++ + F+ G+CY
Sbjct: 221 RIEEDNR---NAERIGAISMAKANFVEHGPKKKKKKLGPKG-GISKKQTKFQ---GKCYN 273
Query: 274 CGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQKWVVD 333
C K GHKA CR K K +AN+ + + D ++ A V+ + ++ ++W +D
Sbjct: 274 CDKMGHKASDCRLPK-----KKREANVVENITQHVSDINLSAVVLEVNMVGSNPREWWID 328
Query: 334 SGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALSDVLN 393
+GATRH+C+ + FTS+ + +++++G+S T+ + G+GKV LK+TSGK L L++VL
Sbjct: 329 TGATRHVCSDKGLFTSF-EAASNREKLFMGNSATSEIEGQGKVALKMTSGKELTLNNVLY 387
Query: 394 VPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
VP IR NL+S +LLNK G ++ FESD +V+SKN ++VG GY + GLF L+
Sbjct: 388 VPEIRKNLVSGSLLNKHGFRMVFESDMVVLSKNGMYVGKGYVNDGLFKLN 437
>UniRef100_Q6L3L8 Putative polyprotein [Solanum demissum]
Length = 1279
Score = 240 bits (612), Expect = 7e-62
Identities = 145/442 (32%), Positives = 231/442 (51%), Gaps = 51/442 (11%)
Query: 37 DVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKP----------DEAKSDAKQIEK 86
D+++ F G +F+RW +V L + V+ VLT+ P DE S + +EK
Sbjct: 14 DLNKPFRFNGNHFKRWKGKVLFYLSLLNVSYVLTEKNPNKVDDSSMEDDELISHHENVEK 73
Query: 87 WNNANKVCRHTILSTISDALFDVYC-SYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*W 145
+N + CR+ +L+ +SD +D Y +Y AK IW + KY EEA +K+ + +
Sbjct: 74 YNGDSYKCRYYLLNCLSDNFYDYYDRTYSSAKKIWKALQSKYDTEEAGAKKYAASRFFRF 133
Query: 146 QMKEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQM 205
QM ++K + Q ++ ++ EL++E + + D + +V+KL SW ++++ ++HK K+
Sbjct: 134 QMVDNKSVVDQAQDFIMIVGELRSEEVKIGDNLIVCGIVDKLPPSWKEFQKTMRHKQKET 193
Query: 206 SLNDLIRHIIIEDTSRKECGAARAKA--LESRANLIQNNARKQQRYENKSDHALKVTNPN 263
SL LI I +E+ +R + + + + ++ NLI +N + ++N S LK
Sbjct: 194 SLETLIMKIRMEEEARGQDALLQTEENNITTKVNLITSNNATPESHKNTS---LKPKKKK 250
Query: 264 FKANLGE----------------------CYVCGKKGHKAYHCRYRKVNGQPPKPKANLT 301
FK N G C+VCGK GH A CR+RK + P P+AN+T
Sbjct: 251 FKKNNGRPPKKNNGENIQAQNQQVQDKGPCFVCGKSGHIARFCRFRK---RGPNPQANVT 307
Query: 302 QGDDKKYDDDDVIAAVISKVHLVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVY 361
+ + AVI+ +++V +V W DSGA RH+C ++ F Y ++ +
Sbjct: 308 E---------EPFVAVITDINMVENVDGWWADSGANRHVCYDKDWFKKYTHF-EEPKTIM 357
Query: 362 LGDSRTTAVNGKGKVILKLTSGKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKM 421
LGDS TT V G G V L TSG+ L L DVL P++R L+S LLNK G K ES++
Sbjct: 358 LGDSHTTQVLGTGDVELCFTSGRVLTLKDVLYTPSMRKKLMSSFLLNKAGFKQIIESNQY 417
Query: 422 VMSKNNVFVGNGYCDQGLFVLS 443
V+ K +FVG GY G+F L+
Sbjct: 418 VIVKKGIFVGKGYACDGMFKLN 439
>UniRef100_Q7XBD8 Putative retrotransposon RIRE1 poly protein [Zea mays]
Length = 1309
Score = 228 bits (582), Expect = 2e-58
Identities = 140/421 (33%), Positives = 218/421 (51%), Gaps = 29/421 (6%)
Query: 37 DVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEK---------W 87
DV + E F G +F+RW + L + V+T P A D+ K W
Sbjct: 23 DVIKPEAFDGASFKRWQIKTRMWLTDLKLFWVVTSAVPQAASDDSDDAAKAAALAEKAKW 82
Query: 88 NNANKVCRHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQM 147
+ AN+ C +L+ +S+ LFDVY ++ AK +W ++ +++ + + F NYL ++M
Sbjct: 83 DEANEACLSRLLNVLSNRLFDVYSAFTSAKGLWTDLENEFSEVDNGNESFTTENYLNYKM 142
Query: 148 KEDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSL 207
E + + Q+ E L+ +L +LPD F A++ KL SW D+ +H KQM+L
Sbjct: 143 VEGRSVMEQLQEIQLLVRDLVQYGCVLPDSFQVNAILAKLPPSWRDFVTSRRHMKKQMTL 202
Query: 208 NDLIRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKAN 267
+L I +E+ +R ++ + + +A++++ ++ + + K+ + P K
Sbjct: 203 TELSAAINVEERAR----SSNKPSQQLQAHVVEKGGDRKFQKKKKNSPQKNLNQPKSKKM 258
Query: 268 LGE----CYVCGKKGHKAYHCRYRKVNGQPPKPK-ANLTQGDDKKYDDDDVIAAVISKVH 322
+ CYVCG GH A C+ RK G PP+ K N+ Y +
Sbjct: 259 KKKEDFICYVCGVSGHTARRCKLRKGKGPPPQRKEGNVVVNSTPGY---------ALQAF 309
Query: 323 LVNDVQKWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTS 382
+ + W +DSGAT HICA R F+S+ G + V +G+ AV G G+V LKLTS
Sbjct: 310 MASPSDDWWMDSGATVHICADRSMFSSF--QGFNSAPVLMGNGVPAAVRGTGQVYLKLTS 367
Query: 383 GKTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVL 442
GKTL L DVL VP++ NLISV+LL + G+K+ FES+K+V+SK FVG Y GLF L
Sbjct: 368 GKTLVLKDVLYVPSMSRNLISVSLLCRQGLKLVFESNKVVLSKFGTFVGKSYESGGLFRL 427
Query: 443 S 443
S
Sbjct: 428 S 428
>UniRef100_Q5XWL7 Pol polyprotein-like [Solanum tuberosum]
Length = 557
Score = 217 bits (552), Expect = 6e-55
Identities = 123/354 (34%), Positives = 202/354 (56%), Gaps = 34/354 (9%)
Query: 114 KVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIKVQINEYHQLIEELKAENI- 172
K +K++WD + KY E+A +KF+V +L ++M + K I Q+ E +I +L AE I
Sbjct: 2 KTSKELWDALEKKYKTEDAGMKKFIVAKFLDYKMIDSKTIVTQVQELQVIIHDLLAEGIN 61
Query: 173 -----------------------ILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLND 209
++ D F AA++EKL W D+K LKHK K+M++ D
Sbjct: 62 LFNTYVGNIKFILNTHIIFNVGLVVNDAFQMAAIIEKLPPLWKDFKNYLKHKRKEMTVED 121
Query: 210 LIRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKANLG 269
L+ + IE+ ++ +R + S N+++ + K ++ + S +NP K G
Sbjct: 122 LMVRLRIEEDNKAAEKRSRGNSAISGVNIVEEDPTKSKKRKKTSGPK---SNPPKKKFNG 178
Query: 270 ECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQK 329
C+ CGK+GHK CR K + + K +ANL + K + DD + A++S+ +LV + ++
Sbjct: 179 SCFNCGKRGHKTTKCRGPKKDKK--KDQANLAES---KGEMDD-LCAMLSECNLVGNPRE 232
Query: 330 WVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLALS 389
W +DSGA+ H+CA +E F++Y +E ++++ +S V G GK++LK+TSGK + L+
Sbjct: 233 WWIDSGASCHVCANKELFSTYTPAPTNE-KLFMANSAVAKVEGTGKILLKMTSGKVVTLN 291
Query: 390 DVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
V VP +R NL+S+ +L K G K F SDK+V+SKN ++VG GY +GLF L+
Sbjct: 292 RVSYVPELRKNLVSIPVLTKNGFKCVFVSDKVVVSKNEMYVGKGYLTEGLFKLN 345
>UniRef100_Q7X7X3 OSJNBb0112E13.7 protein [Oryza sativa]
Length = 1342
Score = 205 bits (521), Expect = 2e-51
Identities = 138/420 (32%), Positives = 220/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFRVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A+++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKSDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGLGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q7DNB8 OSJNBb0046K02.5 protein [Oryza sativa]
Length = 1356
Score = 205 bits (521), Expect = 2e-51
Identities = 138/420 (32%), Positives = 220/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFRVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A+++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKSDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGLGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q7XML8 OSJNBa0040D17.9 protein [Oryza sativa]
Length = 1319
Score = 204 bits (519), Expect = 4e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTMWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPSVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q6L408 Putative polyprotein [Solanum demissum]
Length = 1415
Score = 204 bits (519), Expect = 4e-51
Identities = 115/295 (38%), Positives = 170/295 (56%), Gaps = 36/295 (12%)
Query: 149 EDKEIKVQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLN 208
+ K + Q+ E + +L AE++++ + F AA++EKL SW+D+K LKHK K+M L
Sbjct: 266 DSKTVGSQVQELQLIFHDLIAEDMVVNEAFQVAAMIEKLPPSWNDFKNYLKHKRKEMKLE 325
Query: 209 DLIRHIIIEDTSRKECGAARAKALESRANLIQNNARKQQRYENKSDHALKVTNPNFKANL 268
DL+ + IE+ ++ A K+ R E FK N
Sbjct: 326 DLVIRLKIEEDNK--------------------TAEKKSRQEK------------FKGN- 352
Query: 269 GECYVCGKKGHKAYHCRYRKVNGQPPKPKANLTQGDDKKYDDDDVIAAVISKVHLVNDVQ 328
CY CGK GHK+ CR + + K K ++ +D D + +IS+ +LV + +
Sbjct: 353 --CYNCGKAGHKSSDCRAPRKDKDKDKGKGKSQANIVEEMEDADDLCGMISECNLVGNPK 410
Query: 329 KWVVDSGATRHICAKREAFTSYISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSGKTLAL 388
+W +DSGATRHIC+ +E F +Y V DED +++G++ TT + G GKV+LK+TSGK L L
Sbjct: 411 EWFLDSGATRHICSAKEVFATYTHVEVDED-LFMGNTTTTRIAGTGKVMLKMTSGKVLTL 469
Query: 389 SDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
++VL+VPTIR NL+ VALL K G K SDK V+SKN +F+G GY +GLF L+
Sbjct: 470 NNVLHVPTIRKNLVYVALLVKNGFKCVLVSDKTVISKNEMFLGKGYLTEGLFKLN 524
>UniRef100_Q6F2M4 Putative polyprotein [Oryza sativa]
Length = 1320
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q6AT91 Putative polyprotein [Oryza sativa]
Length = 1320
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q8RYS5 Putative polyprotein [Oryza sativa]
Length = 1320
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q8RU93 Putative polyprotein from transposon TNT 1-94 [Oryza sativa]
Length = 1302
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q7XUB8 OSJNBb0078D11.12 protein [Oryza sativa]
Length = 1320
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q8H7V4 Putative polyprotein from transposon TNT [Oryza sativa]
Length = 1911
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q7XW90 OSJNBb0043H09.7 protein [Oryza sativa]
Length = 1302
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q75J10 Putative gag and pol protein [Oryza sativa]
Length = 1302
Score = 204 bits (518), Expect = 5e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q7XM54 OSJNBb0018J12.5 protein [Oryza sativa]
Length = 1320
Score = 203 bits (517), Expect = 7e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQIGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q6L5B9 Putative polyprotein [Oryza sativa]
Length = 1302
Score = 203 bits (517), Expect = 7e-51
Identities = 138/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 4 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 61
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY A ++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 62 VGCILSVLGDRLVEVYMHMTDANELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 121
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SSW + LKHK ++ S+ LI +
Sbjct: 122 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPSSWRSFGTALKHKRQEYSVEGLIASL 181
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 182 DVEEKAREKDAASKDDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 241
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 242 CFVCGQVGHLARKCSQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 294
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W VD+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 295 NQSTNWWVDTGANVHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 351
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 352 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 411
>UniRef100_Q6AUM4 Putative polyprotein [Oryza sativa]
Length = 1861
Score = 203 bits (517), Expect = 7e-51
Identities = 137/420 (32%), Positives = 219/420 (51%), Gaps = 23/420 (5%)
Query: 35 FPDVSRIEVFKGQNFRRWNERVFTLLDVHGVASVLTDVKPDEAKSDAKQIEKWNNANKVC 94
F D R + F G +F+RW RV L V T KP E A+Q +++ A +
Sbjct: 418 FADALRPDKFTGVHFKRWQIRVTLWLTAMKCFWVSTG-KP-EGVLTAEQQKQFEEATTLF 475
Query: 95 RHTILSTISDALFDVYCSYKVAKDIWDNMNVKYTAEEATKQKFVVGNYL*WQMKEDKEIK 154
ILS + D L +VY AK++WD +N K+ A +A+ +++ + ++M +++ +
Sbjct: 476 VGCILSVLGDRLVEVYMHMTDAKELWDALNTKFGATDASNDLYIMEQFHDYKMADNRSVV 535
Query: 155 VQINEYHQLIEELKAENIILPDVFVAAALVEKLRSSWDDYKQQLKHKHKQMSLNDLIRHI 214
Q +E + +EL+ +LPD FVA ++ KL SW + LKHK ++ S+ LI +
Sbjct: 536 EQAHEIQTMAKELELLKCVLPDKFVAGCIIAKLPPSWRSFGTALKHKRQEYSVEGLIASL 595
Query: 215 IIEDTSRKECGAARAKALESRANLI---QNNAR-KQQRYENKSDHALKVTNPNFKANLGE 270
+E+ +R++ A++ +S AN++ QN ++ K + + + K N N +
Sbjct: 596 DVEEKAREKDAASKGDGGQSSANVVHKAQNKSKGKYKAQQTTNFKKQKKNNNNPNQDERT 655
Query: 271 CYVCGKKGHKAYHCRYRKVNGQP---PKPKANLT---QGDDKKYDDDDVIAAVISKVHLV 324
C+VCG+ GH A C RK P AN+T GD Y + + V V
Sbjct: 656 CFVCGQVGHLARKCPQRKGMKAPAGQTSKSANVTIGNTGDGSGYGN-------LPTVFSV 708
Query: 325 NDVQKWVVDSGATRHICAKREAFTSY-ISVGDDEDQVYLGDSRTTAVNGKGKVILKLTSG 383
N W +D+GA H+CA F+SY ++ G V +G+ +V+G G V LK TSG
Sbjct: 709 NQSTNWWIDTGANIHVCADISLFSSYQVARG---STVLMGNGSHASVHGVGTVDLKFTSG 765
Query: 384 KTLALSDVLNVPTIRTNLISVALLNKVGVKVSFESDKMVMSKNNVFVGNGYCDQGLFVLS 443
K + L +V +VP+I NL+S + L + G K+ FES+K+V+SK+ F+G GY GLF S
Sbjct: 766 KIVQLKNVQHVPSIDRNLVSGSRLTRDGFKLVFESNKVVVSKHGYFIGKGYECGGLFRFS 825
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.384
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 736,648,109
Number of Sequences: 2790947
Number of extensions: 30573806
Number of successful extensions: 95939
Number of sequences better than 10.0: 619
Number of HSP's better than 10.0 without gapping: 303
Number of HSP's successfully gapped in prelim test: 317
Number of HSP's that attempted gapping in prelim test: 94565
Number of HSP's gapped (non-prelim): 875
length of query: 443
length of database: 848,049,833
effective HSP length: 130
effective length of query: 313
effective length of database: 485,226,723
effective search space: 151875964299
effective search space used: 151875964299
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC139343.2


