

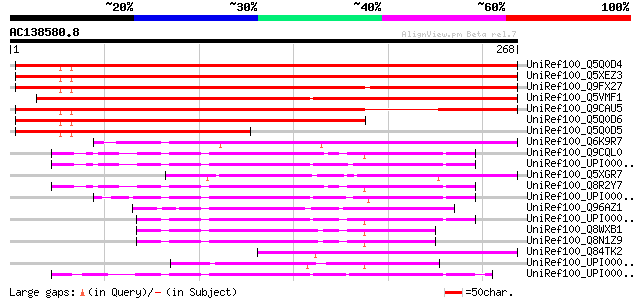
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138580.8 - phase: 0
(268 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q5Q0D4 Hypothetical protein [Arabidopsis thaliana] 360 3e-98
UniRef100_Q5XEZ3 At1g73320 [Arabidopsis thaliana] 360 3e-98
UniRef100_Q9FX27 Tumor-related protein, putative [Arabidopsis th... 352 6e-96
UniRef100_Q5VMF1 Tumor-related protein-like [Oryza sativa] 306 5e-82
UniRef100_Q9CAU5 Hypothetical protein T18K17.1 [Arabidopsis thal... 296 5e-79
UniRef100_Q5Q0D6 Hypothetical protein [Arabidopsis thaliana] 247 2e-64
UniRef100_Q5Q0D5 Hypothetical protein [Arabidopsis thaliana] 162 1e-38
UniRef100_Q6K9R7 DNA ligase-like [Oryza sativa] 137 4e-31
UniRef100_Q9CQL0 Mus musculus 10 days embryo whole body cDNA, RI... 106 7e-22
UniRef100_UPI0000180E91 UPI0000180E91 UniRef100 entry 105 1e-21
UniRef100_Q5XGR7 Hypothetical protein [Xenopus laevis] 105 1e-21
UniRef100_Q8R2Y7 RIKEN cDNA 2310038H17 [Mus musculus] 103 5e-21
UniRef100_UPI000024A8E7 UPI000024A8E7 UniRef100 entry 99 1e-19
UniRef100_Q96AZ1 Hepatocellularcarcinoma-associated antigen HCA5... 97 3e-19
UniRef100_UPI00003697CC UPI00003697CC UniRef100 entry 97 6e-19
UniRef100_Q8WXB1 Hepatocellular carcinoma-associated antigen HCA... 96 1e-18
UniRef100_Q8N1Z9 Hypothetical protein FLJ36493 [Homo sapiens] 94 5e-18
UniRef100_Q84TK2 Hypothetical protein At1g08125 [Arabidopsis tha... 91 4e-17
UniRef100_UPI000035FA3E UPI000035FA3E UniRef100 entry 89 2e-16
UniRef100_UPI00003AE5D2 UPI00003AE5D2 UniRef100 entry 89 2e-16
>UniRef100_Q5Q0D4 Hypothetical protein [Arabidopsis thaliana]
Length = 316
Score = 360 bits (923), Expect = 3e-98
Identities = 182/275 (66%), Positives = 213/275 (77%), Gaps = 10/275 (3%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 27 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 86
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 87 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 146
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTW 173
KKIVELGSGCGLVGCIAALLGG +LTDLPDR+RLL+KNI+TN+ + RGS EL W
Sbjct: 147 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 206
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYF 233
GDDPD +LI+P PDY+LGSDV+YSE AV L++TL QL TTIFL+GELRNDA+LEYF
Sbjct: 207 GDDPDPDLIEPFPDYVLGSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRNDAVLEYF 266
Query: 234 LEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
LE A+ DF IGRV+QT WHPDY S+RVVLYVL KK
Sbjct: 267 LETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 301
>UniRef100_Q5XEZ3 At1g73320 [Arabidopsis thaliana]
Length = 292
Score = 360 bits (923), Expect = 3e-98
Identities = 182/275 (66%), Positives = 213/275 (77%), Gaps = 10/275 (3%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 3 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 62
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 63 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 122
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTW 173
KKIVELGSGCGLVGCIAALLGG +LTDLPDR+RLL+KNI+TN+ + RGS EL W
Sbjct: 123 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 182
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYF 233
GDDPD +LI+P PDY+LGSDV+YSE AV L++TL QL TTIFL+GELRNDA+LEYF
Sbjct: 183 GDDPDPDLIEPFPDYVLGSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRNDAVLEYF 242
Query: 234 LEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
LE A+ DF IGRV+QT WHPDY S+RVVLYVL KK
Sbjct: 243 LETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 277
>UniRef100_Q9FX27 Tumor-related protein, putative [Arabidopsis thaliana]
Length = 314
Score = 352 bits (903), Expect = 6e-96
Identities = 181/275 (65%), Positives = 211/275 (75%), Gaps = 12/275 (4%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 27 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 86
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 87 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 146
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTW 173
KKIVELGSGCGLVGCIAALLGG +LTDLPDR+RLL+KNI+TN+ + RGS EL W
Sbjct: 147 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 206
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYF 233
GDDPD +LI+P PDY GSDV+YSE AV L++TL QL TTIFL+GELRNDA+LEYF
Sbjct: 207 GDDPDPDLIEPFPDY--GSDVIYSEEAVHHLVKTLLQLCSDQTTIFLSGELRNDAVLEYF 264
Query: 234 LEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
LE A+ DF IGRV+QT WHPDY S+RVVLYVL KK
Sbjct: 265 LETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 299
>UniRef100_Q5VMF1 Tumor-related protein-like [Oryza sativa]
Length = 271
Score = 306 bits (783), Expect = 5e-82
Identities = 149/254 (58%), Positives = 188/254 (73%), Gaps = 1/254 (0%)
Query: 15 LGSYGGSVMLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQLRLDSCGHSLSILQ 74
+G+Y G V V G+ AA ETMLLW + QP + NAFV + L LD+CG LS+LQ
Sbjct: 10 MGAYSGPVRPVGDGDGGAAGETMLLWALGQPAAQRHNAFVRHGAHSLTLDACGRRLSLLQ 69
Query: 75 SPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLG 134
SPSS+ PGVTG+V+WDSG+VL KFLEH+VDSG+L L+ + +ELG+GCGL GC+AALLG
Sbjct: 70 SPSSMSTPGVTGAVVWDSGVVLAKFLEHAVDSGLLTLRAARALELGAGCGLAGCVAALLG 129
Query: 135 GEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTPDYILGSDV 194
V+LTDLPDR++LLRKNI+ N+ RGS +L W DDP +L++P DY+LGSDV
Sbjct: 130 AHVLLTDLPDRLKLLRKNIDLNVGD-DARGSARVAQLVWADDPHPDLLNPPLDYVLGSDV 188
Query: 195 VYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQTLWHPD 254
+YSE AV DLL TL LS P+TTI LA ELRNDA+LE FLEAAM DF +G ++Q WHPD
Sbjct: 189 IYSEEAVDDLLLTLKHLSAPHTTIILAAELRNDAVLECFLEAAMADFQVGCIEQQQWHPD 248
Query: 255 YHSNRVVLYVLVKK 268
+ S RV L++L+KK
Sbjct: 249 FRSTRVALFILLKK 262
>UniRef100_Q9CAU5 Hypothetical protein T18K17.1 [Arabidopsis thaliana]
Length = 273
Score = 296 bits (757), Expect = 5e-79
Identities = 157/275 (57%), Positives = 183/275 (66%), Gaps = 48/275 (17%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 22 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 81
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 82 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 141
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTW 173
KKIVELGSGCGLVGCIAALLGG +LTDLPDR+RLL+KNI+TN+ + RGS EL W
Sbjct: 142 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 201
Query: 174 GDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYF 233
GDDPD +LI+P PDY DA+LEYF
Sbjct: 202 GDDPDPDLIEPFPDY--------------------------------------DAVLEYF 223
Query: 234 LEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
LE A+ DF IGRV+QT WHPDY S+RVVLYVL KK
Sbjct: 224 LETALKDFAIGRVEQTQWHPDYRSHRVVLYVLEKK 258
>UniRef100_Q5Q0D6 Hypothetical protein [Arabidopsis thaliana]
Length = 247
Score = 247 bits (631), Expect = 2e-64
Identities = 125/195 (64%), Positives = 147/195 (75%), Gaps = 10/195 (5%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 27 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 86
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 87 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 146
Query: 114 KKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTW 173
KKIVELGSGCGLVGCIAALLGG +LTDLPDR+RLL+KNI+TN+ + RGS EL W
Sbjct: 147 KKIVELGSGCGLVGCIAALLGGNAVLTDLPDRLRLLKKNIQTNLHRGNTRGSAIVQELVW 206
Query: 174 GDDPDQELIDPTPDY 188
GDDPD +LI+P PDY
Sbjct: 207 GDDPDPDLIEPFPDY 221
>UniRef100_Q5Q0D5 Hypothetical protein [Arabidopsis thaliana]
Length = 170
Score = 162 bits (409), Expect = 1e-38
Identities = 85/134 (63%), Positives = 99/134 (73%), Gaps = 10/134 (7%)
Query: 4 EEENATAAMVKLGSYGGSVMLV--VPGEES--------AAEETMLLWGIQQPTLSKPNAF 53
+E+ A + ++ YGG V++V P ES AA E M++W IQ PT PN
Sbjct: 27 KEDEAVEEIRRMSGYGGDVIVVGGFPASESESESESDLAAAEIMVIWAIQGPTSFAPNTL 86
Query: 54 VAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQG 113
VAQSSL+LRLD+CGHSLSILQSP SL PGVTGSVMWDSG+VLGKFLEHSVDS +L L+G
Sbjct: 87 VAQSSLELRLDACGHSLSILQSPCSLNTPGVTGSVMWDSGVVLGKFLEHSVDSKVLSLEG 146
Query: 114 KKIVELGSGCGLVG 127
KKIVELGSGCGLVG
Sbjct: 147 KKIVELGSGCGLVG 160
>UniRef100_Q6K9R7 DNA ligase-like [Oryza sativa]
Length = 489
Score = 137 bits (344), Expect = 4e-31
Identities = 84/234 (35%), Positives = 126/234 (52%), Gaps = 20/234 (8%)
Query: 45 PTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSV 104
P L+ P S+ + L+ GH L I Q P+S G+ +WD+ +V KFLE +
Sbjct: 19 PKLNSP------STSAISLELLGHRLHISQDPNSKH----LGTTVWDASMVFVKFLEKNS 68
Query: 105 DSGMLV---LQGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHIS 161
G L+GK+++ELG+GCGL G ALLG +V+ TD + + LL +N+E N IS
Sbjct: 69 RKGRFSPSKLKGKRVIELGAGCGLAGLGMALLGCDVVTTDQVEVLPLLLRNVERNKSWIS 128
Query: 162 LR-------GSITATELTWGDDPDQELIDPTPDYILGSDVVYSEGAVVDLLETLGQLSGP 214
GS+T EL WG+ +DP DYI+G+DVVYSE + L+ET+ LSGP
Sbjct: 129 QSNSDSGSIGSVTVAELDWGNKDHIRAVDPPFDYIIGTDVVYSEHLLQPLMETIIALSGP 188
Query: 215 NTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQTLWHPDYHSNRVVLYVLVKK 268
T I L E+R+ + E ++ ++F + V ++ Y + LY++ K
Sbjct: 189 KTKIMLGYEIRSTTVHEQMMQMWKSNFNVKTVSKSKMDVKYQHPSIHLYIMDPK 242
>UniRef100_Q9CQL0 Mus musculus 10 days embryo whole body cDNA, RIKEN full-length
enriched library, clone:2610017G06
product:HEPATOCELLULAR CARCINOMA- ASSOCIATED ANTIGEN
HCA557B homolog [Mus musculus]
Length = 218
Score = 106 bits (264), Expect = 7e-22
Identities = 84/228 (36%), Positives = 118/228 (50%), Gaps = 36/228 (15%)
Query: 23 MLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKP 82
M +VP EESAA G+Q+ KP A + ++ H++ I Q LG
Sbjct: 1 MALVPYEESAAI------GLQK--FHKPLATFSFAN---------HTIQIRQDWRQLG-- 41
Query: 83 GVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDL 142
+V+WD+ +VL +LE G + L+G VELG+G GLVG +AALLG +V +TD
Sbjct: 42 --VAAVVWDAAVVLSMYLE----MGAVELRGCSAVELGAGTGLVGIVAALLGAQVTITDR 95
Query: 143 PDRMRLLRKNIETNM-KHISLRGSITATELTWGDDPDQELIDPTP---DYILGSDVVYSE 198
+ L+ N+E N+ HI + + ELTWG Q L +P D ILG+DV+Y E
Sbjct: 96 KVALEFLKSNVEANLPPHIQPKAVV--KELTWG----QNLESFSPGEFDLILGADVIYLE 149
Query: 199 GAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 246
DLL+TLG L N+ I LA +R + FL FT+ +V
Sbjct: 150 DTFTDLLQTLGHLCSNNSVILLACRIRYERD-SNFLTMLERQFTVSKV 196
>UniRef100_UPI0000180E91 UPI0000180E91 UniRef100 entry
Length = 218
Score = 105 bits (262), Expect = 1e-21
Identities = 80/225 (35%), Positives = 115/225 (50%), Gaps = 30/225 (13%)
Query: 23 MLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKP 82
M +VP EES + G+Q+ L KP A + ++ H++ I Q LG
Sbjct: 1 MALVPYEESMSI------GLQK--LHKPLATFSFAN---------HTIQIRQDWGRLG-- 41
Query: 83 GVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDL 142
+V+WD+ IVL +LE G + L+G VELG+G GLVG +AALLG V +TD
Sbjct: 42 --VAAVVWDAAIVLSTYLE----MGAVELRGCSAVELGAGTGLVGIVAALLGAHVTITDR 95
Query: 143 PDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTP-DYILGSDVVYSEGAV 201
+ L+ N+E N+ ++ + ELTWG + D P D ILG+D++Y E
Sbjct: 96 KVALEFLKSNVEANLPP-HIQPKVVVKELTWGQNLDS--FSPGEFDLILGADIIYLEDTF 152
Query: 202 VDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 246
DLL+TLG L N+ I LA +R + FL FT+ +V
Sbjct: 153 TDLLQTLGHLCSNNSVILLACRIRYERD-NNFLTMLERQFTVSKV 196
>UniRef100_Q5XGR7 Hypothetical protein [Xenopus laevis]
Length = 229
Score = 105 bits (262), Expect = 1e-21
Identities = 74/193 (38%), Positives = 103/193 (53%), Gaps = 12/193 (6%)
Query: 83 GVTGSVMWDSGIVLGKFLEHS---VDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVIL 139
G G V+WD+ IVL KFLE G L GK ++ELG+G G+VG +AA G V++
Sbjct: 41 GDVGCVVWDAAIVLSKFLESREFMCPEGHR-LSGKCVLELGAGTGIVGIMAATQGANVMV 99
Query: 140 TDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTPDYILGSDVVYSEG 199
TDL D L++ NIE+N I RGS A L WG++ +EL+ P PDYIL +D +Y E
Sbjct: 100 TDLEDLQELMKTNIESNSHFI--RGSCQAKVLKWGEEV-KELV-PKPDYILLADCIYYEE 155
Query: 200 AVVDLLETLGQLSGPNTTIFLAGELR----NDAILEYFLEAAMNDFTIGRVDQTLWHPDY 255
++ LL+TL L+G +T I E R N I + F E F V +Y
Sbjct: 156 SLEPLLKTLRDLTGSDTCILCCYEQRTMGKNPQIEKRFFELLAEHFKYEEVPLEKHDKEY 215
Query: 256 HSNRVVLYVLVKK 268
S + + + +K
Sbjct: 216 RSEDIHILQIYRK 228
>UniRef100_Q8R2Y7 RIKEN cDNA 2310038H17 [Mus musculus]
Length = 218
Score = 103 bits (257), Expect = 5e-21
Identities = 83/228 (36%), Positives = 117/228 (50%), Gaps = 36/228 (15%)
Query: 23 MLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKP 82
M +VP EESAA G+Q+ KP A + ++ H++ I Q LG
Sbjct: 1 MALVPYEESAAI------GLQK--FHKPLATFSFAN---------HTIQIRQDWRQLG-- 41
Query: 83 GVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDL 142
+V+WD+ +VL +LE G + L+G VELG+G GLVG +AAL G +V +TD
Sbjct: 42 --VAAVVWDAAVVLSMYLE----MGAVELRGCSAVELGAGTGLVGIVAALPGAQVTITDR 95
Query: 143 PDRMRLLRKNIETNM-KHISLRGSITATELTWGDDPDQELIDPTP---DYILGSDVVYSE 198
+ L+ N+E N+ HI + + ELTWG Q L +P D ILG+DV+Y E
Sbjct: 96 KVALEFLKSNVEANLPPHIQPKAVV--KELTWG----QNLESFSPGEFDLILGADVIYLE 149
Query: 199 GAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRV 246
DLL+TLG L N+ I LA +R + FL FT+ +V
Sbjct: 150 DTFTDLLQTLGHLCSNNSVILLACRIRYERD-SNFLTMLERQFTVSKV 196
>UniRef100_UPI000024A8E7 UPI000024A8E7 UniRef100 entry
Length = 218
Score = 99.0 bits (245), Expect = 1e-19
Identities = 72/204 (35%), Positives = 108/204 (52%), Gaps = 21/204 (10%)
Query: 45 PTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSV 104
P LSK + QSS + L H + + Q LG +V+WD+ +VL FLE
Sbjct: 12 PALSK----LHQSSAEFTL--ANHRIRLSQDWKRLG----VAAVVWDAAVVLCMFLE--- 58
Query: 105 DSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRG 164
G + L+GK+++ELG+G GLVG +AALLG V +TD + L N+ N+ +
Sbjct: 59 -MGKVDLKGKRVIELGAGTGLVGIVAALLGANVTITDREPALEFLTANVHENIPQ-GRQK 116
Query: 165 SITATELTWGDDPDQELIDPTPDY--ILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAG 222
++ +ELTWG++ D + P Y ILG+D+VY E LL+TL LS +T + L+
Sbjct: 117 AVQVSELTWGENLD---LYPQGGYDLILGADIVYLEETFPALLQTLEHLSSGDTVVLLSC 173
Query: 223 ELRNDAILEYFLEAAMNDFTIGRV 246
+R + E FL F++ V
Sbjct: 174 RIRYERD-ERFLTELRQRFSVQEV 196
>UniRef100_Q96AZ1 Hepatocellularcarcinoma-associated antigen HCA557a, isoform a [Homo
sapiens]
Length = 226
Score = 97.4 bits (241), Expect = 3e-19
Identities = 62/170 (36%), Positives = 94/170 (54%), Gaps = 12/170 (7%)
Query: 66 CGHSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGL 125
CGH L+I Q+ S + GV V WD+ + L + E S + +GKK++ELG+G G+
Sbjct: 36 CGHVLTITQNFGS--RLGVAARV-WDAALSLCNYFE----SQNVDFRGKKVIELGAGTGI 88
Query: 126 VGCIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPT 185
VG +AAL GG+V +TDLP + ++ N++ N + G L+WG D +
Sbjct: 89 VGILAALQGGDVTITDLPLALEQIQGNVQAN---VPAGGQAQVRALSWG--IDHHVFPAN 143
Query: 186 PDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLE 235
D +LG+D+VY E LL TL L P+ TI+LA ++R + E F +
Sbjct: 144 YDLVLGADIVYLEPTFPLLLGTLQHLCRPHGTIYLASKMRKEHGTESFFQ 193
>UniRef100_UPI00003697CC UPI00003697CC UniRef100 entry
Length = 218
Score = 96.7 bits (239), Expect = 6e-19
Identities = 66/182 (36%), Positives = 96/182 (52%), Gaps = 17/182 (9%)
Query: 68 HSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVG 127
H++ I Q LG +V+WD+ IVL +LE G + L+G+ VELG+G GLVG
Sbjct: 29 HTIQIRQDWRHLG----VAAVVWDAAIVLSTYLE----MGAVELRGRSAVELGAGTGLVG 80
Query: 128 CIAALLGGEVILTDLPDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTP- 186
+AALLG V +TD + L+ N++ N+ ++ ELTWG Q L +P
Sbjct: 81 IVAALLGAHVTITDRKVALEFLKSNVQANLPP-HIQPKTVVKELTWG----QNLGSFSPG 135
Query: 187 --DYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIG 244
D ILG+D++Y E DLL+TL L ++ I LA +R + FL FT+
Sbjct: 136 EFDLILGADIIYLEETFTDLLQTLEHLCSNHSVILLACRIRYERD-NNFLAMLERQFTVR 194
Query: 245 RV 246
+V
Sbjct: 195 KV 196
>UniRef100_Q8WXB1 Hepatocellular carcinoma-associated antigen HCA557b [Homo sapiens]
Length = 218
Score = 95.5 bits (236), Expect = 1e-18
Identities = 63/162 (38%), Positives = 89/162 (54%), Gaps = 18/162 (11%)
Query: 68 HSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVG 127
H++ I Q LG +V+WD+ IVL +LE G + L+G+ VELG+G GLVG
Sbjct: 29 HTIQIRQDWRHLG----VAAVVWDAAIVLSTYLE----MGAVELRGRSAVELGAGTGLVG 80
Query: 128 CIAALLGGEVILTDLPDRMRLLRKNIETNM-KHISLRGSITATELTWGDDPDQELIDPTP 186
+AALLG V +TD + L+ N++ N+ HI + ELTWG Q L +P
Sbjct: 81 IVAALLGAHVTITDRKVALEFLKSNVQANLPPHIQTK--TVVKELTWG----QNLGSFSP 134
Query: 187 ---DYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELR 225
D ILG+D++Y E DLL+TL L ++ I LA +R
Sbjct: 135 GEFDLILGADIIYLEETFTDLLQTLEHLCSNHSVILLACRIR 176
>UniRef100_Q8N1Z9 Hypothetical protein FLJ36493 [Homo sapiens]
Length = 218
Score = 93.6 bits (231), Expect = 5e-18
Identities = 62/162 (38%), Positives = 88/162 (54%), Gaps = 18/162 (11%)
Query: 68 HSLSILQSPSSLGKPGVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVG 127
H++ Q LG +V+WD+ IVL +LE G + L+G+ VELG+G GLVG
Sbjct: 29 HTIQTRQDWRHLG----VAAVVWDAAIVLSTYLE----MGAVELRGRSAVELGAGTGLVG 80
Query: 128 CIAALLGGEVILTDLPDRMRLLRKNIETNM-KHISLRGSITATELTWGDDPDQELIDPTP 186
+AALLG V +TD + L+ N++ N+ HI + ELTWG Q L +P
Sbjct: 81 IVAALLGAHVTITDRKVALEFLKSNVQANLPPHIQTK--TVVKELTWG----QNLGSFSP 134
Query: 187 ---DYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELR 225
D ILG+D++Y E DLL+TL L ++ I LA +R
Sbjct: 135 GEFDLILGADIIYLEETFTDLLQTLEHLCSNHSVILLACRIR 176
>UniRef100_Q84TK2 Hypothetical protein At1g08125 [Arabidopsis thaliana]
Length = 232
Score = 90.5 bits (223), Expect = 4e-17
Identities = 47/143 (32%), Positives = 81/143 (55%), Gaps = 6/143 (4%)
Query: 132 LLGGEVILTDLPDRMRLLRKNIETNMKHI------SLRGSITATELTWGDDPDQELIDPT 185
+LG +V+ TD + + LL++N+E N I S GS+ EL WG++ ++P
Sbjct: 1 MLGCDVVTTDQKEVLPLLKRNVEWNTSRIVQMNPGSAFGSLRVAELDWGNEDHITAVEPP 60
Query: 186 PDYILGSDVVYSEGAVVDLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGR 245
DY++G+DVVYSE + LL T+ LSGP TT+ L E+R+ + E L+ ++F +
Sbjct: 61 FDYVIGTDVVYSEQLLEPLLRTILALSGPKTTVMLGYEIRSTVVHEKMLQMWKDNFEVKT 120
Query: 246 VDQTLWHPDYHSNRVVLYVLVKK 268
+ ++ +Y + LY++ +K
Sbjct: 121 IPRSKMDGEYQDPSIHLYIMAQK 143
>UniRef100_UPI000035FA3E UPI000035FA3E UniRef100 entry
Length = 220
Score = 88.6 bits (218), Expect = 2e-16
Identities = 59/148 (39%), Positives = 81/148 (53%), Gaps = 13/148 (8%)
Query: 86 GSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDLPDR 145
G+V+W S +VL FLE + D L+ K ++ELG+G GLV +++LLG +V TDLPD
Sbjct: 44 GAVLWPSAMVLCHFLETNQDK--FSLRDKNVIELGAGTGLVTIVSSLLGAKVTSTDLPDV 101
Query: 146 MRLLRKNIETN----MKHISLRGSITATELTWGDDPDQELIDPTP--DYILGSDVVYSEG 199
+ L+ N+ N K+I L TELTWG + +Q T DYIL +DVVYS
Sbjct: 102 LGNLQYNVTRNTKGRCKYIPL-----VTELTWGQEVEQRFPRDTHCFDYILAADVVYSHP 156
Query: 200 AVVDLLETLGQLSGPNTTIFLAGELRND 227
+ +L+ T L T I A R D
Sbjct: 157 YLEELMATFDHLCQETTEILWAMRFRLD 184
>UniRef100_UPI00003AE5D2 UPI00003AE5D2 UniRef100 entry
Length = 215
Score = 88.6 bits (218), Expect = 2e-16
Identities = 71/233 (30%), Positives = 106/233 (45%), Gaps = 35/233 (15%)
Query: 23 MLVVPGEESAAEETMLLWGIQQPTLSKPNAFVAQSSLQLRLDSCGHSLSILQSPSSLGKP 82
M +VP EE A L G+++PT + A G ++ I Q G
Sbjct: 1 MALVPYEEGRA-----LQGLRRPTATYRFA--------------GRTVRIRQDWERRG-- 39
Query: 83 GVTGSVMWDSGIVLGKFLEHSVDSGMLVLQGKKIVELGSGCGLVGCIAALLGGEVILTDL 142
+V+WD+ +VL +LE G + L+ + ++ELG+G GL+G +A LLG V +TD
Sbjct: 40 --VAAVVWDAAVVLSAYLE----MGGIDLRDRSVIELGAGTGLLGIVATLLGARVTITDR 93
Query: 143 PDRMRLLRKNIETNMKHISLRGSITATELTWGDDPDQELIDPTPDYILGSDVVYSEGAVV 202
+ L N+ N+ L ELTWG D D+ILG+D++Y E
Sbjct: 94 EPALEFLESNVWANLPS-ELHARAVVKELTWGKDLG-SFPPGAFDFILGADIIYLEETFA 151
Query: 203 DLLETLGQLSGPNTTIFLAGELRNDAILEYFLEAAMNDFTIGRVDQTLWHPDY 255
+LL TL L T I L+ +R + FL+ F++ V H DY
Sbjct: 152 ELLRTLEHLCSEQTVILLSCRIRYERD-NNFLKMLKGRFSVSEV-----HYDY 198
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.316 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 445,377,592
Number of Sequences: 2790947
Number of extensions: 18224558
Number of successful extensions: 46620
Number of sequences better than 10.0: 284
Number of HSP's better than 10.0 without gapping: 98
Number of HSP's successfully gapped in prelim test: 186
Number of HSP's that attempted gapping in prelim test: 46288
Number of HSP's gapped (non-prelim): 287
length of query: 268
length of database: 848,049,833
effective HSP length: 125
effective length of query: 143
effective length of database: 499,181,458
effective search space: 71382948494
effective search space used: 71382948494
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC138580.8


