

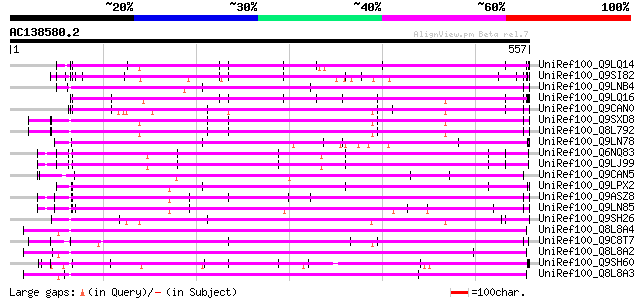
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138580.2 + phase: 0
(557 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LQ14 F16P17.7 protein [Arabidopsis thaliana] 374 e-102
UniRef100_Q9SI82 F23N19.4 [Arabidopsis thaliana] 369 e-100
UniRef100_Q9LNB4 F5O11.4 [Arabidopsis thaliana] 368 e-100
UniRef100_Q9LQ16 F16P17.5 protein [Arabidopsis thaliana] 366 e-100
UniRef100_Q9CAN0 Hypothetical protein F16M19.5 [Arabidopsis thal... 357 7e-97
UniRef100_Q9SXD8 T3P18.15 [Arabidopsis thaliana] 355 2e-96
UniRef100_Q8L792 Putative membrane-associated salt-inducible pro... 355 2e-96
UniRef100_Q9LN78 T12C24.22 [Arabidopsis thaliana] 353 1e-95
UniRef100_Q6NQ83 Hypothetical protein At3g22470 [Arabidopsis tha... 352 2e-95
UniRef100_Q9LJ99 Gb|AAD43616.1 [Arabidopsis thaliana] 352 2e-95
UniRef100_Q9CAN5 Hypothetical protein F16M19.17 [Arabidopsis tha... 352 2e-95
UniRef100_Q9LPX2 F13K23.2 protein [Arabidopsis thaliana] 351 4e-95
UniRef100_Q9ASZ8 At1g12620/T12C24_25 [Arabidopsis thaliana] 348 3e-94
UniRef100_Q9LN85 T12C24.15 [Arabidopsis thaliana] 348 3e-94
UniRef100_Q9SH26 F2K11.22 [Arabidopsis thaliana] 346 9e-94
UniRef100_Q8L8A4 Fertility restorer [Petunia hybrida] 333 8e-90
UniRef100_Q9C8T7 Hypothetical protein F9N12.5 [Arabidopsis thali... 332 1e-89
UniRef100_Q8L8A2 Fertility restorer-like protein [Petunia hybrida] 327 6e-88
UniRef100_Q9SH60 F22C12.14 [Arabidopsis thaliana] 327 7e-88
UniRef100_Q8L8A3 Fertility restorer-like protein [Petunia hybrida] 321 4e-86
>UniRef100_Q9LQ14 F16P17.7 protein [Arabidopsis thaliana]
Length = 613
Score = 374 bits (959), Expect = e-102
Identities = 201/527 (38%), Positives = 309/527 (58%), Gaps = 35/527 (6%)
Query: 66 PTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSF 125
P P I++FNK+LS++ K +SL ++M+ I D +++NILINCF + L+
Sbjct: 60 PLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNILINCFCRRSQLPLAL 119
Query: 126 SVFGKILKKGFDP-----------------------------------NAITFNTLIKGL 150
+V GK++K G++P N +TFNTLI GL
Sbjct: 120 AVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVMEYQPNTVTFNTLIHGL 179
Query: 151 CLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNA 210
L +A+ D++VA+G D +YGT++NGLCK G I AL LLK+++ ++ +
Sbjct: 180 FLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADV 239
Query: 211 VMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHK 270
V+Y IID +C K VNDA +L+++M K I P+ T NSLI C G+ +A LL
Sbjct: 240 VIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSD 299
Query: 271 MILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKE 330
MI INP + TFS L+DAF KEGK+ EA+ + +K+ I D+ TY+SL++G+C+
Sbjct: 300 MIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDR 359
Query: 331 INKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNS 390
+++AK +F+ M S+ NV +Y T+I G CK K V+E + LF EM R ++ N VTYN+
Sbjct: 360 LDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNT 419
Query: 391 LIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQG 450
LI GL + G K+ +M G PP+IITY+ +LD LCK ++KA+ + L+
Sbjct: 420 LIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSK 479
Query: 451 IRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAAL 510
+ PD+YTY ++I+G+C++GK+ED +F L +KG +V YT MI GFC KGL + A
Sbjct: 480 MEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEAD 539
Query: 511 ALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGLL 557
AL +M+++G +PN+ TY +I + + + +L++EM + G +
Sbjct: 540 ALFREMKEDGTLPNSGTYNTLIRARLRDGDKAASAELIKEMRSCGFV 586
Score = 218 bits (556), Expect = 3e-55
Identities = 120/406 (29%), Positives = 212/406 (51%)
Query: 127 VFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLC 186
+FG++++ P+ + FN L+ + ++ +++ D SY LIN C
Sbjct: 51 LFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNILINCFC 110
Query: 187 KVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDF 246
+ ++ AL +L ++ +P+ V + +++ C K +++A L QM P+
Sbjct: 111 RRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVMEYQPNTV 170
Query: 247 TCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVT 306
T N+LI+G + + EAV L+ +M+ P ++T+ +V+ CK G + A +L
Sbjct: 171 TFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLALSLLKKM 230
Query: 307 MKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMV 366
K I DVV Y +++D C K +N A ++F M ++G+ NV +Y ++I LC
Sbjct: 231 EKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRW 290
Query: 367 DEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSI 426
+A L +M RKI PNVVT+++LID K GK+ KL DEM R P+I TY+S+
Sbjct: 291 SDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSL 350
Query: 427 LDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGY 486
++ C + +D+A + + + P++ TY LIKG C++ ++E+ ++F ++ +G
Sbjct: 351 INGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGL 410
Query: 487 NLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVI 532
+ TY +IQG G D A + KM +G P+ TY I++
Sbjct: 411 VGNTVTYNTLIQGLFQAGDCDMAQKIFKKMVSDGVPPDIITYSILL 456
Score = 184 bits (466), Expect = 8e-45
Identities = 109/367 (29%), Positives = 187/367 (50%), Gaps = 35/367 (9%)
Query: 226 VNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSI 285
++DA DL+ +MV R P N L+ M + + L +M I+ +Y+++I
Sbjct: 45 LDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYNI 104
Query: 286 LVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEI-------------- 331
L++ FC+ ++ A +LG MK D+VT +SL++GYC K I
Sbjct: 105 LINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVME 164
Query: 332 ------------------NKAKD---IFDSMASRGVIANVQSYTTMINGLCKIKMVDEAV 370
NKA + + D M +RG ++ +Y T++NGLCK +D A+
Sbjct: 165 YQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLAL 224
Query: 371 NLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDAL 430
+L ++M KI +VV Y ++ID L ++ L L EM ++G PN++TYNS++ L
Sbjct: 225 SLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCL 284
Query: 431 CKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDV 490
C A LL+++ ++ I P++ T++ LI + GKL +A+K++++++ + + D+
Sbjct: 285 CNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDI 344
Query: 491 YTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLRE 550
+TY+ +I GFC+ D A + M C PN TY +I + + +L RE
Sbjct: 345 FTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVEEGMELFRE 404
Query: 551 MIARGLL 557
M RGL+
Sbjct: 405 MSQRGLV 411
Score = 142 bits (359), Expect = 2e-32
Identities = 86/352 (24%), Positives = 177/352 (49%), Gaps = 3/352 (0%)
Query: 51 NNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNI 110
N+ ++ F + +K P ++ +N ++ L S A L M I + TF+
Sbjct: 256 NDALNLFTEM-DNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSA 314
Query: 111 LINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQG 170
LI+ F + G + ++ +++K+ DP+ T+++LI G C+ + +A + + ++++
Sbjct: 315 LIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKD 374
Query: 171 FHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAF 230
+ V+Y TLI G CK R+ ++L + + + + N V YN +I + +A + A
Sbjct: 375 CFPNVVTYNTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYNTLIQGLFQAGDCDMAQ 434
Query: 231 DLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAF 290
++ +MV+ + PD T + L+ G C G+L++A+ + + + P +YT++I+++
Sbjct: 435 KIFKKMVSDGVPPDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGM 494
Query: 291 CKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANV 350
CK GKV++ + K + +V+ Y +++ G+C +A +F M G + N
Sbjct: 495 CKAGKVEDGWDLFCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNS 554
Query: 351 QSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGL--GKLGK 400
+Y T+I + + L +EMR + + T + +I+ L G+L K
Sbjct: 555 GTYNTLIRARLRDGDKAASAELIKEMRSCGFVGDASTISMVINMLHDGRLEK 606
Score = 112 bits (281), Expect = 2e-23
Identities = 67/260 (25%), Positives = 131/260 (49%)
Query: 295 KVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYT 354
K+ +A + G ++ + +V +N L+ + + + + + M + + ++ SY
Sbjct: 44 KLDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYN 103
Query: 355 TMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDR 414
+IN C+ + A+ + +M P++VT +SL++G +IS + LVD+M
Sbjct: 104 ILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVM 163
Query: 415 GQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDA 474
PN +T+N+++ L ++ +A+AL+ + +G +PD++TY ++ GLC+ G ++ A
Sbjct: 164 EYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLA 223
Query: 475 QKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILS 534
+ + + DV YT +I C + AL L ++M++ G PN TY +I
Sbjct: 224 LSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRC 283
Query: 535 LFEKDENDMAEKLLREMIAR 554
L A +LL +MI R
Sbjct: 284 LCNYGRWSDASRLLSDMIER 303
Score = 102 bits (255), Expect = 2e-20
Identities = 57/227 (25%), Positives = 115/227 (50%)
Query: 330 EINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYN 389
+++ A D+F M + ++ + +++ + K+ D ++L E M+ +I ++ +YN
Sbjct: 44 KLDDAVDLFGEMVQSRPLPSIVEFNKLLSAIAKMNKFDLVISLGERMQNLRISYDLYSYN 103
Query: 390 SLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQ 449
LI+ + ++ L ++ +M G P+I+T +S+L+ C + +A+AL+ +
Sbjct: 104 ILINCFCRRSQLPLALAVLGKMMKLGYEPDIVTLSSLLNGYCHGKRISEAVALVDQMFVM 163
Query: 450 GIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAA 509
+P+ T+ LI GL K +A + + ++ +G D++TY ++ G C +G D A
Sbjct: 164 EYQPNTVTFNTLIHGLFLHNKASEAVALIDRMVARGCQPDLFTYGTVVNGLCKRGDIDLA 223
Query: 510 LALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGL 556
L+LL KME + Y +I +L + A L EM +G+
Sbjct: 224 LSLLKKMEKGKIEADVVIYTTIIDALCNYKNVNDALNLFTEMDNKGI 270
Score = 60.5 bits (145), Expect = 1e-07
Identities = 41/167 (24%), Positives = 77/167 (45%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P II ++ +L L K AL + + ++ + + D +T+NI+I + G + +
Sbjct: 447 PDIITYSILLDGLCKYGKLEKALVVFEYLQKSKMEPDIYTYNIMIEGMCKAGKVEDGWDL 506
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F + KG PN I + T+I G C KG +A ++ G + +Y TLI +
Sbjct: 507 FCSLSLKGVKPNVIIYTTMISGFCRKGLKEEADALFREMKEDGTLPNSGTYNTLIRARLR 566
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
G A+ +L+K + +A +M+I+ + +L ++ S
Sbjct: 567 DGDKAASAELIKEMRSCGFVGDASTISMVINMLHDGRLEKSYLEMLS 613
>UniRef100_Q9SI82 F23N19.4 [Arabidopsis thaliana]
Length = 1244
Score = 369 bits (946), Expect = e-100
Identities = 195/525 (37%), Positives = 307/525 (58%), Gaps = 35/525 (6%)
Query: 66 PTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSF 125
P P II+F+K+LS++ K +SL +QM+ GI + +T++ILINCF + L+
Sbjct: 691 PFPSIIEFSKLLSAIAKMNKFDVVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLAL 750
Query: 126 SVFGKILKKGFDPN-----------------------------------AITFNTLIKGL 150
+V GK++K G++PN +TFNTLI GL
Sbjct: 751 AVLGKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGL 810
Query: 151 CLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNA 210
L +A+ D++VA+G D V+YG ++NGLCK G A LL +++ ++P
Sbjct: 811 FLHNKASEAMALIDRMVAKGCQPDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGV 870
Query: 211 VMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHK 270
++YN IID +CK K ++DA +L+ +M K I P+ T +SLI C G+ +A LL
Sbjct: 871 LIYNTIIDGLCKYKHMDDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSDASRLLSD 930
Query: 271 MILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKE 330
MI INP ++TFS L+DAF KEGK+ EA+ + +K+ I +VTY+SL++G+C+
Sbjct: 931 MIERKINPDVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDR 990
Query: 331 INKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNS 390
+++AK +F+ M S+ +V +Y T+I G CK K V+E + +F EM R ++ N VTYN
Sbjct: 991 LDEAKQMFEFMVSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNI 1050
Query: 391 LIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQG 450
LI GL + G ++ EM G PPNI+TYN++LD LCKN ++KA+ + L+
Sbjct: 1051 LIQGLFQAGDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSK 1110
Query: 451 IRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAAL 510
+ P +YTY ++I+G+C++GK+ED +F +L +KG DV Y MI GFC KG + A
Sbjct: 1111 MEPTIYTYNIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEAD 1170
Query: 511 ALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARG 555
AL +M+++G +PN+ Y +I + + + + +L++EM + G
Sbjct: 1171 ALFKEMKEDGTLPNSGCYNTLIRARLRDGDREASAELIKEMRSCG 1215
Score = 314 bits (805), Expect = 4e-84
Identities = 170/474 (35%), Positives = 270/474 (56%), Gaps = 36/474 (7%)
Query: 51 NNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNI 110
N+ I F+ ++ + P P I+ FN++LS++VK K + +SL ++ME+ GI +D +TFNI
Sbjct: 170 NDAIDLFSDMVKSR-PFPSIVDFNRLLSAIVKLKKYDVVISLGKKMEVLGIRNDLYTFNI 228
Query: 111 LINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQG 170
+ I C + AL+ K++ G
Sbjct: 229 V-----------------------------------INCFCCCFQVSLALSILGKMLKLG 253
Query: 171 FHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAF 230
+ D+V+ G+L+NG C+ R++ A+ L+ ++ +P+ V YN IID++CK K VNDAF
Sbjct: 254 YEPDRVTIGSLVNGFCRRNRVSDAVSLVDKMVEIGYKPDIVAYNAIIDSLCKTKRVNDAF 313
Query: 231 DLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAF 290
D + ++ K I P+ T +L+ G C + +A LL MI + I P + T+S L+DAF
Sbjct: 314 DFFKEIERKGIRPNVVTYTALVNGLCNSSRWSDAARLLSDMIKKKITPNVITYSALLDAF 373
Query: 291 CKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANV 350
K GKV EAK + ++ I D+VTY+SL++G CL I++A +FD M S+G +A+V
Sbjct: 374 VKNGKVLEAKELFEEMVRMSIDPDIVTYSSLINGLCLHDRIDEANQMFDLMVSKGCLADV 433
Query: 351 QSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDE 410
SY T+ING CK K V++ + LF EM R ++ N VTYN+LI G + G + + +
Sbjct: 434 VSYNTLINGFCKAKRVEDGMKLFREMSQRGLVSNTVTYNTLIQGFFQAGDVDKAQEFFSQ 493
Query: 411 MHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGK 470
M G P+I TYN +L LC N ++KA+ + +++ + + D+ TYT +I+G+C++GK
Sbjct: 494 MDFFGISPDIWTYNILLGGLCDNGELEKALVIFEDMQKREMDLDIVTYTTVIRGMCKTGK 553
Query: 471 LEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPN 524
+E+A +F L +KG D+ TYT M+ G C KGL AL +KM+ G + N
Sbjct: 554 VEEAWSLFCSLSLKGLKPDIVTYTTMMSGLCTKGLLHEVEALYTKMKQEGLMKN 607
Score = 232 bits (592), Expect = 2e-59
Identities = 140/543 (25%), Positives = 263/543 (47%), Gaps = 52/543 (9%)
Query: 61 LHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGL 120
+ K P ++ + +++ L + S A L M I + T++ L++ F + G
Sbjct: 319 IERKGIRPNVVTYTALVNGLCNSSRWSDAARLLSDMIKKKITPNVITYSALLDAFVKNGK 378
Query: 121 NSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGT 180
+ +F ++++ DP+ +T+++LI GLCL I +A D +V++G D VSY T
Sbjct: 379 VLEAKELFEEMVRMSIDPDIVTYSSLINGLCLHDRIDEANQMFDLMVSKGCLADVVSYNT 438
Query: 181 LINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKR 240
LING CK R+ ++L + + + + N V YN +I +A V+ A + +SQM
Sbjct: 439 LINGFCKAKRVEDGMKLFREMSQRGLVSNTVTYNTLIQGFFQAGDVDKAQEFFSQMDFFG 498
Query: 241 ISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAK 300
ISPD +T N L+ G C G+L++A+ + M ++ + T++ ++ CK GKV+EA
Sbjct: 499 ISPDIWTYNILLGGLCDNGELEKALVIFEDMQKREMDLDIVTYTTVIRGMCKTGKVEEAW 558
Query: 301 MMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIAN----------- 349
+ K + D+VTY ++M G C +++ + ++ M G++ N
Sbjct: 559 SLFCSLSLKGLKPDIVTYTTMMSGLCTKGLLHEVEALYTKMKQEGLMKNDCTLSDGDITL 618
Query: 350 -VQSYTTMI-------------NGLCKIKM---------------------------VDE 368
+ M+ +G+CK + +D+
Sbjct: 619 SAELIKKMLSCGYAPSLLKDIKSGVCKKALSLLRAFSGKTSYDYREKLSRNGLSELKLDD 678
Query: 369 AVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILD 428
AV LF EM + P+++ ++ L+ + K+ K V+ L ++M + G P N TY+ +++
Sbjct: 679 AVALFGEMVKSRPFPSIIEFSKLLSAIAKMNKFDVVISLGEQMQNLGIPHNHYTYSILIN 738
Query: 429 ALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNL 488
C+ + A+A+L + G P++ T + L+ G C S ++ +A + + + V GY
Sbjct: 739 CFCRRSQLPLALAVLGKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQMFVTGYQP 798
Query: 489 DVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLL 548
+ T+ +I G + A+AL+ +M GC P+ TY +V+ L ++ + D+A LL
Sbjct: 799 NTVTFNTLIHGLFLHNKASEAMALIDRMVAKGCQPDLVTYGVVVNGLCKRGDTDLAFNLL 858
Query: 549 REM 551
+M
Sbjct: 859 NKM 861
Score = 220 bits (561), Expect = 7e-56
Identities = 122/434 (28%), Positives = 225/434 (51%)
Query: 124 SFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLIN 183
+ +F ++K P+ + FN L+ + ++ K+ G D ++ +IN
Sbjct: 172 AIDLFSDMVKSRPFPSIVDFNRLLSAIVKLKKYDVVISLGKKMEVLGIRNDLYTFNIVIN 231
Query: 184 GLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISP 243
C +++ AL +L ++ +P+ V +++ C+ V+DA L +MV P
Sbjct: 232 CFCCCFQVSLALSILGKMLKLGYEPDRVTIGSLVNGFCRRNRVSDAVSLVDKMVEIGYKP 291
Query: 244 DDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMML 303
D N++I C ++ +A ++ + I P + T++ LV+ C + +A +L
Sbjct: 292 DIVAYNAIIDSLCKTKRVNDAFDFFKEIERKGIRPNVVTYTALVNGLCNSSRWSDAARLL 351
Query: 304 GVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKI 363
+KK I +V+TY++L+D + ++ +AK++F+ M + ++ +Y+++INGLC
Sbjct: 352 SDMIKKKITPNVITYSALLDAFVKNGKVLEAKELFEEMVRMSIDPDIVTYSSLINGLCLH 411
Query: 364 KMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITY 423
+DEA +F+ M + + +VV+YN+LI+G K ++ +KL EM RG N +TY
Sbjct: 412 DRIDEANQMFDLMVSKGCLADVVSYNTLINGFCKAKRVEDGMKLFREMSQRGLVSNTVTY 471
Query: 424 NSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLV 483
N+++ + VDKA + + GI PD++TY +L+ GLC +G+LE A +FED+
Sbjct: 472 NTLIQGFFQAGDVDKAQEFFSQMDFFGISPDIWTYNILLGGLCDNGELEKALVIFEDMQK 531
Query: 484 KGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDM 543
+ +LD+ TYT +I+G C G + A +L + G P+ TY ++ L K
Sbjct: 532 REMDLDIVTYTTVIRGMCKTGKVEEAWSLFCSLSLKGLKPDIVTYTTMMSGLCTKGLLHE 591
Query: 544 AEKLLREMIARGLL 557
E L +M GL+
Sbjct: 592 VEALYTKMKQEGLM 605
Score = 217 bits (553), Expect = 6e-55
Identities = 127/466 (27%), Positives = 241/466 (51%), Gaps = 15/466 (3%)
Query: 79 SLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDP 138
SL++A T+ +++ NG+ S+L L+ + ++FG+++K P
Sbjct: 649 SLLRAFSGKTSYDYREKLSRNGL--------------SELKLDD-AVALFGEMVKSRPFP 693
Query: 139 NAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLL 198
+ I F+ L+ + ++ +++ G + +Y LIN C+ ++ AL +L
Sbjct: 694 SIIEFSKLLSAIAKMNKFDVVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLALAVL 753
Query: 199 KRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIM 258
++ +PN V + +++ C +K +++A L QM P+ T N+LI+G +
Sbjct: 754 GKMMKLGYEPNIVTLSSLLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFLH 813
Query: 259 GQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTY 318
+ EA+ L+ +M+ + P + T+ ++V+ CK G A +L + + V+ Y
Sbjct: 814 NKASEAMALIDRMVAKGCQPDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGVLIY 873
Query: 319 NSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRC 378
N+++DG C K ++ A ++F M ++G+ NV +Y+++I+ LC +A L +M
Sbjct: 874 NTIIDGLCKYKHMDDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIE 933
Query: 379 RKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDK 438
RKI P+V T+++LID K GK+ KL DEM R P+I+TY+S+++ C + +D+
Sbjct: 934 RKINPDVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDE 993
Query: 439 AIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQ 498
A + + + PD+ TY LIKG C+ ++E+ +VF ++ +G + TY ++IQ
Sbjct: 994 AKQMFEFMVSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQ 1053
Query: 499 GFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMA 544
G G D A + +M +G PN TY ++ L + + + A
Sbjct: 1054 GLFQAGDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKA 1099
Score = 214 bits (546), Expect = 4e-54
Identities = 141/538 (26%), Positives = 248/538 (45%), Gaps = 56/538 (10%)
Query: 71 IQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGK 130
+ +N ++ +A A QM+ GI D +T+NIL+ G + +F
Sbjct: 469 VTYNTLIQGFFQAGDVDKAQEFFSQMDFFGISPDIWTYNILLGGLCDNGELEKALVIFED 528
Query: 131 ILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGR 190
+ K+ D + +T+ T+I+G+C G + +A + + +G D V+Y T+++GLC G
Sbjct: 529 MQKREMDLDIVTYTTVIRGMCKTGKVEEAWSLFCSLSLKGLKPDIVTYTTMMSGLCTKGL 588
Query: 191 ---------------------------ITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKA 223
IT + +L+K++ P+ + I +CK
Sbjct: 589 LHEVEALYTKMKQEGLMKNDCTLSDGDITLSAELIKKMLSCGYAPS--LLKDIKSGVCKK 646
Query: 224 KL---------------------------VNDAFDLYSQMVAKRISPDDFTCNSLIYGFC 256
L ++DA L+ +MV R P + L+
Sbjct: 647 ALSLLRAFSGKTSYDYREKLSRNGLSELKLDDAVALFGEMVKSRPFPSIIEFSKLLSAIA 706
Query: 257 IMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVV 316
M + + L +M I YT+SIL++ FC+ ++ A +LG MK ++V
Sbjct: 707 KMNKFDVVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLALAVLGKMMKLGYEPNIV 766
Query: 317 TYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEM 376
T +SL++GYC K I++A + D M G N ++ T+I+GL EA+ L + M
Sbjct: 767 TLSSLLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFLHNKASEAMALIDRM 826
Query: 377 RCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHV 436
+ P++VTY +++GL K G L+++M P ++ YN+I+D LCK H+
Sbjct: 827 VAKGCQPDLVTYGVVVNGLCKRGDTDLAFNLLNKMEQGKLEPGVLIYNTIIDGLCKYKHM 886
Query: 437 DKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVM 496
D A+ L ++ +GIRP++ TY+ LI LC G+ DA ++ D++ + N DV+T++ +
Sbjct: 887 DDALNLFKEMETKGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIERKINPDVFTFSAL 946
Query: 497 IQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIAR 554
I F +G A L +M P+ TY +I D D A+++ M+++
Sbjct: 947 IDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMFEFMVSK 1004
Score = 214 bits (545), Expect = 5e-54
Identities = 142/540 (26%), Positives = 255/540 (46%), Gaps = 52/540 (9%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P I+ +N I+ SL K K + A +++E GI + T+ L+N S + +
Sbjct: 291 PDIVAYNAIIDSLCKTKRVNDAFDFFKEIERKGIRPNVVTYTALVNGLCNSSRWSDAARL 350
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
++KK PN IT++ L+ G + +A +++V D V+Y +LINGLC
Sbjct: 351 LSDMIKKKITPNVITYSALLDAFVKNGKVLEAKELFEEMVRMSIDPDIVTYSSLINGLCL 410
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
RI A Q+ + K + V YN +I+ CKAK V D L+ +M + + + T
Sbjct: 411 HDRIDEANQMFDLMVSKGCLADVVSYNTLINGFCKAKRVEDGMKLFREMSQRGLVSNTVT 470
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTM 307
N+LI GF G + +A +M I+P ++T++IL+ C G++++A ++
Sbjct: 471 YNTLIQGFFQAGDVDKAQEFFSQMDFFGISPDIWTYNILLGGLCDNGELEKALVIFEDMQ 530
Query: 308 KKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVD 367
K+++ LD+VTY +++ G C ++ +A +F S++ +G+ ++ +YTTM++GLC ++
Sbjct: 531 KREMDLDIVTYTTVIRGMCKTGKVEEAWSLFCSLSLKGLKPDIVTYTTMMSGLCTKGLLH 590
Query: 368 EAVNLFEEMRCRKIIPNVVTYN--------SLIDGLGKLGKISCVLK------------- 406
E L+ +M+ ++ N T + LI + G +LK
Sbjct: 591 EVEALYTKMKQEGLMKNDCTLSDGDITLSAELIKKMLSCGYAPSLLKDIKSGVCKKALSL 650
Query: 407 -------------------------------LVDEMHDRGQPPNIITYNSILDALCKNHH 435
L EM P+II ++ +L A+ K +
Sbjct: 651 LRAFSGKTSYDYREKLSRNGLSELKLDDAVALFGEMVKSRPFPSIIEFSKLLSAIAKMNK 710
Query: 436 VDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTV 495
D I+L +++ GI + YTY++LI C+ +L A V ++ GY ++ T +
Sbjct: 711 FDVVISLGEQMQNLGIPHNHYTYSILINCFCRRSQLPLALAVLGKMMKLGYEPNIVTLSS 770
Query: 496 MIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARG 555
++ G+C A+AL+ +M G PN T+ +I LF ++ A L+ M+A+G
Sbjct: 771 LLNGYCHSKRISEAVALVDQMFVTGYQPNTVTFNTLIHGLFLHNKASEAMALIDRMVAKG 830
Score = 189 bits (480), Expect = 2e-46
Identities = 106/326 (32%), Positives = 175/326 (53%)
Query: 226 VNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSI 285
+NDA DL+S MV R P N L+ + + + L KM + I +YTF+I
Sbjct: 169 LNDAIDLFSDMVKSRPFPSIVDFNRLLSAIVKLKKYDVVISLGKKMEVLGIRNDLYTFNI 228
Query: 286 LVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRG 345
+++ FC +V A +LG +K D VT SL++G+C ++ A + D M G
Sbjct: 229 VINCFCCCFQVSLALSILGKMLKLGYEPDRVTIGSLVNGFCRRNRVSDAVSLVDKMVEIG 288
Query: 346 VIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVL 405
++ +Y +I+ LCK K V++A + F+E+ + I PNVVTY +L++GL + S
Sbjct: 289 YKPDIVAYNAIIDSLCKTKRVNDAFDFFKEIERKGIRPNVVTYTALVNGLCNSSRWSDAA 348
Query: 406 KLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGL 465
+L+ +M + PN+ITY+++LDA KN V +A L + I PD+ TY+ LI GL
Sbjct: 349 RLLSDMIKKKITPNVITYSALLDAFVKNGKVLEAKELFEEMVRMSIDPDIVTYSSLINGL 408
Query: 466 CQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNA 525
C ++++A ++F+ ++ KG DV +Y +I GFC + + L +M G + N
Sbjct: 409 CLHDRIDEANQMFDLMVSKGCLADVVSYNTLINGFCKAKRVEDGMKLFREMSQRGLVSNT 468
Query: 526 KTYEIVILSLFEKDENDMAEKLLREM 551
TY +I F+ + D A++ +M
Sbjct: 469 VTYNTLIQGFFQAGDVDKAQEFFSQM 494
Score = 157 bits (397), Expect = 8e-37
Identities = 88/334 (26%), Positives = 171/334 (50%), Gaps = 4/334 (1%)
Query: 44 HHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVS 103
+ H D+ NL + K P ++ ++ ++S L S A L M I
Sbjct: 883 YKHMDDALNLFKEMET----KGIRPNVVTYSSLISCLCNYGRWSDASRLLSDMIERKINP 938
Query: 104 DFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFH 163
D FTF+ LI+ F + G + ++ +++K+ DP+ +T+++LI G C+ + +A
Sbjct: 939 DVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQMF 998
Query: 164 DKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKA 223
+ +V++ D V+Y TLI G CK R+ +++ + + + + N V YN++I + +A
Sbjct: 999 EFMVSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQGLFQA 1058
Query: 224 KLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTF 283
+ A +++ +MV+ + P+ T N+L+ G C G+L++A+ + + + P +YT+
Sbjct: 1059 GDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTIYTY 1118
Query: 284 SILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMAS 343
+I+++ CK GKV++ + K + DVV YN+++ G+C +A +F M
Sbjct: 1119 NIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKE 1178
Query: 344 RGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMR 377
G + N Y T+I + + + L +EMR
Sbjct: 1179 DGTLPNSGCYNTLIRARLRDGDREASAELIKEMR 1212
Score = 125 bits (313), Expect = 4e-27
Identities = 73/263 (27%), Positives = 132/263 (49%)
Query: 295 KVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYT 354
K+ +A + +K +V +N L+ +K+ + + M G+ ++ ++
Sbjct: 168 KLNDAIDLFSDMVKSRPFPSIVDFNRLLSAIVKLKKYDVVISLGKKMEVLGIRNDLYTFN 227
Query: 355 TMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDR 414
+IN C V A+++ +M P+ VT SL++G + ++S + LVD+M +
Sbjct: 228 IVINCFCCCFQVSLALSILGKMLKLGYEPDRVTIGSLVNGFCRRNRVSDAVSLVDKMVEI 287
Query: 415 GQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDA 474
G P+I+ YN+I+D+LCK V+ A ++ +GIRP++ TYT L+ GLC S + DA
Sbjct: 288 GYKPDIVAYNAIIDSLCKTKRVNDAFDFFKEIERKGIRPNVVTYTALVNGLCNSSRWSDA 347
Query: 475 QKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILS 534
++ D++ K +V TY+ ++ F G A L +M P+ TY +I
Sbjct: 348 ARLLSDMIKKKITPNVITYSALLDAFVKNGKVLEAKELFEEMVRMSIDPDIVTYSSLING 407
Query: 535 LFEKDENDMAEKLLREMIARGLL 557
L D D A ++ M+++G L
Sbjct: 408 LCLHDRIDEANQMFDLMVSKGCL 430
Score = 110 bits (275), Expect = 1e-22
Identities = 66/293 (22%), Positives = 134/293 (45%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P + F+ ++ + VK A L+ +M I T++ LIN F + +
Sbjct: 938 PDVFTFSALIDAFVKEGKLVEAEKLYDEMVKRSIDPSIVTYSSLINGFCMHDRLDEAKQM 997
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F ++ K P+ +T+NTLIKG C + + + ++ +G + V+Y LI GL +
Sbjct: 998 FEFMVSKHCFPDVVTYNTLIKGFCKYKRVEEGMEVFREMSQRGLVGNTVTYNILIQGLFQ 1057
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
G A ++ K + V PN + YN ++D +CK + A ++ + ++ P +T
Sbjct: 1058 AGDCDMAQEIFKEMVSDGVPPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTIYT 1117
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTM 307
N +I G C G++++ L + L+ + P + ++ ++ FC++G +EA +
Sbjct: 1118 YNIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMK 1177
Query: 308 KKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGL 360
+ + + YN+L+ + + ++ M S G + + + N L
Sbjct: 1178 EDGTLPNSGCYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGLVTNML 1230
Score = 56.6 bits (135), Expect = 2e-06
Identities = 37/167 (22%), Positives = 74/167 (44%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P I+ +N +L L K A+ + + ++ + + +T+NI+I + G + +
Sbjct: 1078 PNIMTYNTLLDGLCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDL 1137
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F + KG P+ + +NT+I G C KG +A ++ G + Y TLI +
Sbjct: 1138 FCNLSLKGVKPDVVAYNTMISGFCRKGSKEEADALFKEMKEDGTLPNSGCYNTLIRARLR 1197
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
G A+ +L+K + +A ++ + + +L D+ S
Sbjct: 1198 DGDREASAELIKEMRSCGFAGDASTIGLVTNMLHDGRLDKSFLDMLS 1244
Score = 42.4 bits (98), Expect = 0.036
Identities = 29/125 (23%), Positives = 54/125 (43%)
Query: 61 LHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGL 120
L P I +N ++ + KA L + L G+ D +N +I+ F + G
Sbjct: 1106 LQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDLFCNLSLKGVKPDVVAYNTMISGFCRKGS 1165
Query: 121 NSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGT 180
+ ++F ++ + G PN+ +NTLI+ G + ++ + GF D + G
Sbjct: 1166 KEEADALFKEMKEDGTLPNSGCYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGL 1225
Query: 181 LINGL 185
+ N L
Sbjct: 1226 VTNML 1230
>UniRef100_Q9LNB4 F5O11.4 [Arabidopsis thaliana]
Length = 975
Score = 368 bits (945), Expect = e-100
Identities = 191/536 (35%), Positives = 309/536 (57%), Gaps = 36/536 (6%)
Query: 51 NNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNI 110
++ I F ++H + P P +I F+++ S++ K K + L+L +QMEL GI + +T +I
Sbjct: 168 DDAIDLFRDMIHSR-PLPTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYTLSI 226
Query: 111 LINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQG 170
+INCF + L+FS GKI+K G++PN ITF+TLI GLCL+G + +AL D++V G
Sbjct: 227 MINCFCRCRKLCLAFSAMGKIIKLGYEPNTITFSTLINGLCLEGRVSEALELVDRMVEMG 286
Query: 171 FHLDQVSYGTLINGLC-----------------------------------KVGRITAAL 195
D ++ TL+NGLC K G+ A+
Sbjct: 287 HKPDLITINTLVNGLCLSGKEAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCKSGQTALAM 346
Query: 196 QLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGF 255
+LL++++ + ++ +AV Y++IID +CK +++AF+L+++M K I+ + T N LI GF
Sbjct: 347 ELLRKMEERNIKLDAVKYSIIIDGLCKHGSLDNAFNLFNEMEMKGITTNIITYNILIGGF 406
Query: 256 CIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDV 315
C G+ + LL MI INP + TFS+L+D+F KEGK++EA+ + + + I D
Sbjct: 407 CNAGRWDDGAKLLRDMIKRKINPNVVTFSVLIDSFVKEGKLREAEELHKEMIHRGIAPDT 466
Query: 316 VTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEE 375
+TY SL+DG+C ++KA + D M S+G N++++ +ING CK +D+ + LF +
Sbjct: 467 ITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFNILINGYCKANRIDDGLELFRK 526
Query: 376 MRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHH 435
M R ++ + VTYN+LI G +LGK++ +L EM R PPNI+TY +LD LC N
Sbjct: 527 MSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKVPPNIVTYKILLDGLCDNGE 586
Query: 436 VDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTV 495
+KA+ + ++ + D+ Y ++I G+C + K++DA +F L +KG V TY +
Sbjct: 587 SEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPGVKTYNI 646
Query: 496 MIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREM 551
MI G C KG A L KME++G P+ TY I+I + + + KL+ E+
Sbjct: 647 MIGGLCKKGPLSEAELLFRKMEEDGHAPDGWTYNILIRAHLGDGDATKSVKLIEEL 702
Score = 175 bits (444), Expect = 3e-42
Identities = 105/350 (30%), Positives = 178/350 (50%)
Query: 208 PNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGL 267
P + ++ + + K K + L QM K I+ + +T + +I FC +L A
Sbjct: 184 PTVIDFSRLFSAIAKTKQYDLVLALCKQMELKGIAHNLYTLSIMINCFCRCRKLCLAFSA 243
Query: 268 LHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCL 327
+ K+I P TFS L++ C EG+V EA ++ ++ D++T N+L++G CL
Sbjct: 244 MGKIIKLGYEPNTITFSTLINGLCLEGRVSEALELVDRMVEMGHKPDLITINTLVNGLCL 303
Query: 328 VKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVT 387
+ +A + D M G N +Y ++N +CK A+ L +M R I + V
Sbjct: 304 SGKEAEAMLLIDKMVEYGCQPNAVTYGPVLNVMCKSGQTALAMELLRKMEERNIKLDAVK 363
Query: 388 YNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLK 447
Y+ +IDGL K G + L +EM +G NIITYN ++ C D LL ++
Sbjct: 364 YSIIIDGLCKHGSLDNAFNLFNEMEMKGITTNIITYNILIGGFCNAGRWDDGAKLLRDMI 423
Query: 448 DQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFD 507
+ I P++ T++VLI + GKL +A+++ ++++ +G D TYT +I GFC + D
Sbjct: 424 KRKINPNVVTFSVLIDSFVKEGKLREAEELHKEMIHRGIAPDTITYTSLIDGFCKENHLD 483
Query: 508 AALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGLL 557
A ++ M GC PN +T+ I+I + + D +L R+M RG++
Sbjct: 484 KANQMVDLMVSKGCDPNIRTFNILINGYCKANRIDDGLELFRKMSLRGVV 533
Score = 120 bits (301), Expect = 1e-25
Identities = 72/261 (27%), Positives = 128/261 (48%)
Query: 63 HKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNS 122
H+ P I + ++ K H A + M G + TFNILIN + +
Sbjct: 459 HRGIAPDTITYTSLIDGFCKENHLDKANQMVDLMVSKGCDPNIRTFNILINGYCKANRID 518
Query: 123 LSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLI 182
+F K+ +G + +T+NTLI+G C G ++ A ++V++ + V+Y L+
Sbjct: 519 DGLELFRKMSLRGVVADTVTYNTLIQGFCELGKLNVAKELFQEMVSRKVPPNIVTYKILL 578
Query: 183 NGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRIS 242
+GLC G AL++ ++++ ++ + +YN+II MC A V+DA+DL+ + K +
Sbjct: 579 DGLCDNGESEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVK 638
Query: 243 PDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMM 302
P T N +I G C G L EA L KM + P +T++IL+ A +G ++ +
Sbjct: 639 PGVKTYNIMIGGLCKKGPLSEAELLFRKMEEDGHAPDGWTYNILIRAHLGDGDATKSVKL 698
Query: 303 LGVTMKKDIILDVVTYNSLMD 323
+ + +D T ++D
Sbjct: 699 IEELKRCGFSVDASTIKMVID 719
>UniRef100_Q9LQ16 F16P17.5 protein [Arabidopsis thaliana]
Length = 632
Score = 366 bits (940), Expect = e-100
Identities = 197/525 (37%), Positives = 309/525 (58%), Gaps = 35/525 (6%)
Query: 66 PTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSF 125
P P I++FNK+LS++ K +SL +QM+ GI D +T++I INCF + SL+
Sbjct: 79 PFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYTYSIFINCFCRRSQLSLAL 138
Query: 126 SVFGKILKKGFDPNAIT-----------------------------------FNTLIKGL 150
+V K++K G++P+ +T F TLI GL
Sbjct: 139 AVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQMVEMGYKPDTFTFTTLIHGL 198
Query: 151 CLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNA 210
L +A+ D++V +G D V+YGT++NGLCK G I AL LLK+++ ++ +
Sbjct: 199 FLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNGLCKRGDIDLALSLLKKMEKGKIEADV 258
Query: 211 VMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHK 270
V+YN IID +CK K ++DA +L+++M K I PD FT +SLI C G+ +A LL
Sbjct: 259 VIYNTIIDGLCKYKHMDDALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSD 318
Query: 271 MILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKE 330
MI INP + TFS L+DAF KEGK+ EA+ + +K+ I D+ TY+SL++G+C+
Sbjct: 319 MIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDR 378
Query: 331 INKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNS 390
+++AK +F+ M S+ NV +Y+T+I G CK K V+E + LF EM R ++ N VTY +
Sbjct: 379 LDEAKHMFELMISKDCFPNVVTYSTLIKGFCKAKRVEEGMELFREMSQRGLVGNTVTYTT 438
Query: 391 LIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQG 450
LI G + + +M G PNI+TYN +LD LCKN + KA+ + L+
Sbjct: 439 LIHGFFQARDCDNAQMVFKQMVSVGVHPNILTYNILLDGLCKNGKLAKAMVVFEYLQRST 498
Query: 451 IRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAAL 510
+ PD+YTY ++I+G+C++GK+ED ++F +L +KG + +V Y MI GFC KG + A
Sbjct: 499 MEPDIYTYNIMIEGMCKAGKVEDGWELFCNLSLKGVSPNVIAYNTMISGFCRKGSKEEAD 558
Query: 511 ALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARG 555
+LL KM+++G +PN+ TY +I + + + + +L++EM + G
Sbjct: 559 SLLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKEMRSCG 603
Score = 222 bits (566), Expect = 2e-56
Identities = 127/423 (30%), Positives = 223/423 (52%)
Query: 110 ILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQ 169
IL N S + + +FG ++K P+ + FN L+ + ++ +++
Sbjct: 53 ILRNRLSDIIKVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTL 112
Query: 170 GFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDA 229
G D +Y IN C+ +++ AL +L ++ +P+ V + +++ C +K ++DA
Sbjct: 113 GISHDLYTYSIFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDA 172
Query: 230 FDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDA 289
L QMV PD FT +LI+G + + EAV L+ +M+ P + T+ +V+
Sbjct: 173 VALVDQMVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNG 232
Query: 290 FCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIAN 349
CK G + A +L K I DVV YN+++DG C K ++ A ++F M ++G+ +
Sbjct: 233 LCKRGDIDLALSLLKKMEKGKIEADVVIYNTIIDGLCKYKHMDDALNLFTEMDNKGIRPD 292
Query: 350 VQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVD 409
V +Y+++I+ LC +A L +M RKI PNVVT+++LID K GK+ KL D
Sbjct: 293 VFTYSSLISCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYD 352
Query: 410 EMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSG 469
EM R P+I TY+S+++ C + +D+A + + + P++ TY+ LIKG C++
Sbjct: 353 EMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYSTLIKGFCKAK 412
Query: 470 KLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYE 529
++E+ ++F ++ +G + TYT +I GF D A + +M G PN TY
Sbjct: 413 RVEEGMELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSVGVHPNILTYN 472
Query: 530 IVI 532
I++
Sbjct: 473 ILL 475
Score = 194 bits (493), Expect = 6e-48
Identities = 113/367 (30%), Positives = 185/367 (49%), Gaps = 35/367 (9%)
Query: 226 VNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSI 285
V+DA DL+ MV R P N L+ M + + + L +M I+ +YT+SI
Sbjct: 64 VDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYTYSI 123
Query: 286 LVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRG 345
++ FC+ ++ A +L MK D+VT +SL++GYC K I+ A + D M G
Sbjct: 124 FINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQMVEMG 183
Query: 346 VIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVL 405
+ ++TT+I+GL EAV L ++M R P++VTY ++++GL K G I L
Sbjct: 184 YKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNGLCKRGDIDLAL 243
Query: 406 KLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGL 465
L+ +M +++ YN+I+D LCK H+D A+ L T + ++GIRPD++TY+ LI L
Sbjct: 244 SLLKKMEKGKIEADVVIYNTIIDGLCKYKHMDDALNLFTEMDNKGIRPDVFTYSSLISCL 303
Query: 466 C-----------------------------------QSGKLEDAQKVFEDLLVKGYNLDV 490
C + GKL +A+K++++++ + + D+
Sbjct: 304 CNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDI 363
Query: 491 YTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLRE 550
+TY+ +I GFC+ D A + M C PN TY +I + + +L RE
Sbjct: 364 FTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYSTLIKGFCKAKRVEEGMELFRE 423
Query: 551 MIARGLL 557
M RGL+
Sbjct: 424 MSQRGLV 430
Score = 118 bits (295), Expect = 5e-25
Identities = 59/194 (30%), Positives = 106/194 (54%)
Query: 358 NGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQP 417
N L I VD+AV+LF +M + P++V +N L+ + K+ K V+ L ++M G
Sbjct: 56 NRLSDIIKVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGIS 115
Query: 418 PNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKV 477
++ TY+ ++ C+ + A+A+L + G PD+ T + L+ G C S ++ DA +
Sbjct: 116 HDLYTYSIFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVAL 175
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
+ ++ GY D +T+T +I G + A+AL+ +M GC P+ TY V+ L +
Sbjct: 176 VDQMVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNGLCK 235
Query: 538 KDENDMAEKLLREM 551
+ + D+A LL++M
Sbjct: 236 RGDIDLALSLLKKM 249
Score = 116 bits (290), Expect = 2e-24
Identities = 68/260 (26%), Positives = 129/260 (49%)
Query: 295 KVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYT 354
KV +A + G +K +V +N L+ + + + + M + G+ ++ +Y+
Sbjct: 63 KVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYTYS 122
Query: 355 TMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDR 414
IN C+ + A+ + +M P++VT +SL++G +IS + LVD+M +
Sbjct: 123 IFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQMVEM 182
Query: 415 GQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDA 474
G P+ T+ +++ L ++ +A+AL+ + +G +PD+ TY ++ GLC+ G ++ A
Sbjct: 183 GYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNGLCKRGDIDLA 242
Query: 475 QKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILS 534
+ + + DV Y +I G C D AL L ++M++ G P+ TY +I
Sbjct: 243 LSLLKKMEKGKIEADVVIYNTIIDGLCKYKHMDDALNLFTEMDNKGIRPDVFTYSSLISC 302
Query: 535 LFEKDENDMAEKLLREMIAR 554
L A +LL +MI R
Sbjct: 303 LCNYGRWSDASRLLSDMIER 322
Score = 112 bits (279), Expect = 4e-23
Identities = 60/227 (26%), Positives = 117/227 (51%)
Query: 330 EINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYN 389
+++ A D+F M ++ + +++ + K+ + ++L E+M+ I ++ TY+
Sbjct: 63 KVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHDLYTYS 122
Query: 390 SLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQ 449
I+ + ++S L ++ +M G P+I+T +S+L+ C + + A+AL+ + +
Sbjct: 123 IFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVDQMVEM 182
Query: 450 GIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAA 509
G +PD +T+T LI GL K +A + + ++ +G D+ TY ++ G C +G D A
Sbjct: 183 GYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRGCQPDLVTYGTVVNGLCKRGDIDLA 242
Query: 510 LALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGL 556
L+LL KME + Y +I L + D A L EM +G+
Sbjct: 243 LSLLKKMEKGKIEADVVIYNTIIDGLCKYKHMDDALNLFTEMDNKGI 289
Score = 77.4 bits (189), Expect = 1e-12
Identities = 44/161 (27%), Positives = 82/161 (50%)
Query: 395 LGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPD 454
L + K+ + L +M P+I+ +N +L A+ K + + I+L ++ GI D
Sbjct: 58 LSDIIKVDDAVDLFGDMVKSRPFPSIVEFNKLLSAVAKMNKFELVISLGEQMQTLGISHD 117
Query: 455 MYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLS 514
+YTY++ I C+ +L A V ++ GY D+ T + ++ G+C A+AL+
Sbjct: 118 LYTYSIFINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLSSLLNGYCHSKRISDAVALVD 177
Query: 515 KMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARG 555
+M + G P+ T+ +I LF ++ A L+ +M+ RG
Sbjct: 178 QMVEMGYKPDTFTFTTLIHGLFLHNKASEAVALVDQMVQRG 218
Score = 63.9 bits (154), Expect = 1e-08
Identities = 41/167 (24%), Positives = 78/167 (46%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P I+ +N +L L K + A+ + + ++ + + D +T+NI+I + G + +
Sbjct: 466 PNILTYNILLDGLCKNGKLAKAMVVFEYLQRSTMEPDIYTYNIMIEGMCKAGKVEDGWEL 525
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F + KG PN I +NT+I G C KG +A + K+ G + +Y TLI +
Sbjct: 526 FCNLSLKGVSPNVIAYNTMISGFCRKGSKEEADSLLKKMKEDGPLPNSGTYNTLIRARLR 585
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
G A+ +L+K + +A ++ + + +L D+ S
Sbjct: 586 DGDREASAELIKEMRSCGFAGDASTIGLVTNMLHDGRLDKSFLDMLS 632
Score = 42.7 bits (99), Expect = 0.027
Identities = 30/125 (24%), Positives = 54/125 (43%)
Query: 61 LHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGL 120
L P I +N ++ + KA L + L G+ + +N +I+ F + G
Sbjct: 494 LQRSTMEPDIYTYNIMIEGMCKAGKVEDGWELFCNLSLKGVSPNVIAYNTMISGFCRKGS 553
Query: 121 NSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGT 180
+ S+ K+ + G PN+ T+NTLI+ G + ++ + GF D + G
Sbjct: 554 KEEADSLLKKMKEDGPLPNSGTYNTLIRARLRDGDREASAELIKEMRSCGFAGDASTIGL 613
Query: 181 LINGL 185
+ N L
Sbjct: 614 VTNML 618
>UniRef100_Q9CAN0 Hypothetical protein F16M19.5 [Arabidopsis thaliana]
Length = 630
Score = 357 bits (915), Expect = 7e-97
Identities = 198/521 (38%), Positives = 305/521 (58%), Gaps = 35/521 (6%)
Query: 66 PTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCF---SQLGL-- 120
P P I++F+K+LS++ K +SL +QM+ GI + +T++ILINCF SQL L
Sbjct: 77 PFPSIVEFSKLLSAIAKMNKFDLVISLGEQMQNLGISHNLYTYSILINCFCRRSQLSLAL 136
Query: 121 ------------------NSL------------SFSVFGKILKKGFDPNAITFNTLIKGL 150
NSL + S+ G++++ G+ P++ TFNTLI GL
Sbjct: 137 AVLAKMMKLGYEPDIVTLNSLLNGFCHGNRISDAVSLVGQMVEMGYQPDSFTFNTLIHGL 196
Query: 151 CLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNA 210
+A+ D++V +G D V+YG ++NGLCK G I AL LLK+++ ++P
Sbjct: 197 FRHNRASEAVALVDRMVVKGCQPDLVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIEPGV 256
Query: 211 VMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHK 270
V+YN IID +C K VNDA +L+++M K I P+ T NSLI C G+ +A LL
Sbjct: 257 VIYNTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSD 316
Query: 271 MILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKE 330
MI INP + TFS L+DAF KEGK+ EA+ + +K+ I D+ TY+SL++G+C+
Sbjct: 317 MIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDR 376
Query: 331 INKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNS 390
+++AK +F+ M S+ NV +Y T+I G CK K VDE + LF EM R ++ N VTY +
Sbjct: 377 LDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVDEGMELFREMSQRGLVGNTVTYTT 436
Query: 391 LIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQG 450
LI G + + + +M G P+I+TY+ +LD LC N V+ A+ + L+
Sbjct: 437 LIHGFFQARECDNAQIVFKQMVSDGVLPDIMTYSILLDGLCNNGKVETALVVFEYLQRSK 496
Query: 451 IRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAAL 510
+ PD+YTY ++I+G+C++GK+ED +F L +KG +V TYT M+ GFC KGL + A
Sbjct: 497 MEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEAD 556
Query: 511 ALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREM 551
AL +M++ G +P++ TY +I + + + +L+REM
Sbjct: 557 ALFREMKEEGPLPDSGTYNTLIRAHLRDGDKAASAELIREM 597
Score = 224 bits (570), Expect = 7e-57
Identities = 126/423 (29%), Positives = 225/423 (52%), Gaps = 1/423 (0%)
Query: 110 ILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQ 169
I IN + L L+ + ++FG ++K P+ + F+ L+ + ++ +++
Sbjct: 52 ISINRLNDLKLDD-AVNLFGDMVKSRPFPSIVEFSKLLSAIAKMNKFDLVISLGEQMQNL 110
Query: 170 GFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDA 229
G + +Y LIN C+ +++ AL +L ++ +P+ V N +++ C ++DA
Sbjct: 111 GISHNLYTYSILINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCHGNRISDA 170
Query: 230 FDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDA 289
L QMV PD FT N+LI+G + EAV L+ +M+++ P + T+ I+V+
Sbjct: 171 VSLVGQMVEMGYQPDSFTFNTLIHGLFRHNRASEAVALVDRMVVKGCQPDLVTYGIVVNG 230
Query: 290 FCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIAN 349
CK G + A +L + I VV YN+++D C K +N A ++F M ++G+ N
Sbjct: 231 LCKRGDIDLALSLLKKMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDNKGIRPN 290
Query: 350 VQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVD 409
V +Y ++I LC +A L +M RKI PNVVT+++LID K GK+ KL D
Sbjct: 291 VVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYD 350
Query: 410 EMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSG 469
EM R P+I TY+S+++ C + +D+A + + + P++ TY LIKG C++
Sbjct: 351 EMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAK 410
Query: 470 KLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYE 529
++++ ++F ++ +G + TYT +I GF D A + +M +G +P+ TY
Sbjct: 411 RVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVFKQMVSDGVLPDIMTYS 470
Query: 530 IVI 532
I++
Sbjct: 471 ILL 473
Score = 182 bits (461), Expect = 3e-44
Identities = 110/380 (28%), Positives = 183/380 (47%), Gaps = 35/380 (9%)
Query: 213 YNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMI 272
Y I N ++DA +L+ MV R P + L+ M + + L +M
Sbjct: 49 YRKISINRLNDLKLDDAVNLFGDMVKSRPFPSIVEFSKLLSAIAKMNKFDLVISLGEQMQ 108
Query: 273 LENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEIN 332
I+ +YT+SIL++ FC+ ++ A +L MK D+VT NSL++G+C I+
Sbjct: 109 NLGISHNLYTYSILINCFCRRSQLSLALAVLAKMMKLGYEPDIVTLNSLLNGFCHGNRIS 168
Query: 333 KAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLI 392
A + M G + ++ T+I+GL + EAV L + M + P++VTY ++
Sbjct: 169 DAVSLVGQMVEMGYQPDSFTFNTLIHGLFRHNRASEAVALVDRMVVKGCQPDLVTYGIVV 228
Query: 393 DGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIR 452
+GL K G I L L+ +M P ++ YN+I+DALC +V+ A+ L T + ++GIR
Sbjct: 229 NGLCKRGDIDLALSLLKKMEQGKIEPGVVIYNTIIDALCNYKNVNDALNLFTEMDNKGIR 288
Query: 453 PDMYTYTVLIKGLC-----------------------------------QSGKLEDAQKV 477
P++ TY LI+ LC + GKL +A+K+
Sbjct: 289 PNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKL 348
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
+++++ + + D++TY+ +I GFC+ D A + M C PN TY +I +
Sbjct: 349 YDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCK 408
Query: 538 KDENDMAEKLLREMIARGLL 557
D +L REM RGL+
Sbjct: 409 AKRVDEGMELFREMSQRGLV 428
Score = 166 bits (419), Expect = 2e-39
Identities = 103/383 (26%), Positives = 187/383 (47%), Gaps = 35/383 (9%)
Query: 64 KNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSL 123
K P ++ + +++ L K ALSL ++ME I +N +I+ +
Sbjct: 215 KGCQPDLVTYGIVVNGLCKRGDIDLALSLLKKMEQGKIEPGVVIYNTIIDALCNYKNVND 274
Query: 124 SFSVFGKILKKGFDPNAITFNTLIKGLC-------------------------------- 151
+ ++F ++ KG PN +T+N+LI+ LC
Sbjct: 275 ALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRLLSDMIERKINPNVVTFSALID 334
Query: 152 ---LKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQP 208
+G + +A +D+++ + D +Y +LING C R+ A + + + K P
Sbjct: 335 AFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFP 394
Query: 209 NAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLL 268
N V YN +I CKAK V++ +L+ +M + + + T +LI+GF + A +
Sbjct: 395 NVVTYNTLIKGFCKAKRVDEGMELFREMSQRGLVGNTVTYTTLIHGFFQARECDNAQIVF 454
Query: 269 HKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLV 328
+M+ + + P + T+SIL+D C GKV+ A ++ + + D+ TYN +++G C
Sbjct: 455 KQMVSDGVLPDIMTYSILLDGLCNNGKVETALVVFEYLQRSKMEPDIYTYNIMIEGMCKA 514
Query: 329 KEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTY 388
++ D+F S++ +GV NV +YTTM++G C+ + +EA LF EM+ +P+ TY
Sbjct: 515 GKVEDGWDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKEEADALFREMKEEGPLPDSGTY 574
Query: 389 NSLIDGLGKLGKISCVLKLVDEM 411
N+LI + G + +L+ EM
Sbjct: 575 NTLIRAHLRDGDKAASAELIREM 597
Score = 145 bits (365), Expect = 4e-33
Identities = 93/366 (25%), Positives = 169/366 (45%), Gaps = 39/366 (10%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P ++ +N I+ +L K+ + AL+L +M+ GI + T+N LI C G S + +
Sbjct: 254 PGVVIYNTIIDALCNYKNVNDALNLFTEMDNKGIRPNVVTYNSLIRCLCNYGRWSDASRL 313
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
++++ +PN +TF+ LI +G + +A +D+++ + D +Y +LING C
Sbjct: 314 LSDMIERKINPNVVTFSALIDAFVKEGKLVEAEKLYDEMIKRSIDPDIFTYSSLINGFCM 373
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLY-------------- 233
R+ A + + + K PN V YN +I CKAK V++ +L+
Sbjct: 374 HDRLDEAKHMFELMISKDCFPNVVTYNTLIKGFCKAKRVDEGMELFREMSQRGLVGNTVT 433
Query: 234 ---------------------SQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMI 272
QMV+ + PD T + L+ G C G+++ A+ + +
Sbjct: 434 YTTLIHGFFQARECDNAQIVFKQMVSDGVLPDIMTYSILLDGLCNNGKVETALVVFEYLQ 493
Query: 273 LENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEIN 332
+ P +YT++I+++ CK GKV++ + K + +VVTY ++M G+C
Sbjct: 494 RSKMEPDIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPNVVTYTTMMSGFCRKGLKE 553
Query: 333 KAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTY---- 388
+A +F M G + + +Y T+I + + L EMR + + + T
Sbjct: 554 EADALFREMKEEGPLPDSGTYNTLIRAHLRDGDKAASAELIREMRSCRFVGDASTIGLVT 613
Query: 389 NSLIDG 394
N L DG
Sbjct: 614 NMLHDG 619
>UniRef100_Q9SXD8 T3P18.15 [Arabidopsis thaliana]
Length = 634
Score = 355 bits (911), Expect = 2e-96
Identities = 193/541 (35%), Positives = 309/541 (56%), Gaps = 36/541 (6%)
Query: 46 HQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDF 105
H + ++ I F ++ + P P I++FNK+LS++ K K +SL ++M+ IV
Sbjct: 62 HDMKLDDAIGLFGGMVKSR-PLPSIVEFNKLLSAIAKMKKFDVVISLGEKMQRLEIVHGL 120
Query: 106 FTFNILINCFSQLGLNSLSFSVFGKILKKGFD---------------------------- 137
+T+NILINCF + SL+ ++ GK++K G++
Sbjct: 121 YTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRISDAVALVDQ 180
Query: 138 -------PNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGR 190
P+ ITF TLI GL L +A+ D++V +G + V+YG ++NGLCK G
Sbjct: 181 MVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVVNGLCKRGD 240
Query: 191 ITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNS 250
AL LL +++ ++ + V++N IID++CK + V+DA +L+ +M K I P+ T +S
Sbjct: 241 TDLALNLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETKGIRPNVVTYSS 300
Query: 251 LIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKD 310
LI C G+ +A LL MI + INP + TF+ L+DAF KEGK EA+ + +K+
Sbjct: 301 LISCLCSYGRWSDASQLLSDMIEKKINPNLVTFNALIDAFVKEGKFVEAEKLYDDMIKRS 360
Query: 311 IILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAV 370
I D+ TYNSL++G+C+ ++KAK +F+ M S+ +V +Y T+I G CK K V++
Sbjct: 361 IDPDIFTYNSLVNGFCMHDRLDKAKQMFEFMVSKDCFPDVVTYNTLIKGFCKSKRVEDGT 420
Query: 371 NLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDAL 430
LF EM R ++ + VTY +LI GL G K+ +M G PP+I+TY+ +LD L
Sbjct: 421 ELFREMSHRGLVGDTVTYTTLIQGLFHDGDCDNAQKVFKQMVSDGVPPDIMTYSILLDGL 480
Query: 431 CKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDV 490
C N ++KA+ + ++ I+ D+Y YT +I+G+C++GK++D +F L +KG +V
Sbjct: 481 CNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNV 540
Query: 491 YTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLRE 550
TY MI G C K L A ALL KM+++G +PN+ TY +I + + + +L+RE
Sbjct: 541 VTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPNSGTYNTLIRAHLRDGDKAASAELIRE 600
Query: 551 M 551
M
Sbjct: 601 M 601
Score = 183 bits (464), Expect = 1e-44
Identities = 111/380 (29%), Positives = 182/380 (47%), Gaps = 35/380 (9%)
Query: 213 YNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMI 272
Y I+ N ++DA L+ MV R P N L+ M + + L KM
Sbjct: 53 YREILRNGLHDMKLDDAIGLFGGMVKSRPLPSIVEFNKLLSAIAKMKKFDVVISLGEKMQ 112
Query: 273 LENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEIN 332
I +YT++IL++ FC+ ++ A +LG MK +VT +SL++GYC K I+
Sbjct: 113 RLEIVHGLYTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRIS 172
Query: 333 KAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLI 392
A + D M G + ++TT+I+GL EAV L + M R PN+VTY ++
Sbjct: 173 DAVALVDQMVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVV 232
Query: 393 DGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIR 452
+GL K G L L+++M +++ +N+I+D+LCK HVD A+ L ++ +GIR
Sbjct: 233 NGLCKRGDTDLALNLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETKGIR 292
Query: 453 PDMYTYTVLIKGLC-----------------------------------QSGKLEDAQKV 477
P++ TY+ LI LC + GK +A+K+
Sbjct: 293 PNVVTYSSLISCLCSYGRWSDASQLLSDMIEKKINPNLVTFNALIDAFVKEGKFVEAEKL 352
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
++D++ + + D++TY ++ GFC+ D A + M C P+ TY +I +
Sbjct: 353 YDDMIKRSIDPDIFTYNSLVNGFCMHDRLDKAKQMFEFMVSKDCFPDVVTYNTLIKGFCK 412
Query: 538 KDENDMAEKLLREMIARGLL 557
+ +L REM RGL+
Sbjct: 413 SKRVEDGTELFREMSHRGLV 432
Score = 137 bits (346), Expect = 6e-31
Identities = 89/355 (25%), Positives = 166/355 (46%), Gaps = 8/355 (2%)
Query: 44 HHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVS 103
+ H D+ NL + K P ++ ++ ++S L S A L M I
Sbjct: 273 YRHVDDALNLFKEMET----KGIRPNVVTYSSLISCLCSYGRWSDASQLLSDMIEKKINP 328
Query: 104 DFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFH 163
+ TFN LI+ F + G + ++ ++K+ DP+ T+N+L+ G C+ + +A
Sbjct: 329 NLVTFNALIDAFVKEGKFVEAEKLYDDMIKRSIDPDIFTYNSLVNGFCMHDRLDKAKQMF 388
Query: 164 DKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKA 223
+ +V++ D V+Y TLI G CK R+ +L + + + + + V Y +I +
Sbjct: 389 EFMVSKDCFPDVVTYNTLIKGFCKSKRVEDGTELFREMSHRGLVGDTVTYTTLIQGLFHD 448
Query: 224 KLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTF 283
++A ++ QMV+ + PD T + L+ G C G+L++A+ + M I +Y +
Sbjct: 449 GDCDNAQKVFKQMVSDGVPPDIMTYSILLDGLCNNGKLEKALEVFDYMQKSEIKLDIYIY 508
Query: 284 SILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMAS 343
+ +++ CK GKV + + K + +VVTYN+++ G C + + +A + M
Sbjct: 509 TTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNVVTYNTMISGLCSKRLLQEAYALLKKMKE 568
Query: 344 RGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTY----NSLIDG 394
G + N +Y T+I + + L EMR + + + T N L DG
Sbjct: 569 DGPLPNSGTYNTLIRAHLRDGDKAASAELIREMRSCRFVGDASTIGLVANMLHDG 623
Score = 57.4 bits (137), Expect = 1e-06
Identities = 47/214 (21%), Positives = 91/214 (41%), Gaps = 2/214 (0%)
Query: 21 FQQSPNPNFIGLFPSSISLYSQLHHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSL 80
F++ + +G + +L L H D +N F ++ P P I+ ++ +L L
Sbjct: 423 FREMSHRGLVGDTVTYTTLIQGLFHDGDCDNAQ-KVFKQMVSDGVP-PDIMTYSILLDGL 480
Query: 81 VKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNA 140
AL + M+ + I D + + +I + G + +F + KG PN
Sbjct: 481 CNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNV 540
Query: 141 ITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKR 200
+T+NT+I GLC K + +A K+ G + +Y TLI + G A+ +L++
Sbjct: 541 VTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPNSGTYNTLIRAHLRDGDKAASAELIRE 600
Query: 201 VDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
+ +A ++ + + +L D+ S
Sbjct: 601 MRSCRFVGDASTIGLVANMLHDGRLDKSFLDMLS 634
>UniRef100_Q8L792 Putative membrane-associated salt-inducible protein [Arabidopsis
thaliana]
Length = 596
Score = 355 bits (911), Expect = 2e-96
Identities = 193/541 (35%), Positives = 309/541 (56%), Gaps = 36/541 (6%)
Query: 46 HQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDF 105
H + ++ I F ++ + P P I++FNK+LS++ K K +SL ++M+ IV
Sbjct: 24 HDMKLDDAIGLFGGMVKSR-PLPSIVEFNKLLSAIAKMKKFDVVISLGEKMQRLEIVHGL 82
Query: 106 FTFNILINCFSQLGLNSLSFSVFGKILKKGFD---------------------------- 137
+T+NILINCF + SL+ ++ GK++K G++
Sbjct: 83 YTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRISDAVALVDQ 142
Query: 138 -------PNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGR 190
P+ ITF TLI GL L +A+ D++V +G + V+YG ++NGLCK G
Sbjct: 143 MVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVVNGLCKRGD 202
Query: 191 ITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNS 250
AL LL +++ ++ + V++N IID++CK + V+DA +L+ +M K I P+ T +S
Sbjct: 203 TDLALNLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETKGIRPNVVTYSS 262
Query: 251 LIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKD 310
LI C G+ +A LL MI + INP + TF+ L+DAF KEGK EA+ + +K+
Sbjct: 263 LISCLCSYGRWSDASQLLSDMIEKKINPNLVTFNALIDAFVKEGKFVEAEKLYDDMIKRS 322
Query: 311 IILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAV 370
I D+ TYNSL++G+C+ ++KAK +F+ M S+ +V +Y T+I G CK K V++
Sbjct: 323 IDPDIFTYNSLVNGFCMHDRLDKAKQMFEFMVSKDCFPDVVTYNTLIKGFCKSKRVEDGT 382
Query: 371 NLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDAL 430
LF EM R ++ + VTY +LI GL G K+ +M G PP+I+TY+ +LD L
Sbjct: 383 ELFREMSHRGLVGDTVTYTTLIQGLFHDGDCDNAQKVFKQMVSDGVPPDIMTYSILLDGL 442
Query: 431 CKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDV 490
C N ++KA+ + ++ I+ D+Y YT +I+G+C++GK++D +F L +KG +V
Sbjct: 443 CNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNV 502
Query: 491 YTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLRE 550
TY MI G C K L A ALL KM+++G +PN+ TY +I + + + +L+RE
Sbjct: 503 VTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPNSGTYNTLIRAHLRDGDKAASAELIRE 562
Query: 551 M 551
M
Sbjct: 563 M 563
Score = 183 bits (464), Expect = 1e-44
Identities = 111/380 (29%), Positives = 182/380 (47%), Gaps = 35/380 (9%)
Query: 213 YNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMI 272
Y I+ N ++DA L+ MV R P N L+ M + + L KM
Sbjct: 15 YREILRNGLHDMKLDDAIGLFGGMVKSRPLPSIVEFNKLLSAIAKMKKFDVVISLGEKMQ 74
Query: 273 LENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEIN 332
I +YT++IL++ FC+ ++ A +LG MK +VT +SL++GYC K I+
Sbjct: 75 RLEIVHGLYTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRIS 134
Query: 333 KAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLI 392
A + D M G + ++TT+I+GL EAV L + M R PN+VTY ++
Sbjct: 135 DAVALVDQMVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVV 194
Query: 393 DGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIR 452
+GL K G L L+++M +++ +N+I+D+LCK HVD A+ L ++ +GIR
Sbjct: 195 NGLCKRGDTDLALNLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETKGIR 254
Query: 453 PDMYTYTVLIKGLC-----------------------------------QSGKLEDAQKV 477
P++ TY+ LI LC + GK +A+K+
Sbjct: 255 PNVVTYSSLISCLCSYGRWSDASQLLSDMIEKKINPNLVTFNALIDAFVKEGKFVEAEKL 314
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
++D++ + + D++TY ++ GFC+ D A + M C P+ TY +I +
Sbjct: 315 YDDMIKRSIDPDIFTYNSLVNGFCMHDRLDKAKQMFEFMVSKDCFPDVVTYNTLIKGFCK 374
Query: 538 KDENDMAEKLLREMIARGLL 557
+ +L REM RGL+
Sbjct: 375 SKRVEDGTELFREMSHRGLV 394
Score = 137 bits (346), Expect = 6e-31
Identities = 89/355 (25%), Positives = 166/355 (46%), Gaps = 8/355 (2%)
Query: 44 HHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVS 103
+ H D+ NL + K P ++ ++ ++S L S A L M I
Sbjct: 235 YRHVDDALNLFKEMET----KGIRPNVVTYSSLISCLCSYGRWSDASQLLSDMIEKKINP 290
Query: 104 DFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFH 163
+ TFN LI+ F + G + ++ ++K+ DP+ T+N+L+ G C+ + +A
Sbjct: 291 NLVTFNALIDAFVKEGKFVEAEKLYDDMIKRSIDPDIFTYNSLVNGFCMHDRLDKAKQMF 350
Query: 164 DKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKA 223
+ +V++ D V+Y TLI G CK R+ +L + + + + + V Y +I +
Sbjct: 351 EFMVSKDCFPDVVTYNTLIKGFCKSKRVEDGTELFREMSHRGLVGDTVTYTTLIQGLFHD 410
Query: 224 KLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTF 283
++A ++ QMV+ + PD T + L+ G C G+L++A+ + M I +Y +
Sbjct: 411 GDCDNAQKVFKQMVSDGVPPDIMTYSILLDGLCNNGKLEKALEVFDYMQKSEIKLDIYIY 470
Query: 284 SILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMAS 343
+ +++ CK GKV + + K + +VVTYN+++ G C + + +A + M
Sbjct: 471 TTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNVVTYNTMISGLCSKRLLQEAYALLKKMKE 530
Query: 344 RGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTY----NSLIDG 394
G + N +Y T+I + + L EMR + + + T N L DG
Sbjct: 531 DGPLPNSGTYNTLIRAHLRDGDKAASAELIREMRSCRFVGDASTIGLVANMLHDG 585
Score = 57.4 bits (137), Expect = 1e-06
Identities = 47/214 (21%), Positives = 91/214 (41%), Gaps = 2/214 (0%)
Query: 21 FQQSPNPNFIGLFPSSISLYSQLHHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSL 80
F++ + +G + +L L H D +N F ++ P P I+ ++ +L L
Sbjct: 385 FREMSHRGLVGDTVTYTTLIQGLFHDGDCDNAQ-KVFKQMVSDGVP-PDIMTYSILLDGL 442
Query: 81 VKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNA 140
AL + M+ + I D + + +I + G + +F + KG PN
Sbjct: 443 CNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNV 502
Query: 141 ITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKR 200
+T+NT+I GLC K + +A K+ G + +Y TLI + G A+ +L++
Sbjct: 503 VTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPNSGTYNTLIRAHLRDGDKAASAELIRE 562
Query: 201 VDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
+ +A ++ + + +L D+ S
Sbjct: 563 MRSCRFVGDASTIGLVANMLHDGRLDKSFLDMLS 596
>UniRef100_Q9LN78 T12C24.22 [Arabidopsis thaliana]
Length = 1245
Score = 353 bits (905), Expect = 1e-95
Identities = 176/507 (34%), Positives = 300/507 (58%), Gaps = 1/507 (0%)
Query: 49 EENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTF 108
++++ I+ F ++ + P P ++ F++ S++ + K + L +Q+ELNGI + +T
Sbjct: 58 KKDDAIALFQEMIRSR-PLPSLVDFSRFFSAIARTKQFNLVLDFCKQLELNGIAHNIYTL 116
Query: 109 NILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVA 168
NI+INCF + ++SV GK++K G++P+ TFNTLIKGL L+G + +A+ D++V
Sbjct: 117 NIMINCFCRCCKTCFAYSVLGKVMKLGYEPDTTTFNTLIKGLFLEGKVSEAVVLVDRMVE 176
Query: 169 QGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVND 228
G D V+Y +++NG+C+ G + AL LL++++ + V+ + Y+ IID++C+ ++
Sbjct: 177 NGCQPDVVTYNSIVNGICRSGDTSLALDLLRKMEERNVKADVFTYSTIIDSLCRDGCIDA 236
Query: 229 AFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVD 288
A L+ +M K I T NSL+ G C G+ + LL M+ I P + TF++L+D
Sbjct: 237 AISLFKEMETKGIKSSVVTYNSLVRGLCKAGKWNDGALLLKDMVSREIVPNVITFNVLLD 296
Query: 289 AFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIA 348
F KEGK++EA + + + I +++TYN+LMDGYC+ +++A ++ D M
Sbjct: 297 VFVKEGKLQEANELYKEMITRGISPNIITYNTLMDGYCMQNRLSEANNMLDLMVRNKCSP 356
Query: 349 NVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLV 408
++ ++T++I G C +K VD+ + +F + R ++ N VTY+ L+ G + GKI +L
Sbjct: 357 DIVTFTSLIKGYCMVKRVDDGMKVFRNISKRGLVANAVTYSILVQGFCQSGKIKLAEELF 416
Query: 409 DEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQS 468
EM G P+++TY +LD LC N ++KA+ + +L+ + + YT +I+G+C+
Sbjct: 417 QEMVSHGVLPDVMTYGILLDGLCDNGKLEKALEIFEDLQKSKMDLGIVMYTTIIEGMCKG 476
Query: 469 GKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTY 528
GK+EDA +F L KG +V TYTVMI G C KG A LL KME++G PN TY
Sbjct: 477 GKVEDAWNLFCSLPCKGVKPNVMTYTVMISGLCKKGSLSEANILLRKMEEDGNAPNDCTY 536
Query: 529 EIVILSLFEKDENDMAEKLLREMIARG 555
+I + + + KL+ EM + G
Sbjct: 537 NTLIRAHLRDGDLTASAKLIEEMKSCG 563
Score = 190 bits (483), Expect = 8e-47
Identities = 108/350 (30%), Positives = 188/350 (52%)
Query: 208 PNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGL 267
P+ V ++ + + K N D Q+ I+ + +T N +I FC + A +
Sbjct: 76 PSLVDFSRFFSAIARTKQFNLVLDFCKQLELNGIAHNIYTLNIMINCFCRCCKTCFAYSV 135
Query: 268 LHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCL 327
L K++ P TF+ L+ EGKV EA +++ ++ DVVTYNS+++G C
Sbjct: 136 LGKVMKLGYEPDTTTFNTLIKGLFLEGKVSEAVVLVDRMVENGCQPDVVTYNSIVNGICR 195
Query: 328 VKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVT 387
+ + A D+ M R V A+V +Y+T+I+ LC+ +D A++LF+EM + I +VVT
Sbjct: 196 SGDTSLALDLLRKMEERNVKADVFTYSTIIDSLCRDGCIDAAISLFKEMETKGIKSSVVT 255
Query: 388 YNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLK 447
YNSL+ GL K GK + L+ +M R PN+IT+N +LD K + +A L +
Sbjct: 256 YNSLVRGLCKAGKWNDGALLLKDMVSREIVPNVITFNVLLDVFVKEGKLQEANELYKEMI 315
Query: 448 DQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFD 507
+GI P++ TY L+ G C +L +A + + ++ + D+ T+T +I+G+C+ D
Sbjct: 316 TRGISPNIITYNTLMDGYCMQNRLSEANNMLDLMVRNKCSPDIVTFTSLIKGYCMVKRVD 375
Query: 508 AALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGLL 557
+ + + G + NA TY I++ + + +AE+L +EM++ G+L
Sbjct: 376 DGMKVFRNISKRGLVANAVTYSILVQGFCQSGKIKLAEELFQEMVSHGVL 425
Score = 134 bits (338), Expect = 5e-30
Identities = 117/478 (24%), Positives = 202/478 (41%), Gaps = 65/478 (13%)
Query: 67 TPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFS 126
+P II +N ++ S A ++ M N D TF LI + +
Sbjct: 320 SPNIITYNTLMDGYCMQNRLSEANNMLDLMVRNKCSPDIVTFTSLIKGYCMVKRVDDGMK 379
Query: 127 VFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLC 186
VF I K+G NA+T++ L++G C G I A ++V+ G D ++YG L++GLC
Sbjct: 380 VFRNISKRGLVANAVTYSILVQGFCQSGKIKLAEELFQEMVSHGVLPDVMTYGILLDGLC 439
Query: 187 KVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDF 246
G++ AL++ + + + VMY II+ MCK V DA++L+ + K + P+
Sbjct: 440 DNGKLEKALEIFEDLQKSKMDLGIVMYTTIIEGMCKGGKVEDAWNLFCSLPCKGVKPNVM 499
Query: 247 TCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMML--- 303
T +I G C G L EA LL KM + P T++ L+ A ++G + + ++
Sbjct: 500 TYTVMISGLCKKGSLSEANILLRKMEEDGNAPNDCTYNTLIRAHLRDGDLTASAKLIEEM 559
Query: 304 ----------GVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSY 353
+ M D++L + +L YCL K +D+ + S + + ++
Sbjct: 560 KSCGFSADASSIKMVIDMLLSAMKRLTLR--YCLSKGSKSRQDLLELSGSEKIRLSSLTF 617
Query: 354 ---------TTMING-------------------------LCKIKMVDEAVNL------- 372
TT +N L K K D V +
Sbjct: 618 VKMFPCNTITTSLNVNTIEARGMNSAELNRDLRKLRRSSVLKKFKNRDVRVLVTNELLTW 677
Query: 373 -FEEMRCRKIIP-----NVVTYNSLIDGLGKLGKISCVL---KLVDEMHDRGQPPNIITY 423
E+ C ++ + V Y + + G+ V+ +L EM RG PN ITY
Sbjct: 678 GLEDAECDLMVDLELPTDAVHYAHRAGRMRRPGRKMTVVTAEELHKEMIQRGIAPNTITY 737
Query: 424 NSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDL 481
+S++D CK + +D+A +L + +G Y L++ + + E + + +DL
Sbjct: 738 SSLIDGFCKENRLDEANQMLDLMVTKGDSDIRYLLAGLMRKKRKGSETEGWENLPDDL 795
Score = 94.4 bits (233), Expect = 8e-18
Identities = 48/221 (21%), Positives = 108/221 (48%)
Query: 337 IFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLG 396
+F M + ++ ++ + + + K + ++ +++ I N+ T N +I+
Sbjct: 65 LFQEMIRSRPLPSLVDFSRFFSAIARTKQFNLVLDFCKQLELNGIAHNIYTLNIMINCFC 124
Query: 397 KLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMY 456
+ K ++ ++ G P+ T+N+++ L V +A+ L+ + + G +PD+
Sbjct: 125 RCCKTCFAYSVLGKVMKLGYEPDTTTFNTLIKGLFLEGKVSEAVVLVDRMVENGCQPDVV 184
Query: 457 TYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKM 516
TY ++ G+C+SG A + + + DV+TY+ +I C G DAA++L +M
Sbjct: 185 TYNSIVNGICRSGDTSLALDLLRKMEERNVKADVFTYSTIIDSLCRDGCIDAAISLFKEM 244
Query: 517 EDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGLL 557
E G + TY ++ L + + + LL++M++R ++
Sbjct: 245 ETKGIKSSVVTYNSLVRGLCKAGKWNDGALLLKDMVSREIV 285
Score = 94.0 bits (232), Expect = 1e-17
Identities = 59/199 (29%), Positives = 98/199 (48%), Gaps = 1/199 (0%)
Query: 358 NGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQP 417
+G+ IK D+A+ LF+EM + +P++V ++ + + + + VL ++ G
Sbjct: 52 SGIVDIKK-DDAIALFQEMIRSRPLPSLVDFSRFFSAIARTKQFNLVLDFCKQLELNGIA 110
Query: 418 PNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKV 477
NI T N +++ C+ A ++L + G PD T+ LIKGL GK+ +A +
Sbjct: 111 HNIYTLNIMINCFCRCCKTCFAYSVLGKVMKLGYEPDTTTFNTLIKGLFLEGKVSEAVVL 170
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
+ ++ G DV TY ++ G C G AL LL KME+ + TY +I SL
Sbjct: 171 VDRMVENGCQPDVVTYNSIVNGICRSGDTSLALDLLRKMEERNVKADVFTYSTIIDSLCR 230
Query: 538 KDENDMAEKLLREMIARGL 556
D A L +EM +G+
Sbjct: 231 DGCIDAAISLFKEMETKGI 249
>UniRef100_Q6NQ83 Hypothetical protein At3g22470 [Arabidopsis thaliana]
Length = 619
Score = 352 bits (903), Expect = 2e-95
Identities = 186/517 (35%), Positives = 292/517 (55%), Gaps = 36/517 (6%)
Query: 51 NNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNI 110
N+ I F S++ + P P I FN++ S++ + K + L + MELNGI D +T I
Sbjct: 52 NDAIDLFESMIQSR-PLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNGIEHDMYTMTI 110
Query: 111 LINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTL------------------------ 146
+INC+ + +FSV G+ K G++P+ ITF+TL
Sbjct: 111 MINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRMVEMK 170
Query: 147 -----------IKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAAL 195
I GLCLKG + +AL D++V GF D+V+YG ++N LCK G AL
Sbjct: 171 QRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSALAL 230
Query: 196 QLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGF 255
L ++++ + ++ + V Y+++ID++CK +DA L+++M K I D T +SLI G
Sbjct: 231 DLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGL 290
Query: 256 CIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDV 315
C G+ + +L +MI NI P + TFS L+D F KEGK+ EAK + + + I D
Sbjct: 291 CNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDT 350
Query: 316 VTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEE 375
+TYNSL+DG+C +++A +FD M S+G ++ +Y+ +IN CK K VD+ + LF E
Sbjct: 351 ITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFRE 410
Query: 376 MRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHH 435
+ + +IPN +TYN+L+ G + GK++ +L EM RG PP+++TY +LD LC N
Sbjct: 411 ISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGE 470
Query: 436 VDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTV 495
++KA+ + ++ + + Y ++I G+C + K++DA +F L KG DV TY V
Sbjct: 471 LNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNV 530
Query: 496 MIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVI 532
MI G C KG A L KM+++GC P+ TY I+I
Sbjct: 531 MIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILI 567
Score = 233 bits (594), Expect = 1e-59
Identities = 127/447 (28%), Positives = 242/447 (53%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P ++ + +++ L S AL L +M G D T+ ++N + G ++L+ +
Sbjct: 173 PDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSALALDL 232
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F K+ ++ + + ++ +I LC G AL+ +++ +G D V+Y +LI GLC
Sbjct: 233 FRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGLCN 292
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
G+ ++L+ + G+ + P+ V ++ +ID K + +A +LY++M+ + I+PD T
Sbjct: 293 DGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTIT 352
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTM 307
NSLI GFC L EA + M+ + P + T+SIL++++CK +V + +
Sbjct: 353 YNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFREIS 412
Query: 308 KKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVD 367
K +I + +TYN+L+ G+C ++N AK++F M SRGV +V +Y +++GLC ++
Sbjct: 413 SKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELN 472
Query: 368 EAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSIL 427
+A+ +FE+M+ ++ + YN +I G+ K+ L + D+G P+++TYN ++
Sbjct: 473 KALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNVMI 532
Query: 428 DALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYN 487
LCK + +A L +K+ G PD +TY +LI+ L + ++ E++ V G++
Sbjct: 533 GGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELIEEMKVCGFS 592
Query: 488 LDVYTYTVMIQGFCVKGLFDAALALLS 514
D T ++I + L + L +LS
Sbjct: 593 ADSSTIKMVIDMLSDRRLDKSFLDMLS 619
Score = 173 bits (438), Expect = 1e-41
Identities = 112/411 (27%), Positives = 197/411 (47%), Gaps = 36/411 (8%)
Query: 181 LINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKR 240
L NG+ + ++ A+ L + + P + +N + + + K + M
Sbjct: 42 LRNGIVDI-KVNDAIDLFESMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNG 100
Query: 241 ISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAK 300
I D +T +I +C +L A +L + P TFS LV+ FC EG+V EA
Sbjct: 101 IEHDMYTMTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAV 160
Query: 301 MMLGVTMKKDIILDVVTYNSLMDGYCLVKEINK--------------------------- 333
++ ++ D+VT ++L++G CL +++
Sbjct: 161 ALVDRMVEMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRL 220
Query: 334 --------AKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNV 385
A D+F M R + A+V Y+ +I+ LCK D+A++LF EM + I +V
Sbjct: 221 CKSGNSALALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADV 280
Query: 386 VTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTN 445
VTY+SLI GL GK K++ EM R P+++T+++++D K + +A L
Sbjct: 281 VTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNE 340
Query: 446 LKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGL 505
+ +GI PD TY LI G C+ L +A ++F+ ++ KG D+ TY+++I +C
Sbjct: 341 MITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKR 400
Query: 506 FDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGL 556
D + L ++ G IPN TY ++L + + + A++L +EM++RG+
Sbjct: 401 VDDGMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGV 451
Score = 124 bits (311), Expect = 7e-27
Identities = 68/297 (22%), Positives = 146/297 (48%)
Query: 64 KNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSL 123
+N P ++ F+ ++ VK A L+ +M GI D T+N LI+ F +
Sbjct: 309 RNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTITYNSLIDGFCKENCLHE 368
Query: 124 SFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLIN 183
+ +F ++ KG +P+ +T++ LI C + + ++ ++G + ++Y TL+
Sbjct: 369 ANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLVL 428
Query: 184 GLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISP 243
G C+ G++ AA +L + + + V P+ V Y +++D +C +N A +++ +M R++
Sbjct: 429 GFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELNKALEIFEKMQKSRMTL 488
Query: 244 DDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMML 303
N +I+G C ++ +A L + + + P + T+++++ CK+G + EA M+
Sbjct: 489 GIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNVMIGGLCKKGSLSEADMLF 548
Query: 304 GVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGL 360
+ D TYN L+ + + + ++ + M G A+ + +I+ L
Sbjct: 549 RKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELIEEMKVCGFSADSSTIKMVIDML 605
Score = 110 bits (274), Expect = 1e-22
Identities = 69/260 (26%), Positives = 132/260 (50%), Gaps = 3/260 (1%)
Query: 31 GLFPSSISLYSQLHHHQDEENNL--ISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHST 88
G+ P +I+ Y+ L +EN L + L+ K P I+ ++ +++S KAK
Sbjct: 345 GIAPDTIT-YNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDD 403
Query: 89 ALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIK 148
+ L +++ G++ + T+N L+ F Q G + + +F +++ +G P+ +T+ L+
Sbjct: 404 GMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLD 463
Query: 149 GLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQP 208
GLC G +++AL +K+ L Y +I+G+C ++ A L + K V+P
Sbjct: 464 GLCDNGELNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKP 523
Query: 209 NAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLL 268
+ V YN++I +CK +++A L+ +M +PDDFT N LI L +V L+
Sbjct: 524 DVVTYNVMIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELI 583
Query: 269 HKMILENINPRMYTFSILVD 288
+M + + T +++D
Sbjct: 584 EEMKVCGFSADSSTIKMVID 603
>UniRef100_Q9LJ99 Gb|AAD43616.1 [Arabidopsis thaliana]
Length = 648
Score = 352 bits (903), Expect = 2e-95
Identities = 186/517 (35%), Positives = 292/517 (55%), Gaps = 36/517 (6%)
Query: 51 NNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNI 110
N+ I F S++ + P P I FN++ S++ + K + L + MELNGI D +T I
Sbjct: 81 NDAIDLFESMIQSR-PLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNGIEHDMYTMTI 139
Query: 111 LINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTL------------------------ 146
+INC+ + +FSV G+ K G++P+ ITF+TL
Sbjct: 140 MINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAVALVDRMVEMK 199
Query: 147 -----------IKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAAL 195
I GLCLKG + +AL D++V GF D+V+YG ++N LCK G AL
Sbjct: 200 QRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSALAL 259
Query: 196 QLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGF 255
L ++++ + ++ + V Y+++ID++CK +DA L+++M K I D T +SLI G
Sbjct: 260 DLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGL 319
Query: 256 CIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDV 315
C G+ + +L +MI NI P + TFS L+D F KEGK+ EAK + + + I D
Sbjct: 320 CNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDT 379
Query: 316 VTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEE 375
+TYNSL+DG+C +++A +FD M S+G ++ +Y+ +IN CK K VD+ + LF E
Sbjct: 380 ITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFRE 439
Query: 376 MRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHH 435
+ + +IPN +TYN+L+ G + GK++ +L EM RG PP+++TY +LD LC N
Sbjct: 440 ISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGE 499
Query: 436 VDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTV 495
++KA+ + ++ + + Y ++I G+C + K++DA +F L KG DV TY V
Sbjct: 500 LNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNV 559
Query: 496 MIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVI 532
MI G C KG A L KM+++GC P+ TY I+I
Sbjct: 560 MIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILI 596
Score = 233 bits (594), Expect = 1e-59
Identities = 127/447 (28%), Positives = 242/447 (53%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P ++ + +++ L S AL L +M G D T+ ++N + G ++L+ +
Sbjct: 202 PDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRLCKSGNSALALDL 261
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F K+ ++ + + ++ +I LC G AL+ +++ +G D V+Y +LI GLC
Sbjct: 262 FRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADVVTYSSLIGGLCN 321
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
G+ ++L+ + G+ + P+ V ++ +ID K + +A +LY++M+ + I+PD T
Sbjct: 322 DGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTIT 381
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTM 307
NSLI GFC L EA + M+ + P + T+SIL++++CK +V + +
Sbjct: 382 YNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFREIS 441
Query: 308 KKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVD 367
K +I + +TYN+L+ G+C ++N AK++F M SRGV +V +Y +++GLC ++
Sbjct: 442 SKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELN 501
Query: 368 EAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSIL 427
+A+ +FE+M+ ++ + YN +I G+ K+ L + D+G P+++TYN ++
Sbjct: 502 KALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNVMI 561
Query: 428 DALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYN 487
LCK + +A L +K+ G PD +TY +LI+ L + ++ E++ V G++
Sbjct: 562 GGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELIEEMKVCGFS 621
Query: 488 LDVYTYTVMIQGFCVKGLFDAALALLS 514
D T ++I + L + L +LS
Sbjct: 622 ADSSTIKMVIDMLSDRRLDKSFLDMLS 648
Score = 173 bits (438), Expect = 1e-41
Identities = 112/411 (27%), Positives = 197/411 (47%), Gaps = 36/411 (8%)
Query: 181 LINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKR 240
L NG+ + ++ A+ L + + P + +N + + + K + M
Sbjct: 71 LRNGIVDI-KVNDAIDLFESMIQSRPLPTPIDFNRLCSAVARTKQYDLVLGFCKGMELNG 129
Query: 241 ISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAK 300
I D +T +I +C +L A +L + P TFS LV+ FC EG+V EA
Sbjct: 130 IEHDMYTMTIMINCYCRKKKLLFAFSVLGRAWKLGYEPDTITFSTLVNGFCLEGRVSEAV 189
Query: 301 MMLGVTMKKDIILDVVTYNSLMDGYCLVKEINK--------------------------- 333
++ ++ D+VT ++L++G CL +++
Sbjct: 190 ALVDRMVEMKQRPDLVTVSTLINGLCLKGRVSEALVLIDRMVEYGFQPDEVTYGPVLNRL 249
Query: 334 --------AKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNV 385
A D+F M R + A+V Y+ +I+ LCK D+A++LF EM + I +V
Sbjct: 250 CKSGNSALALDLFRKMEERNIKASVVQYSIVIDSLCKDGSFDDALSLFNEMEMKGIKADV 309
Query: 386 VTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTN 445
VTY+SLI GL GK K++ EM R P+++T+++++D K + +A L
Sbjct: 310 VTYSSLIGGLCNDGKWDDGAKMLREMIGRNIIPDVVTFSALIDVFVKEGKLLEAKELYNE 369
Query: 446 LKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGL 505
+ +GI PD TY LI G C+ L +A ++F+ ++ KG D+ TY+++I +C
Sbjct: 370 MITRGIAPDTITYNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKR 429
Query: 506 FDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGL 556
D + L ++ G IPN TY ++L + + + A++L +EM++RG+
Sbjct: 430 VDDGMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGV 480
Score = 124 bits (311), Expect = 7e-27
Identities = 68/297 (22%), Positives = 146/297 (48%)
Query: 64 KNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSL 123
+N P ++ F+ ++ VK A L+ +M GI D T+N LI+ F +
Sbjct: 338 RNIIPDVVTFSALIDVFVKEGKLLEAKELYNEMITRGIAPDTITYNSLIDGFCKENCLHE 397
Query: 124 SFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLIN 183
+ +F ++ KG +P+ +T++ LI C + + ++ ++G + ++Y TL+
Sbjct: 398 ANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDDGMRLFREISSKGLIPNTITYNTLVL 457
Query: 184 GLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISP 243
G C+ G++ AA +L + + + V P+ V Y +++D +C +N A +++ +M R++
Sbjct: 458 GFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLDGLCDNGELNKALEIFEKMQKSRMTL 517
Query: 244 DDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMML 303
N +I+G C ++ +A L + + + P + T+++++ CK+G + EA M+
Sbjct: 518 GIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKPDVVTYNVMIGGLCKKGSLSEADMLF 577
Query: 304 GVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGL 360
+ D TYN L+ + + + ++ + M G A+ + +I+ L
Sbjct: 578 RKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELIEEMKVCGFSADSSTIKMVIDML 634
Score = 110 bits (274), Expect = 1e-22
Identities = 69/260 (26%), Positives = 132/260 (50%), Gaps = 3/260 (1%)
Query: 31 GLFPSSISLYSQLHHHQDEENNL--ISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHST 88
G+ P +I+ Y+ L +EN L + L+ K P I+ ++ +++S KAK
Sbjct: 374 GIAPDTIT-YNSLIDGFCKENCLHEANQMFDLMVSKGCEPDIVTYSILINSYCKAKRVDD 432
Query: 89 ALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIK 148
+ L +++ G++ + T+N L+ F Q G + + +F +++ +G P+ +T+ L+
Sbjct: 433 GMRLFREISSKGLIPNTITYNTLVLGFCQSGKLNAAKELFQEMVSRGVPPSVVTYGILLD 492
Query: 149 GLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQP 208
GLC G +++AL +K+ L Y +I+G+C ++ A L + K V+P
Sbjct: 493 GLCDNGELNKALEIFEKMQKSRMTLGIGIYNIIIHGMCNASKVDDAWSLFCSLSDKGVKP 552
Query: 209 NAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLL 268
+ V YN++I +CK +++A L+ +M +PDDFT N LI L +V L+
Sbjct: 553 DVVTYNVMIGGLCKKGSLSEADMLFRKMKEDGCTPDDFTYNILIRAHLGGSGLISSVELI 612
Query: 269 HKMILENINPRMYTFSILVD 288
+M + + T +++D
Sbjct: 613 EEMKVCGFSADSSTIKMVID 632
>UniRef100_Q9CAN5 Hypothetical protein F16M19.17 [Arabidopsis thaliana]
Length = 614
Score = 352 bits (903), Expect = 2e-95
Identities = 187/554 (33%), Positives = 312/554 (55%), Gaps = 39/554 (7%)
Query: 33 FPSSISLYSQLHHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSL 92
+ +S + LH DE +L + P P I++F+K+LS++ K K +S
Sbjct: 32 YREKLSRNALLHLKLDEAVDLFGE----MVKSRPFPSIVEFSKLLSAIAKMKKFDLVISF 87
Query: 93 HQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCL 152
++ME+ G+ + +T+NI+INC + S + ++ GK++K G+ P+ +T N+L+ G C
Sbjct: 88 GEKMEILGVSHNLYTYNIMINCLCRRSQLSFALAILGKMMKLGYGPSIVTLNSLLNGFCH 147
Query: 153 KGHIHQALNFHDKVVAQGFHLDQVS----------------------------------- 177
I +A+ D++V G+ D V+
Sbjct: 148 GNRISEAVALVDQMVEMGYQPDTVTFTTLVHGLFQHNKASEAVALVERMVVKGCQPDLVT 207
Query: 178 YGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMV 237
YG +INGLCK G AL LL +++ ++ + V+Y+ +ID++CK + V+DA +L+++M
Sbjct: 208 YGAVINGLCKRGEPDLALNLLNKMEKGKIEADVVIYSTVIDSLCKYRHVDDALNLFTEMD 267
Query: 238 AKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVK 297
K I PD FT +SLI C G+ +A LL M+ INP + TF+ L+DAF KEGK+
Sbjct: 268 NKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMLERKINPNVVTFNSLIDAFAKEGKLI 327
Query: 298 EAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMI 357
EA+ + +++ I ++VTYNSL++G+C+ +++A+ IF M S+ + +V +Y T+I
Sbjct: 328 EAEKLFDEMIQRSIDPNIVTYNSLINGFCMHDRLDEAQQIFTLMVSKDCLPDVVTYNTLI 387
Query: 358 NGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQP 417
NG CK K V + + LF +M R ++ N VTY +LI G + + +M G
Sbjct: 388 NGFCKAKKVVDGMELFRDMSRRGLVGNTVTYTTLIHGFFQASDCDNAQMVFKQMVSDGVH 447
Query: 418 PNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKV 477
PNI+TYN++LD LCKN ++KA+ + L+ + PD+YTY ++ +G+C++GK+ED +
Sbjct: 448 PNIMTYNTLLDGLCKNGKLEKAMVVFEYLQKSKMEPDIYTYNIMSEGMCKAGKVEDGWDL 507
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
F L +KG DV Y MI GFC KGL + A L KM+++G +P++ TY +I +
Sbjct: 508 FCSLSLKGVKPDVIAYNTMISGFCKKGLKEEAYTLFIKMKEDGPLPDSGTYNTLIRAHLR 567
Query: 538 KDENDMAEKLLREM 551
+ + +L++EM
Sbjct: 568 DGDKAASAELIKEM 581
Score = 194 bits (494), Expect = 4e-48
Identities = 108/361 (29%), Positives = 193/361 (52%)
Query: 70 IIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFG 129
++ ++ ++ SL K +H AL+L +M+ GI D FT++ LI+C G S + +
Sbjct: 240 VVIYSTVIDSLCKYRHVDDALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLS 299
Query: 130 KILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVG 189
+L++ +PN +TFN+LI +G + +A D+++ + + V+Y +LING C
Sbjct: 300 DMLERKINPNVVTFNSLIDAFAKEGKLIEAEKLFDEMIQRSIDPNIVTYNSLINGFCMHD 359
Query: 190 RITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCN 249
R+ A Q+ + K P+ V YN +I+ CKAK V D +L+ M + + + T
Sbjct: 360 RLDEAQQIFTLMVSKDCLPDVVTYNTLINGFCKAKKVVDGMELFRDMSRRGLVGNTVTYT 419
Query: 250 SLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKK 309
+LI+GF A + +M+ + ++P + T++ L+D CK GK+++A ++ K
Sbjct: 420 TLIHGFFQASDCDNAQMVFKQMVSDGVHPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQKS 479
Query: 310 DIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEA 369
+ D+ TYN + +G C ++ D+F S++ +GV +V +Y TMI+G CK + +EA
Sbjct: 480 KMEPDIYTYNIMSEGMCKAGKVEDGWDLFCSLSLKGVKPDVIAYNTMISGFCKKGLKEEA 539
Query: 370 VNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDA 429
LF +M+ +P+ TYN+LI + G + +L+ EM + TY + D
Sbjct: 540 YTLFIKMKEDGPLPDSGTYNTLIRAHLRDGDKAASAELIKEMRSCRFAGDASTYGLVTDM 599
Query: 430 L 430
L
Sbjct: 600 L 600
Score = 176 bits (447), Expect = 1e-42
Identities = 118/480 (24%), Positives = 229/480 (47%), Gaps = 39/480 (8%)
Query: 30 IGLFPSSISLYSQLHH--HQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHS 87
+G PS ++L S L+ H + + ++ + ++ P + F ++ L + S
Sbjct: 129 LGYGPSIVTLNSLLNGFCHGNRISEAVALVDQMVE-MGYQPDTVTFTTLVHGLFQHNKAS 187
Query: 88 TALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLI 147
A++L ++M + G D T+ +IN + G L+ ++ K+ K + + + ++T+I
Sbjct: 188 EAVALVERMVVKGCQPDLVTYGAVINGLCKRGEPDLALNLLNKMEKGKIEADVVIYSTVI 247
Query: 148 KGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQ 207
LC H+ ALN ++ +G D +Y +LI+ LC GR + A +LL + + +
Sbjct: 248 DSLCKYRHVDDALNLFTEMDNKGIRPDVFTYSSLISCLCNYGRWSDASRLLSDMLERKIN 307
Query: 208 PNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGL 267
PN V +N +ID K + +A L+ +M+ + I P+ T NSLI GFC+ +L EA +
Sbjct: 308 PNVVTFNSLIDAFAKEGKLIEAEKLFDEMIQRSIDPNIVTYNSLINGFCMHDRLDEAQQI 367
Query: 268 LHKMILENINPRMYTFSILVDAFCKEGKVKE----------------------------- 298
M+ ++ P + T++ L++ FCK KV +
Sbjct: 368 FTLMVSKDCLPDVVTYNTLINGFCKAKKVVDGMELFRDMSRRGLVGNTVTYTTLIHGFFQ 427
Query: 299 ------AKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQS 352
A+M+ + + +++TYN+L+DG C ++ KA +F+ + + ++ +
Sbjct: 428 ASDCDNAQMVFKQMVSDGVHPNIMTYNTLLDGLCKNGKLEKAMVVFEYLQKSKMEPDIYT 487
Query: 353 YTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMH 412
Y M G+CK V++ +LF + + + P+V+ YN++I G K G L +M
Sbjct: 488 YNIMSEGMCKAGKVEDGWDLFCSLSLKGVKPDVIAYNTMISGFCKKGLKEEAYTLFIKMK 547
Query: 413 DRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLE 472
+ G P+ TYN+++ A ++ + L+ ++ D TY L+ + G+L+
Sbjct: 548 EDGPLPDSGTYNTLIRAHLRDGDKAASAELIKEMRSCRFAGDASTYG-LVTDMLHDGRLD 606
>UniRef100_Q9LPX2 F13K23.2 protein [Arabidopsis thaliana]
Length = 644
Score = 351 bits (900), Expect = 4e-95
Identities = 182/540 (33%), Positives = 302/540 (55%), Gaps = 36/540 (6%)
Query: 51 NNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNI 110
++ + F ++ + P P +I FN++ S++ K K + L+L +QME GI +T +I
Sbjct: 70 DDAVDLFRDMIQSR-PLPTVIDFNRLFSAIAKTKQYELVLALCKQMESKGIAHSIYTLSI 128
Query: 111 LINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQG 170
+INCF + S +FS GKI+K G++P+ + FNTL+ GLCL+ + +AL D++V G
Sbjct: 129 MINCFCRCRKLSYAFSTMGKIMKLGYEPDTVIFNTLLNGLCLECRVSEALELVDRMVEMG 188
Query: 171 -----------------------------------FHLDQVSYGTLINGLCKVGRITAAL 195
F ++V+YG ++N +CK G+ A+
Sbjct: 189 HKPTLITLNTLVNGLCLNGKVSDAVVLIDRMVETGFQPNEVTYGPVLNVMCKSGQTALAM 248
Query: 196 QLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGF 255
+LL++++ + ++ +AV Y++IID +CK +++AF+L+++M K D T N+LI GF
Sbjct: 249 ELLRKMEERNIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIITYNTLIGGF 308
Query: 256 CIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDV 315
C G+ + LL MI I+P + TFS+L+D+F KEGK++EA +L M++ I +
Sbjct: 309 CNAGRWDDGAKLLRDMIKRKISPNVVTFSVLIDSFVKEGKLREADQLLKEMMQRGIAPNT 368
Query: 316 VTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEE 375
+TYNSL+DG+C + +A + D M S+G ++ ++ +ING CK +D+ + LF E
Sbjct: 369 ITYNSLIDGFCKENRLEEAIQMVDLMISKGCDPDIMTFNILINGYCKANRIDDGLELFRE 428
Query: 376 MRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHH 435
M R +I N VTYN+L+ G + GK+ KL EM R P+I++Y +LD LC N
Sbjct: 429 MSLRGVIANTVTYNTLVQGFCQSGKLEVAKKLFQEMVSRRVRPDIVSYKILLDGLCDNGE 488
Query: 436 VDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTV 495
++KA+ + ++ + D+ Y ++I G+C + K++DA +F L +KG LD Y +
Sbjct: 489 LEKALEIFGKIEKSKMELDIGIYMIIIHGMCNASKVDDAWDLFCSLPLKGVKLDARAYNI 548
Query: 496 MIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARG 555
MI C K A L KM + G P+ TY I+I + D+ A +L+ EM + G
Sbjct: 549 MISELCRKDSLSKADILFRKMTEEGHAPDELTYNILIRAHLGDDDATTAAELIEEMKSSG 608
Score = 211 bits (538), Expect = 3e-53
Identities = 120/447 (26%), Positives = 230/447 (50%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P +I N +++ L S A+ L +M G + T+ ++N + G +L+ +
Sbjct: 191 PTLITLNTLVNGLCLNGKVSDAVVLIDRMVETGFQPNEVTYGPVLNVMCKSGQTALAMEL 250
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
K+ ++ +A+ ++ +I GLC G + A N +++ +GF D ++Y TLI G C
Sbjct: 251 LRKMEERNIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIITYNTLIGGFCN 310
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
GR +LL+ + + + PN V ++++ID+ K + +A L +M+ + I+P+ T
Sbjct: 311 AGRWDDGAKLLRDMIKRKISPNVVTFSVLIDSFVKEGKLREADQLLKEMMQRGIAPNTIT 370
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTM 307
NSLI GFC +L+EA+ ++ MI + +P + TF+IL++ +CK ++ + +
Sbjct: 371 YNSLIDGFCKENRLEEAIQMVDLMISKGCDPDIMTFNILINGYCKANRIDDGLELFREMS 430
Query: 308 KKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVD 367
+ +I + VTYN+L+ G+C ++ AK +F M SR V ++ SY +++GLC ++
Sbjct: 431 LRGVIANTVTYNTLVQGFCQSGKLEVAKKLFQEMVSRRVRPDIVSYKILLDGLCDNGELE 490
Query: 368 EAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSIL 427
+A+ +F ++ K+ ++ Y +I G+ K+ L + +G + YN ++
Sbjct: 491 KALEIFGKIEKSKMELDIGIYMIIIHGMCNASKVDDAWDLFCSLPLKGVKLDARAYNIMI 550
Query: 428 DALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYN 487
LC+ + KA L + ++G PD TY +LI+ A ++ E++ G+
Sbjct: 551 SELCRKDSLSKADILFRKMTEEGHAPDELTYNILIRAHLGDDDATTAAELIEEMKSSGFP 610
Query: 488 LDVYTYTVMIQGFCVKGLFDAALALLS 514
DV T ++I L + L +LS
Sbjct: 611 ADVSTVKMVINMLSSGELDKSFLDMLS 637
Score = 162 bits (410), Expect = 2e-38
Identities = 97/350 (27%), Positives = 174/350 (49%)
Query: 208 PNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGL 267
P + +N + + K K L QM +K I+ +T + +I FC +L A
Sbjct: 86 PTVIDFNRLFSAIAKTKQYELVLALCKQMESKGIAHSIYTLSIMINCFCRCRKLSYAFST 145
Query: 268 LHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCL 327
+ K++ P F+ L++ C E +V EA ++ ++ ++T N+L++G CL
Sbjct: 146 MGKIMKLGYEPDTVIFNTLLNGLCLECRVSEALELVDRMVEMGHKPTLITLNTLVNGLCL 205
Query: 328 VKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVT 387
+++ A + D M G N +Y ++N +CK A+ L +M R I + V
Sbjct: 206 NGKVSDAVVLIDRMVETGFQPNEVTYGPVLNVMCKSGQTALAMELLRKMEERNIKLDAVK 265
Query: 388 YNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLK 447
Y+ +IDGL K G + L +EM +G +IITYN+++ C D LL ++
Sbjct: 266 YSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIITYNTLIGGFCNAGRWDDGAKLLRDMI 325
Query: 448 DQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFD 507
+ I P++ T++VLI + GKL +A ++ ++++ +G + TY +I GFC + +
Sbjct: 326 KRKISPNVVTFSVLIDSFVKEGKLREADQLLKEMMQRGIAPNTITYNSLIDGFCKENRLE 385
Query: 508 AALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGLL 557
A+ ++ M GC P+ T+ I+I + + D +L REM RG++
Sbjct: 386 EAIQMVDLMISKGCDPDIMTFNILINGYCKANRIDDGLELFREMSLRGVI 435
Score = 120 bits (301), Expect = 1e-25
Identities = 69/294 (23%), Positives = 144/294 (48%)
Query: 67 TPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFS 126
+P ++ F+ ++ S VK A L ++M GI + T+N LI+ F + +
Sbjct: 330 SPNVVTFSVLIDSFVKEGKLREADQLLKEMMQRGIAPNTITYNSLIDGFCKENRLEEAIQ 389
Query: 127 VFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLC 186
+ ++ KG DP+ +TFN LI G C I L ++ +G + V+Y TL+ G C
Sbjct: 390 MVDLMISKGCDPDIMTFNILINGYCKANRIDDGLELFREMSLRGVIANTVTYNTLVQGFC 449
Query: 187 KVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDF 246
+ G++ A +L + + + V+P+ V Y +++D +C + A +++ ++ ++ D
Sbjct: 450 QSGKLEVAKKLFQEMVSRRVRPDIVSYKILLDGLCDNGELEKALEIFGKIEKSKMELDIG 509
Query: 247 TCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVT 306
+I+G C ++ +A L + L+ + ++I++ C++ + +A ++
Sbjct: 510 IYMIIIHGMCNASKVDDAWDLFCSLPLKGVKLDARAYNIMISELCRKDSLSKADILFRKM 569
Query: 307 MKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGL 360
++ D +TYN L+ + + A ++ + M S G A+V + +IN L
Sbjct: 570 TEEGHAPDELTYNILIRAHLGDDDATTAAELIEEMKSSGFPADVSTVKMVINML 623
>UniRef100_Q9ASZ8 At1g12620/T12C24_25 [Arabidopsis thaliana]
Length = 621
Score = 348 bits (892), Expect = 3e-94
Identities = 181/538 (33%), Positives = 304/538 (55%), Gaps = 36/538 (6%)
Query: 49 EENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTF 108
+E++ + F + + P P +I F+++ S + + K + L L +QMEL GI + +T
Sbjct: 52 KEDDAVDLFQEMTRSR-PRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTL 110
Query: 109 NILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVA 168
+I+INC + SL+FS GKI+K G++P+ +TF+TLI GLCL+G + +AL D++V
Sbjct: 111 SIMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVE 170
Query: 169 QG-----------------------------------FHLDQVSYGTLINGLCKVGRITA 193
G F ++V+YG ++ +CK G+
Sbjct: 171 MGHKPTLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTAL 230
Query: 194 ALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIY 253
A++LL++++ + ++ +AV Y++IID +CK +++AF+L+++M K D +LI
Sbjct: 231 AMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIR 290
Query: 254 GFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIIL 313
GFC G+ + LL MI I P + FS L+D F KEGK++EA+ + +++ I
Sbjct: 291 GFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISP 350
Query: 314 DVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLF 373
D VTY SL+DG+C +++KA + D M S+G N++++ +ING CK ++D+ + LF
Sbjct: 351 DTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLELF 410
Query: 374 EEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKN 433
+M R ++ + VTYN+LI G +LGK+ +L EM R P+I++Y +LD LC N
Sbjct: 411 RKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLCDN 470
Query: 434 HHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTY 493
+KA+ + ++ + D+ Y ++I G+C + K++DA +F L +KG DV TY
Sbjct: 471 GEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTY 530
Query: 494 TVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREM 551
+MI G C KG A L KME++G PN TY I+I + + + + KL+ E+
Sbjct: 531 NIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEI 588
Score = 165 bits (417), Expect = 4e-39
Identities = 102/364 (28%), Positives = 182/364 (49%)
Query: 194 ALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIY 253
A+ L + + +P + ++ + + + K + DL QM K I+ + +T + +I
Sbjct: 56 AVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTLSIMIN 115
Query: 254 GFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIIL 313
C +L A + K+I P TFS L++ C EG+V EA ++ ++
Sbjct: 116 CCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVEMGHKP 175
Query: 314 DVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLF 373
++T N+L++G CL +++ A + D M G N +Y ++ +CK A+ L
Sbjct: 176 TLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTALAMELL 235
Query: 374 EEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKN 433
+M RKI + V Y+ +IDGL K G + L +EM +G +II Y +++ C
Sbjct: 236 RKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCYA 295
Query: 434 HHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTY 493
D LL ++ + I PD+ ++ LI + GKL +A+++ ++++ +G + D TY
Sbjct: 296 GRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPDTVTY 355
Query: 494 TVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIA 553
T +I GFC + D A +L M GC PN +T+ I+I + + D +L R+M
Sbjct: 356 TSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLELFRKMSL 415
Query: 554 RGLL 557
RG++
Sbjct: 416 RGVV 419
Score = 124 bits (311), Expect = 7e-27
Identities = 74/257 (28%), Positives = 127/257 (48%)
Query: 67 TPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFS 126
+P + + ++ K A + M G + TFNILIN + + L
Sbjct: 349 SPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLE 408
Query: 127 VFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLC 186
+F K+ +G + +T+NTLI+G C G + A ++V++ D VSY L++GLC
Sbjct: 409 LFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLC 468
Query: 187 KVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDF 246
G AL++ ++++ ++ + +YN+II MC A V+DA+DL+ + K + PD
Sbjct: 469 DNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVK 528
Query: 247 TCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVT 306
T N +I G C G L EA L KM + +P T++IL+ A EG ++ ++
Sbjct: 529 TYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEI 588
Query: 307 MKKDIILDVVTYNSLMD 323
+ +D T ++D
Sbjct: 589 KRCGFSVDASTVKMVVD 605
Score = 101 bits (252), Expect = 5e-20
Identities = 71/271 (26%), Positives = 134/271 (49%), Gaps = 4/271 (1%)
Query: 31 GLFPSSISLYSQLHHHQDEENNLISSFN--SLLHHKNPTPPIIQFNKILSSLVKAKHHST 88
G+ P +++ Y+ L +EN L + + L+ K P I FN +++ KA
Sbjct: 347 GISPDTVT-YTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDD 405
Query: 89 ALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIK 148
L L ++M L G+V+D T+N LI F +LG ++ +F +++ + P+ +++ L+
Sbjct: 406 GLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLD 465
Query: 149 GLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQP 208
GLC G +AL +K+ LD Y +I+G+C ++ A L + K V+P
Sbjct: 466 GLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKP 525
Query: 209 NAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLL 268
+ YN++I +CK +++A L+ +M SP+ T N LI G ++ L+
Sbjct: 526 DVKTYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLI 585
Query: 269 HKMILENINPRMYTFSILVDAFCKEGKVKEA 299
++ + T ++VD +G++K++
Sbjct: 586 EEIKRCGFSVDASTVKMVVD-MLSDGRLKKS 615
Score = 56.6 bits (135), Expect = 2e-06
Identities = 38/167 (22%), Positives = 73/167 (42%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P I+ + +L L AL + +++E + + D +NI+I+ ++ +
Sbjct: 455 PDIVSYKILLDGLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDL 514
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
F + KG P+ T+N +I GLC KG + +A K+ G + +Y LI
Sbjct: 515 FCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLG 574
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
G T + +L++ + +A M++D + +L D+ S
Sbjct: 575 EGDATKSAKLIEEIKRCGFSVDASTVKMVVDMLSDGRLKKSFLDMLS 621
>UniRef100_Q9LN85 T12C24.15 [Arabidopsis thaliana]
Length = 735
Score = 348 bits (892), Expect = 3e-94
Identities = 181/538 (33%), Positives = 304/538 (55%), Gaps = 36/538 (6%)
Query: 49 EENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTF 108
+E++ + F + + P P +I F+++ S + + K + L L +QMEL GI + +T
Sbjct: 52 KEDDAVDLFQEMTRSR-PRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTL 110
Query: 109 NILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVA 168
+I+INC + SL+FS GKI+K G++P+ +TF+TLI GLCL+G + +AL D++V
Sbjct: 111 SIMINCCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVE 170
Query: 169 QG-----------------------------------FHLDQVSYGTLINGLCKVGRITA 193
G F ++V+YG ++ +CK G+
Sbjct: 171 MGHKPTLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTAL 230
Query: 194 ALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIY 253
A++LL++++ + ++ +AV Y++IID +CK +++AF+L+++M K D +LI
Sbjct: 231 AMELLRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIR 290
Query: 254 GFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIIL 313
GFC G+ + LL MI I P + FS L+D F KEGK++EA+ + +++ I
Sbjct: 291 GFCYAGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISP 350
Query: 314 DVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLF 373
D VTY SL+DG+C +++KA + D M S+G N++++ +ING CK ++D+ + LF
Sbjct: 351 DTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLELF 410
Query: 374 EEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKN 433
+M R ++ + VTYN+LI G +LGK+ +L EM R P+I++Y +LD LC N
Sbjct: 411 RKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLCDN 470
Query: 434 HHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTY 493
+KA+ + ++ + D+ Y ++I G+C + K++DA +F L +KG DV TY
Sbjct: 471 GEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTY 530
Query: 494 TVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREM 551
+MI G C KG A L KME++G PN TY I+I + + + + KL+ E+
Sbjct: 531 NIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEI 588
Score = 223 bits (567), Expect = 1e-56
Identities = 134/473 (28%), Positives = 231/473 (48%), Gaps = 24/473 (5%)
Query: 71 IQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGK 130
++++ I+ L K A +L +ME+ G +D + LI F G +
Sbjct: 248 VKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCYAGRWDDGAKLLRD 307
Query: 131 ILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGR 190
++K+ P+ + F+ LI +G + +A H +++ +G D V+Y +LI+G CK +
Sbjct: 308 MIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPDTVTYTSLIDGFCKENQ 367
Query: 191 ITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNS 250
+ A +L + K PN +N++I+ CKA L++D +L+ +M + + D T N+
Sbjct: 368 LDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLELFRKMSLRGVVADTVTYNT 427
Query: 251 LIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKD 310
LI GFC +G+L+ A L +M+ + P + ++ IL+D C G+ ++A + K
Sbjct: 428 LIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLCDNGEPEKALEIFEKIEKSK 487
Query: 311 IILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAV 370
+ LD+ YN ++ G C +++ A D+F S+ +GV +V++Y MI GLCK + EA
Sbjct: 488 MELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTYNIMIGGLCKKGSLSEAD 547
Query: 371 NLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDE-------------------- 410
LF +M PN TYN LI G + KL++E
Sbjct: 548 LLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEIKRCGFSVDASTLRFALSTL 607
Query: 411 --MHDRGQPPNIITYNSILDALC--KNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLC 466
M G P++ T+ ++L C +N V A L N+K G +P++ TY +IKGL
Sbjct: 608 ARMLKAGHEPDVFTFTTLLRPFCLEENASVYDAPTLFKNMKAMGYKPNVVTYNTVIKGLL 667
Query: 467 QSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDN 519
+ V + + +G + T + I G C + L +A+ LL KME++
Sbjct: 668 NGNMISQVPGVLDQMFERGCQPNAVTKSTFISGLCKQDLHGSAILLLRKMEND 720
Score = 220 bits (560), Expect = 1e-55
Identities = 146/543 (26%), Positives = 255/543 (46%), Gaps = 59/543 (10%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P +I N +++ L S A+ L +M G + T+ ++ + G +L+ +
Sbjct: 175 PTLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTALAMEL 234
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
K+ ++ +A+ ++ +I GLC G + A N +++ +GF D + Y TLI G C
Sbjct: 235 LRKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCY 294
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
GR +LL+ + + + P+ V ++ +ID K + +A +L+ +M+ + ISPD T
Sbjct: 295 AGRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPDTVT 354
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKE-------------- 293
SLI GFC QL +A +L M+ + P + TF+IL++ +CK
Sbjct: 355 YTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLELFRKMS 414
Query: 294 ---------------------GKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEIN 332
GK++ AK + + + + D+V+Y L+DG C E
Sbjct: 415 LRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLCDNGEPE 474
Query: 333 KAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLI 392
KA +IF+ + + ++ Y +I+G+C VD+A +LF + + + P+V TYN +I
Sbjct: 475 KALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVKTYNIMI 534
Query: 393 DGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLK----- 447
GL K G +S L +M + G PN TYN ++ A K+ L+ +K
Sbjct: 535 GGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEIKRCGFS 594
Query: 448 -----------------DQGIRPDMYTYTVLIKGLC--QSGKLEDAQKVFEDLLVKGYNL 488
G PD++T+T L++ C ++ + DA +F+++ GY
Sbjct: 595 VDASTLRFALSTLARMLKAGHEPDVFTFTTLLRPFCLEENASVYDAPTLFKNMKAMGYKP 654
Query: 489 DVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLL 548
+V TY +I+G + +L +M + GC PNA T I L ++D + A LL
Sbjct: 655 NVVTYNTVIKGLLNGNMISQVPGVLDQMFERGCQPNAVTKSTFISGLCKQDLHGSAILLL 714
Query: 549 REM 551
R+M
Sbjct: 715 RKM 717
Score = 170 bits (431), Expect = 9e-41
Identities = 110/384 (28%), Positives = 186/384 (47%), Gaps = 15/384 (3%)
Query: 67 TPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFS 126
+P + + ++ K A + M G + TFNILIN + + L
Sbjct: 349 SPDTVTYTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLE 408
Query: 127 VFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLC 186
+F K+ +G + +T+NTLI+G C G + A ++V++ D VSY L++GLC
Sbjct: 409 LFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLDGLC 468
Query: 187 KVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDF 246
G AL++ ++++ ++ + +YN+II MC A V+DA+DL+ + K + PD
Sbjct: 469 DNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKPDVK 528
Query: 247 TCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVT 306
T N +I G C G L EA L KM + +P T++IL+ A EG ++ ++
Sbjct: 529 TYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLIEEI 588
Query: 307 MKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLC--KIK 364
+ +D T + + + D+F ++TT++ C +
Sbjct: 589 KRCGFSVDASTLRFALSTLARMLKAGHEPDVF-------------TFTTLLRPFCLEENA 635
Query: 365 MVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYN 424
V +A LF+ M+ PNVVTYN++I GL IS V ++D+M +RG PN +T +
Sbjct: 636 SVYDAPTLFKNMKAMGYKPNVVTYNTVIKGLLNGNMISQVPGVLDQMFERGCQPNAVTKS 695
Query: 425 SILDALCKNHHVDKAIALLTNLKD 448
+ + LCK AI LL +++
Sbjct: 696 TFISGLCKQDLHGSAILLLRKMEN 719
Score = 165 bits (417), Expect = 4e-39
Identities = 102/364 (28%), Positives = 182/364 (49%)
Query: 194 ALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIY 253
A+ L + + +P + ++ + + + K + DL QM K I+ + +T + +I
Sbjct: 56 AVDLFQEMTRSRPRPRLIDFSRLFSVVARTKQYDLVLDLCKQMELKGIAHNLYTLSIMIN 115
Query: 254 GFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIIL 313
C +L A + K+I P TFS L++ C EG+V EA ++ ++
Sbjct: 116 CCCRCRKLSLAFSAMGKIIKLGYEPDTVTFSTLINGLCLEGRVSEALELVDRMVEMGHKP 175
Query: 314 DVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLF 373
++T N+L++G CL +++ A + D M G N +Y ++ +CK A+ L
Sbjct: 176 TLITLNALVNGLCLNGKVSDAVLLIDRMVETGFQPNEVTYGPVLKVMCKSGQTALAMELL 235
Query: 374 EEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKN 433
+M RKI + V Y+ +IDGL K G + L +EM +G +II Y +++ C
Sbjct: 236 RKMEERKIKLDAVKYSIIIDGLCKDGSLDNAFNLFNEMEIKGFKADIIIYTTLIRGFCYA 295
Query: 434 HHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTY 493
D LL ++ + I PD+ ++ LI + GKL +A+++ ++++ +G + D TY
Sbjct: 296 GRWDDGAKLLRDMIKRKITPDVVAFSALIDCFVKEGKLREAEELHKEMIQRGISPDTVTY 355
Query: 494 TVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIA 553
T +I GFC + D A +L M GC PN +T+ I+I + + D +L R+M
Sbjct: 356 TSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDDGLELFRKMSL 415
Query: 554 RGLL 557
RG++
Sbjct: 416 RGVV 419
Score = 133 bits (334), Expect = 2e-29
Identities = 107/401 (26%), Positives = 191/401 (46%), Gaps = 20/401 (4%)
Query: 31 GLFPSSISLYSQLHHHQDEENNLISSFN--SLLHHKNPTPPIIQFNKILSSLVKAKHHST 88
G+ P +++ Y+ L +EN L + + L+ K P I FN +++ KA
Sbjct: 347 GISPDTVT-YTSLIDGFCKENQLDKANHMLDLMVSKGCGPNIRTFNILINGYCKANLIDD 405
Query: 89 ALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIK 148
L L ++M L G+V+D T+N LI F +LG ++ +F +++ + P+ +++ L+
Sbjct: 406 GLELFRKMSLRGVVADTVTYNTLIQGFCELGKLEVAKELFQEMVSRRVRPDIVSYKILLD 465
Query: 149 GLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQP 208
GLC G +AL +K+ LD Y +I+G+C ++ A L + K V+P
Sbjct: 466 GLCDNGEPEKALEIFEKIEKSKMELDIGIYNIIIHGMCNASKVDDAWDLFCSLPLKGVKP 525
Query: 209 NAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLL 268
+ YN++I +CK +++A L+ +M SP+ T N LI G ++ L+
Sbjct: 526 DVKTYNIMIGGLCKKGSLSEADLLFRKMEEDGHSPNGCTYNILIRAHLGEGDATKSAKLI 585
Query: 269 HKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLV 328
++ + FS VDA ++ A L +K DV T+ +L+ +CL
Sbjct: 586 EEI-------KRCGFS--VDA----STLRFALSTLARMLKAGHEPDVFTFTTLLRPFCLE 632
Query: 329 K--EINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVV 386
+ + A +F +M + G NV +Y T+I GL M+ + + ++M R PN V
Sbjct: 633 ENASVYDAPTLFKNMKAMGYKPNVVTYNTVIKGLLNGNMISQVPGVLDQMFERGCQPNAV 692
Query: 387 TYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSIL 427
T ++ I GL K + L+ +M + + ++ T IL
Sbjct: 693 TKSTFISGLCKQDLHGSAILLLRKMENDNE--DVTTKKKIL 731
>UniRef100_Q9SH26 F2K11.22 [Arabidopsis thaliana]
Length = 577
Score = 346 bits (888), Expect = 9e-94
Identities = 187/517 (36%), Positives = 297/517 (57%), Gaps = 36/517 (6%)
Query: 46 HQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDF 105
H + ++ I F ++ + P P I +FNK+LS++ K K +SL ++M+ GI +
Sbjct: 62 HSMKLDDAIGLFGGMVKSR-PLPSIFEFNKLLSAIAKMKKFDLVISLGEKMQRLGISHNL 120
Query: 106 FTFNILINCFSQLGLNSLSFSVFGKILKKGFD---------------------------- 137
+T+NILINCF + SL+ ++ GK++K G++
Sbjct: 121 YTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRISDAVALVDQ 180
Query: 138 -------PNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGR 190
P+ ITF TLI GL L +A+ D++V +G + V+YG ++NGLCK G
Sbjct: 181 MVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVVNGLCKRGD 240
Query: 191 ITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNS 250
I A LL +++ ++ N V+Y+ +ID++CK + +DA +L+++M K + P+ T +S
Sbjct: 241 IDLAFNLLNKMEAAKIEANVVIYSTVIDSLCKYRHEDDALNLFTEMENKGVRPNVITYSS 300
Query: 251 LIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKD 310
LI C + +A LL MI INP + TF+ L+DAF KEGK+ EA+ + +K+
Sbjct: 301 LISCLCNYERWSDASRLLSDMIERKINPNVVTFNALIDAFVKEGKLVEAEKLYDEMIKRS 360
Query: 311 IILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAV 370
I D+ TY+SL++G+C+ +++AK +F+ M S+ NV +Y T+ING CK K +DE V
Sbjct: 361 IDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLINGFCKAKRIDEGV 420
Query: 371 NLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDAL 430
LF EM R ++ N VTY +LI G + + +M G PNI+TYN++LD L
Sbjct: 421 ELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSDGVHPNIMTYNTLLDGL 480
Query: 431 CKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDV 490
CKN ++KA+ + L+ + P +YTY ++I+G+C++GK+ED +F L +KG DV
Sbjct: 481 CKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMIEGMCKAGKVEDGWDLFCSLSLKGVKPDV 540
Query: 491 YTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKT 527
Y MI GFC KGL + A AL KM ++G +P++ T
Sbjct: 541 IIYNTMISGFCRKGLKEEADALFRKMREDGPLPDSGT 577
Score = 194 bits (494), Expect = 4e-48
Identities = 118/453 (26%), Positives = 217/453 (47%), Gaps = 39/453 (8%)
Query: 119 GLNSL----SFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLD 174
GL+S+ + +FG ++K P+ FN L+ + ++ +K+ G +
Sbjct: 60 GLHSMKLDDAIGLFGGMVKSRPLPSIFEFNKLLSAIAKMKKFDLVISLGEKMQRLGISHN 119
Query: 175 QVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
+Y LIN C+ +I+ AL LL ++ +P+ V + +++ C K ++DA L
Sbjct: 120 LYTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRISDAVALVD 179
Query: 235 QMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEG 294
QMV PD T +LI+G + + EAV L+ +M+ P + T+ ++V+ CK G
Sbjct: 180 QMVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVVNGLCKRG 239
Query: 295 KVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYT 354
+ A +L I +VV Y++++D C + + A ++F M ++GV NV +Y+
Sbjct: 240 DIDLAFNLLNKMEAAKIEANVVIYSTVIDSLCKYRHEDDALNLFTEMENKGVRPNVITYS 299
Query: 355 TMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVT--------------------------- 387
++I+ LC + +A L +M RKI PNVVT
Sbjct: 300 SLISCLCNYERWSDASRLLSDMIERKINPNVVTFNALIDAFVKEGKLVEAEKLYDEMIKR 359
Query: 388 --------YNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKA 439
Y+SLI+G ++ + + M + PN++TYN++++ CK +D+
Sbjct: 360 SIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLINGFCKAKRIDEG 419
Query: 440 IALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQG 499
+ L + +G+ + TYT LI G Q+ ++AQ VF+ ++ G + ++ TY ++ G
Sbjct: 420 VELFREMSQRGLVGNTVTYTTLIHGFFQARDCDNAQMVFKQMVSDGVHPNIMTYNTLLDG 479
Query: 500 FCVKGLFDAALALLSKMEDNGCIPNAKTYEIVI 532
C G + A+ + ++ + P TY I+I
Sbjct: 480 LCKNGKLEKAMVVFEYLQRSKMEPTIYTYNIMI 512
Score = 194 bits (493), Expect = 6e-48
Identities = 114/380 (30%), Positives = 188/380 (49%), Gaps = 35/380 (9%)
Query: 213 YNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMI 272
Y I+ N + ++DA L+ MV R P F N L+ M + + L KM
Sbjct: 53 YREILRNGLHSMKLDDAIGLFGGMVKSRPLPSIFEFNKLLSAIAKMKKFDLVISLGEKMQ 112
Query: 273 LENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEIN 332
I+ +YT++IL++ FC+ ++ A +LG MK +VT +SL++GYC K I+
Sbjct: 113 RLGISHNLYTYNILINCFCRRSQISLALALLGKMMKLGYEPSIVTLSSLLNGYCHGKRIS 172
Query: 333 KAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLI 392
A + D M G + ++TT+I+GL EAV L + M R PN+VTY ++
Sbjct: 173 DAVALVDQMVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVV 232
Query: 393 DGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIR 452
+GL K G I L+++M N++ Y++++D+LCK H D A+ L T ++++G+R
Sbjct: 233 NGLCKRGDIDLAFNLLNKMEAAKIEANVVIYSTVIDSLCKYRHEDDALNLFTEMENKGVR 292
Query: 453 PDMYTYTVLIKGLC-----------------------------------QSGKLEDAQKV 477
P++ TY+ LI LC + GKL +A+K+
Sbjct: 293 PNVITYSSLISCLCNYERWSDASRLLSDMIERKINPNVVTFNALIDAFVKEGKLVEAEKL 352
Query: 478 FEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFE 537
+++++ + + D++TY+ +I GFC+ D A + M C PN TY +I +
Sbjct: 353 YDEMIKRSIDPDIFTYSSLINGFCMHDRLDEAKHMFELMISKDCFPNVVTYNTLINGFCK 412
Query: 538 KDENDMAEKLLREMIARGLL 557
D +L REM RGL+
Sbjct: 413 AKRIDEGVELFREMSQRGLV 432
>UniRef100_Q8L8A4 Fertility restorer [Petunia hybrida]
Length = 592
Score = 333 bits (854), Expect = 8e-90
Identities = 187/545 (34%), Positives = 306/545 (55%), Gaps = 7/545 (1%)
Query: 16 PNFSHFQQSPNPNFIGLFPSSISLYSQLHHHQDEEN-----NLISSFNSLLHHKNPTPPI 70
P FS F S P SIS+ + EN + S F ++ K P P
Sbjct: 14 PFFSFFAYSIAPRHYSTNTCSISVKGNFGVSNEFENVKCLDDAFSLFRQMVTTK-PLPSA 72
Query: 71 IQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGK 130
+ F+K+L +LV KH+S+ +S+ +++ I D F + ++N + L FSV
Sbjct: 73 VSFSKLLKALVHMKHYSSVVSIFREIHKLRIPVDAFALSTVVNSCCLMHRTDLGFSVLAI 132
Query: 131 ILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGF-HLDQVSYGTLINGLCKVG 189
KKG N +TF TLI+GL + + A++ K+V + D+V YGT+++GLCK G
Sbjct: 133 HFKKGIPYNEVTFTTLIRGLFAENKVKDAVHLFKKLVRENICEPDEVMYGTVMDGLCKKG 192
Query: 190 RITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCN 249
A LL+ ++ + +P+ +YN++ID CK +++ A L ++M K I PD T
Sbjct: 193 HTQKAFDLLRLMEQGITKPDTCIYNIVIDAFCKDGMLDGATSLLNEMKQKNIPPDIITYT 252
Query: 250 SLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKK 309
SLI G + Q ++ L +MI NI P + TF+ ++D CKEGKV++A+ ++ ++K
Sbjct: 253 SLIDGLGKLSQWEKVRTLFLEMIHLNIYPDVCTFNSVIDGLCKEGKVEDAEEIMTYMIEK 312
Query: 310 DIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEA 369
+ + +TYN +MDGYCL ++ +A+ IFDSM +G+ ++ SYT +ING + K +D+A
Sbjct: 313 GVEPNEITYNVVMDGYCLRGQMGRARRIFDSMIDKGIEPDIISYTALINGYVEKKKMDKA 372
Query: 370 VNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDA 429
+ LF E+ + P++VT + L+ GL ++G+ C DEM G PN+ T+ ++L
Sbjct: 373 MQLFREISQNGLKPSIVTCSVLLRGLFEVGRTECAKIFFDEMQAAGHIPNLYTHCTLLGG 432
Query: 430 LCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLD 489
KN V++A++ L+ + ++ YT +I GLC++GKL+ A FE L + G + D
Sbjct: 433 YFKNGLVEEAMSHFHKLERRREDTNIQIYTAVINGLCKNGKLDKAHATFEKLPLIGLHPD 492
Query: 490 VYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLR 549
V TYT MI G+C +GL D A +L KMEDNGC+P+ +TY +++ F + + L+
Sbjct: 493 VITYTAMISGYCQEGLLDEAKDMLRKMEDNGCLPDNRTYNVIVRGFFRSSKVSEMKAFLK 552
Query: 550 EMIAR 554
E+ +
Sbjct: 553 EIAGK 557
>UniRef100_Q9C8T7 Hypothetical protein F9N12.5 [Arabidopsis thaliana]
Length = 558
Score = 332 bits (852), Expect = 1e-89
Identities = 179/489 (36%), Positives = 290/489 (58%), Gaps = 4/489 (0%)
Query: 66 PTPPIIQFNKILSSLVKAKHHSTALSLH---QQMELNGIVSDFFTFNILINCFSQLGLNS 122
P P I +FNK+LS++ K K +SL + M+L G T + L+N + S
Sbjct: 38 PLPSIFEFNKLLSAIAKMKKFDLVISLALLGKMMKL-GYEPSIVTLSSLLNGYCHGKRIS 96
Query: 123 LSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLI 182
+ ++ ++++ G+ P+ ITF TLI GL L +A+ D++V +G + V+YG ++
Sbjct: 97 DAVALVDQMVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQRGCQPNLVTYGVVV 156
Query: 183 NGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRIS 242
NGLCK G I A LL +++ ++ + V++N IID++CK + V+DA +L+ +M K I
Sbjct: 157 NGLCKRGDIDLAFNLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDDALNLFKEMETKGIR 216
Query: 243 PDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMM 302
P+ T +SLI C G+ +A LL MI + INP + TF+ L+DAF KEGK EA+ +
Sbjct: 217 PNVVTYSSLISCLCSYGRWSDASQLLSDMIEKKINPNLVTFNALIDAFVKEGKFVEAEKL 276
Query: 303 LGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCK 362
+K+ I D+ TYNSL++G+C+ ++KAK +F+ M S+ ++ +Y T+I G CK
Sbjct: 277 HDDMIKRSIDPDIFTYNSLINGFCMHDRLDKAKQMFEFMVSKDCFPDLDTYNTLIKGFCK 336
Query: 363 IKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIIT 422
K V++ LF EM R ++ + VTY +LI GL G K+ +M G PP+I+T
Sbjct: 337 SKRVEDGTELFREMSHRGLVGDTVTYTTLIQGLFHDGDCDNAQKVFKQMVSDGVPPDIMT 396
Query: 423 YNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLL 482
Y+ +LD LC N ++KA+ + ++ I+ D+Y YT +I+G+C++GK++D +F L
Sbjct: 397 YSILLDGLCNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLS 456
Query: 483 VKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDEND 542
+KG +V TY MI G C K L A ALL KM+++G +P++ TY +I + +
Sbjct: 457 LKGVKPNVVTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPDSGTYNTLIRAHLRDGDKA 516
Query: 543 MAEKLLREM 551
+ +L+REM
Sbjct: 517 ASAELIREM 525
Score = 132 bits (331), Expect = 3e-29
Identities = 88/355 (24%), Positives = 164/355 (45%), Gaps = 8/355 (2%)
Query: 44 HHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVS 103
+ H D+ NL + K P ++ ++ ++S L S A L M I
Sbjct: 197 YRHVDDALNLFKEMET----KGIRPNVVTYSSLISCLCSYGRWSDASQLLSDMIEKKINP 252
Query: 104 DFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFH 163
+ TFN LI+ F + G + + ++K+ DP+ T+N+LI G C+ + +A
Sbjct: 253 NLVTFNALIDAFVKEGKFVEAEKLHDDMIKRSIDPDIFTYNSLINGFCMHDRLDKAKQMF 312
Query: 164 DKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKA 223
+ +V++ D +Y TLI G CK R+ +L + + + + + V Y +I +
Sbjct: 313 EFMVSKDCFPDLDTYNTLIKGFCKSKRVEDGTELFREMSHRGLVGDTVTYTTLIQGLFHD 372
Query: 224 KLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTF 283
++A ++ QMV+ + PD T + L+ G C G+L++A+ + M I +Y +
Sbjct: 373 GDCDNAQKVFKQMVSDGVPPDIMTYSILLDGLCNNGKLEKALEVFDYMQKSEIKLDIYIY 432
Query: 284 SILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMAS 343
+ +++ CK GKV + + K + +VVTYN+++ G C + + +A + M
Sbjct: 433 TTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNVVTYNTMISGLCSKRLLQEAYALLKKMKE 492
Query: 344 RGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTY----NSLIDG 394
G + + +Y T+I + + L EMR + + + T N L DG
Sbjct: 493 DGPLPDSGTYNTLIRAHLRDGDKAASAELIREMRSCRFVGDASTIGLVANMLHDG 547
Score = 103 bits (258), Expect = 1e-20
Identities = 59/193 (30%), Positives = 102/193 (52%), Gaps = 2/193 (1%)
Query: 366 VDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCV--LKLVDEMHDRGQPPNIITY 423
+D+A+ LF M + +P++ +N L+ + K+ K V L L+ +M G P+I+T
Sbjct: 23 LDDAIGLFGGMVKSRPLPSIFEFNKLLSAIAKMKKFDLVISLALLGKMMKLGYEPSIVTL 82
Query: 424 NSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLV 483
+S+L+ C + A+AL+ + + G RPD T+T LI GL K +A + + ++
Sbjct: 83 SSLLNGYCHGKRISDAVALVDQMVEMGYRPDTITFTTLIHGLFLHNKASEAVALVDRMVQ 142
Query: 484 KGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDM 543
+G ++ TY V++ G C +G D A LL+KME + + +I SL + D
Sbjct: 143 RGCQPNLVTYGVVVNGLCKRGDIDLAFNLLNKMEAAKIEADVVIFNTIIDSLCKYRHVDD 202
Query: 544 AEKLLREMIARGL 556
A L +EM +G+
Sbjct: 203 ALNLFKEMETKGI 215
Score = 59.3 bits (142), Expect = 3e-07
Identities = 48/214 (22%), Positives = 91/214 (42%), Gaps = 2/214 (0%)
Query: 21 FQQSPNPNFIGLFPSSISLYSQLHHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSL 80
F++ + +G + +L L H D +N F ++ P P I+ ++ +L L
Sbjct: 347 FREMSHRGLVGDTVTYTTLIQGLFHDGDCDNAQ-KVFKQMVSDGVP-PDIMTYSILLDGL 404
Query: 81 VKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNA 140
AL + M+ + I D + + +I + G + +F + KG PN
Sbjct: 405 CNNGKLEKALEVFDYMQKSEIKLDIYIYTTMIEGMCKAGKVDDGWDLFCSLSLKGVKPNV 464
Query: 141 ITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKR 200
+T+NT+I GLC K + +A K+ G D +Y TLI + G A+ +L++
Sbjct: 465 VTYNTMISGLCSKRLLQEAYALLKKMKEDGPLPDSGTYNTLIRAHLRDGDKAASAELIRE 524
Query: 201 VDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYS 234
+ +A ++ + + +L D+ S
Sbjct: 525 MRSCRFVGDASTIGLVANMLHDGRLDKSFLDMLS 558
>UniRef100_Q8L8A2 Fertility restorer-like protein [Petunia hybrida]
Length = 592
Score = 327 bits (838), Expect = 6e-88
Identities = 183/545 (33%), Positives = 305/545 (55%), Gaps = 7/545 (1%)
Query: 16 PNFSHFQQSPNPNFIGLFPSSISLYSQLHHHQDEEN-----NLISSFNSLLHHKNPTPPI 70
P FS F S P SIS+ + EN + S F ++ K P P +
Sbjct: 14 PFFSFFAYSIAPRHYSTNTRSISVKGNFGVSNEFENVKCLDDAFSLFRQMVRTK-PLPSV 72
Query: 71 IQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGK 130
+ F+K+L +LV KH+S+ +SL +++ I F +I++N + L FSV
Sbjct: 73 VSFSKLLKALVHMKHYSSVVSLFREIHKLRIPVHEFILSIVVNSCCLMHRTDLGFSVLAI 132
Query: 131 ILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGF-HLDQVSYGTLINGLCKVG 189
KKG N + FNTL++GL + + A++ K+V + ++V YGT++NGLCK G
Sbjct: 133 HFKKGIPFNQVIFNTLLRGLFAENKVKDAVHLFKKLVRENICEPNEVMYGTVMNGLCKKG 192
Query: 190 RITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCN 249
A LL+ ++ +PN +Y+++ID CK +++ A L ++M K I PD FT +
Sbjct: 193 HTQKAFDLLRLMEQGSTKPNTCIYSIVIDAFCKDGMLDGATSLLNEMKQKSIPPDIFTYS 252
Query: 250 SLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKK 309
+LI C + Q + L +MI NI P + TF+ ++D CKEGKV++A+ ++ ++K
Sbjct: 253 TLIDALCKLSQWENVRTLFLEMIHLNIYPNVCTFNSVIDGLCKEGKVEDAEEIMRYMIEK 312
Query: 310 DIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEA 369
+ DV+TYN ++DGY L ++++A++IFDSM ++ + N+ SY +ING + K +DEA
Sbjct: 313 GVDPDVITYNMIIDGYGLRGQVDRAREIFDSMINKSIEPNIISYNILINGYARQKKIDEA 372
Query: 370 VNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDA 429
+ + E+ + + P++VT N L+ GL +LG+ DEM G P++ T+ ++L
Sbjct: 373 MQVCREISQKGLKPSIVTCNVLLHGLFELGRTKSAQNFFDEMLSAGHIPDLYTHCTLLGG 432
Query: 430 LCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLD 489
KN V++A++ L+ + ++ YT +I GLC++GKL+ A FE L + G + D
Sbjct: 433 YFKNGLVEEAMSHFHKLERRREDTNIQIYTAVIDGLCKNGKLDKAHATFEKLPLIGLHPD 492
Query: 490 VYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLR 549
V TYT MI G+C +GL D A +L KMEDNGC+ + +TY +++ ++ + L
Sbjct: 493 VITYTAMISGYCQEGLLDEAKDMLRKMEDNGCLADNRTYNVIVRGFLRSNKVSEMKAFLE 552
Query: 550 EMIAR 554
E+ +
Sbjct: 553 EIAGK 557
Score = 160 bits (406), Expect = 7e-38
Identities = 103/451 (22%), Positives = 211/451 (45%), Gaps = 35/451 (7%)
Query: 47 QDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFF 106
+++ + + F L+ P + + +++ L K H A L + ME +
Sbjct: 155 ENKVKDAVHLFKKLVRENICEPNEVMYGTVMNGLCKKGHTQKAFDLLRLMEQGSTKPNTC 214
Query: 107 TFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKV 166
++I+I+ F + G+ + S+ ++ +K P+ T++TLI LC ++
Sbjct: 215 IYSIVIDAFCKDGMLDGATSLLNEMKQKSIPPDIFTYSTLIDALCKLSQWENVRTLFLEM 274
Query: 167 VAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLV 226
+ + + ++ ++I+GLCK G++ A ++++ + K V P+ + YNMIID V
Sbjct: 275 IHLNIYPNVCTFNSVIDGLCKEGKVEDAEEIMRYMIEKGVDPDVITYNMIIDGYGLRGQV 334
Query: 227 NDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSIL 286
+ A +++ M+ K I P+ + N LI G+ ++ EA+ + ++ + + P + T ++L
Sbjct: 335 DRAREIFDSMINKSIEPNIISYNILINGYARQKKIDEAMQVCREISQKGLKPSIVTCNVL 394
Query: 287 VDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGV 346
+ + G+ K A+ + I D+ T+ +L+ GY + +A F + R
Sbjct: 395 LHGLFELGRTKSAQNFFDEMLSAGHIPDLYTHCTLLGGYFKNGLVEEAMSHFHKLERRRE 454
Query: 347 IANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLK 406
N+Q YT +I+GLCK +D+A FE++ + P+V+TY ++I G C
Sbjct: 455 DTNIQIYTAVIDGLCKNGKLDKAHATFEKLPLIGLHPDVITYTAMISGY-------CQEG 507
Query: 407 LVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLC 466
L+DE D +L ++D G D TY V+++G
Sbjct: 508 LLDEAKD----------------------------MLRKMEDNGCLADNRTYNVIVRGFL 539
Query: 467 QSGKLEDAQKVFEDLLVKGYNLDVYTYTVMI 497
+S K+ + + E++ K ++ + T +++
Sbjct: 540 RSNKVSEMKAFLEEIAGKSFSFEAATVELLM 570
>UniRef100_Q9SH60 F22C12.14 [Arabidopsis thaliana]
Length = 661
Score = 327 bits (837), Expect = 7e-88
Identities = 189/556 (33%), Positives = 295/556 (52%), Gaps = 50/556 (8%)
Query: 45 HHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSD 104
H+ ++ I F+ ++ + P + NK++ V+ A+SL+++ME+ I +
Sbjct: 82 HYFKSLDDAIDFFDYMVRSR-PFYTAVDCNKVIGVFVRMNRPDVAISLYRKMEIRRIPLN 140
Query: 105 FFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNA------------------------ 140
++FNILI CF S S S FGK+ K GF P+
Sbjct: 141 IYSFNILIKCFCDCHKLSFSLSTFGKLTKLGFQPDVVTFNTLLHGLCLEDRISEALALFG 200
Query: 141 ---------------------ITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYG 179
ITFNTLI GLCL+G + +A +K+V +G H+D V+YG
Sbjct: 201 YMVETGSLFDQMVEIGLTPVVITFNTLINGLCLEGRVLEAAALVNKMVGKGLHIDVVTYG 260
Query: 180 TLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAK 239
T++NG+CK+G +AL LL +++ ++P+ V+Y+ IID +CK +DA L+S+M+ K
Sbjct: 261 TIVNGMCKMGDTKSALNLLSKMEETHIKPDVVIYSAIIDRLCKDGHHSDAQYLFSEMLEK 320
Query: 240 RISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEA 299
I+P+ FT N +I GFC G+ +A LL MI INP + TF+ L+ A KEGK+ EA
Sbjct: 321 GIAPNVFTYNCMIDGFCSFGRWSDAQRLLRDMIEREINPDVLTFNALISASVKEGKLFEA 380
Query: 300 KMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMING 359
+ + + + I D VTYNS++ G+C + AK +FD MAS V+ ++ T+I+
Sbjct: 381 EKLCDEMLHRCIFPDTVTYNSMIYGFCKHNRFDDAKHMFDLMASPDVV----TFNTIIDV 436
Query: 360 LCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPN 419
C+ K VDE + L E+ R ++ N TYN+LI G ++ ++ L EM G P+
Sbjct: 437 YCRAKRVDEGMQLLREISRRGLVANTTTYNTLIHGFCEVDNLNAAQDLFQEMISHGVCPD 496
Query: 420 IITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFE 479
IT N +L C+N +++A+ L ++ I D Y ++I G+C+ K+++A +F
Sbjct: 497 TITCNILLYGFCENEKLEEALELFEVIQMSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFC 556
Query: 480 DLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKD 539
L + G DV TY VMI GFC K A L KM+DNG P+ TY +I +
Sbjct: 557 SLPIHGVEPDVQTYNVMISGFCGKSAISDANVLFHKMKDNGHEPDNSTYNTLIRGCLKAG 616
Query: 540 ENDMAEKLLREMIARG 555
E D + +L+ EM + G
Sbjct: 617 EIDKSIELISEMRSNG 632
Score = 203 bits (516), Expect = 1e-50
Identities = 130/458 (28%), Positives = 227/458 (49%), Gaps = 14/458 (3%)
Query: 109 NILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVA 168
N +I F ++ ++ S++ K+ + N +FN LIK C + +L+ K+
Sbjct: 110 NKVIGVFVRMNRPDVAISLYRKMEIRRIPLNIYSFNILIKCFCDCHKLSFSLSTFGKLTK 169
Query: 169 QGFHLDQVSYGTLINGLCKVGRITAALQLLKRV--DGKL--------VQPNAVMYNMIID 218
GF D V++ TL++GLC RI+ AL L + G L + P + +N +I+
Sbjct: 170 LGFQPDVVTFNTLLHGLCLEDRISEALALFGYMVETGSLFDQMVEIGLTPVVITFNTLIN 229
Query: 219 NMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINP 278
+C V +A L ++MV K + D T +++ G C MG K A+ LL KM +I P
Sbjct: 230 GLCLEGRVLEAAALVNKMVGKGLHIDVVTYGTIVNGMCKMGDTKSALNLLSKMEETHIKP 289
Query: 279 RMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIF 338
+ +S ++D CK+G +A+ + ++K I +V TYN ++DG+C + A+ +
Sbjct: 290 DVVIYSAIIDRLCKDGHHSDAQYLFSEMLEKGIAPNVFTYNCMIDGFCSFGRWSDAQRLL 349
Query: 339 DSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKL 398
M R + +V ++ +I+ K + EA L +EM R I P+ VTYNS+I G K
Sbjct: 350 RDMIEREINPDVLTFNALISASVKEGKLFEAEKLCDEMLHRCIFPDTVTYNSMIYGFCKH 409
Query: 399 GKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAIALLTNLKDQGIRPDMYTY 458
+ + D M P+++T+N+I+D C+ VD+ + LL + +G+ + TY
Sbjct: 410 NRFDDAKHMFDLMAS----PDVVTFNTIIDVYCRAKRVDEGMQLLREISRRGLVANTTTY 465
Query: 459 TVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQGFCVKGLFDAALALLSKMED 518
LI G C+ L AQ +F++++ G D T +++ GFC + AL L ++
Sbjct: 466 NTLIHGFCEVDNLNAAQDLFQEMISHGVCPDTITCNILLYGFCENEKLEEALELFEVIQM 525
Query: 519 NGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGL 556
+ + Y I+I + + + D A L + G+
Sbjct: 526 SKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGV 563
Score = 190 bits (482), Expect = 1e-46
Identities = 115/374 (30%), Positives = 194/374 (51%), Gaps = 5/374 (1%)
Query: 68 PPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSV 127
P ++ ++ I+ L K HHS A L +M GI + FT+N +I+ F G S + +
Sbjct: 289 PDVVIYSAIIDRLCKDGHHSDAQYLFSEMLEKGIAPNVFTYNCMIDGFCSFGRWSDAQRL 348
Query: 128 FGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCK 187
++++ +P+ +TFN LI +G + +A D+++ + D V+Y ++I G CK
Sbjct: 349 LRDMIEREINPDVLTFNALISASVKEGKLFEAEKLCDEMLHRCIFPDTVTYNSMIYGFCK 408
Query: 188 VGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFT 247
R A K + + P+ V +N IID C+AK V++ L ++ + + + T
Sbjct: 409 HNRFDDA----KHMFDLMASPDVVTFNTIIDVYCRAKRVDEGMQLLREISRRGLVANTTT 464
Query: 248 CNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTM 307
N+LI+GFC + L A L +MI + P T +IL+ FC+ K++EA + V
Sbjct: 465 YNTLIHGFCEVDNLNAAQDLFQEMISHGVCPDTITCNILLYGFCENEKLEEALELFEVIQ 524
Query: 308 KKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVD 367
I LD V YN ++ G C ++++A D+F S+ GV +VQ+Y MI+G C +
Sbjct: 525 MSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGVEPDVQTYNVMISGFCGKSAIS 584
Query: 368 EAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSIL 427
+A LF +M+ P+ TYN+LI G K G+I ++L+ EM G + T +
Sbjct: 585 DANVLFHKMKDNGHEPDNSTYNTLIRGCLKAGEIDKSIELISEMRSNGFSGDAFTIKMVA 644
Query: 428 DALCKNHHVDKAIA 441
D L + +DK+ +
Sbjct: 645 D-LITDGRLDKSFS 657
Score = 169 bits (427), Expect = 3e-40
Identities = 108/358 (30%), Positives = 178/358 (49%), Gaps = 14/358 (3%)
Query: 210 AVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVGLLH 269
AV N +I + + A LY +M +RI + ++ N LI FC +L ++
Sbjct: 106 AVDCNKVIGVFVRMNRPDVAISLYRKMEIRRIPLNIYSFNILIKCFCDCHKLSFSLSTFG 165
Query: 270 KMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKKDIILD----------VVTYN 319
K+ P + TF+ L+ C E ++ EA + G ++ + D V+T+N
Sbjct: 166 KLTKLGFQPDVVTFNTLLHGLCLEDRISEALALFGYMVETGSLFDQMVEIGLTPVVITFN 225
Query: 320 SLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCR 379
+L++G CL + +A + + M +G+ +V +Y T++NG+CK+ A+NL +M
Sbjct: 226 TLINGLCLEGRVLEAAALVNKMVGKGLHIDVVTYGTIVNGMCKMGDTKSALNLLSKMEET 285
Query: 380 KIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKA 439
I P+VV Y+++ID L K G S L EM ++G PN+ TYN ++D C A
Sbjct: 286 HIKPDVVIYSAIIDRLCKDGHHSDAQYLFSEMLEKGIAPNVFTYNCMIDGFCSFGRWSDA 345
Query: 440 IALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDVYTYTVMIQG 499
LL ++ ++ I PD+ T+ LI + GKL +A+K+ +++L + D TY MI G
Sbjct: 346 QRLLRDMIEREINPDVLTFNALISASVKEGKLFEAEKLCDEMLHRCIFPDTVTYNSMIYG 405
Query: 500 FCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLREMIARGLL 557
FC FD A + M P+ T+ +I D +LLRE+ RGL+
Sbjct: 406 FCKHNRFDDAKHMFDLMAS----PDVVTFNTIIDVYCRAKRVDEGMQLLREISRRGLV 459
Score = 112 bits (280), Expect = 3e-23
Identities = 71/246 (28%), Positives = 120/246 (47%), Gaps = 10/246 (4%)
Query: 321 LMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEAVNLFEEMRCRK 380
L G K ++ A D FD M +I ++ D A++L+ +M R+
Sbjct: 77 LKSGSHYFKSLDDAIDFFDYMVRSRPFYTAVDCNKVIGVFVRMNRPDVAISLYRKMEIRR 136
Query: 381 IIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDALCKNHHVDKAI 440
I N+ ++N LI K+S L ++ G P+++T+N++L LC + +A+
Sbjct: 137 IPLNIYSFNILIKCFCDCHKLSFSLSTFGKLTKLGFQPDVVTFNTLLHGLCLEDRISEAL 196
Query: 441 ALL------TNLKDQ----GIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLDV 490
AL +L DQ G+ P + T+ LI GLC G++ +A + ++ KG ++DV
Sbjct: 197 ALFGYMVETGSLFDQMVEIGLTPVVITFNTLINGLCLEGRVLEAAALVNKMVGKGLHIDV 256
Query: 491 YTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLRE 550
TY ++ G C G +AL LLSKME+ P+ Y +I L + + A+ L E
Sbjct: 257 VTYGTIVNGMCKMGDTKSALNLLSKMEETHIKPDVVIYSAIIDRLCKDGHHSDAQYLFSE 316
Query: 551 MIARGL 556
M+ +G+
Sbjct: 317 MLEKGI 322
Score = 106 bits (264), Expect = 2e-21
Identities = 63/262 (24%), Positives = 127/262 (48%), Gaps = 13/262 (4%)
Query: 32 LFPSSISLYSQL-----HHHQDEENNLISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHH 86
+FP +++ S + H+ D+ ++ S P ++ FN I+ +AK
Sbjct: 392 IFPDTVTYNSMIYGFCKHNRFDDAKHMFDLMAS--------PDVVTFNTIIDVYCRAKRV 443
Query: 87 STALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTL 146
+ L +++ G+V++ T+N LI+ F ++ + + +F +++ G P+ IT N L
Sbjct: 444 DEGMQLLREISRRGLVANTTTYNTLIHGFCEVDNLNAAQDLFQEMISHGVCPDTITCNIL 503
Query: 147 IKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLV 206
+ G C + +AL + + LD V+Y +I+G+CK ++ A L + V
Sbjct: 504 LYGFCENEKLEEALELFEVIQMSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGV 563
Query: 207 QPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCNSLIYGFCIMGQLKEAVG 266
+P+ YN++I C ++DA L+ +M PD+ T N+LI G G++ +++
Sbjct: 564 EPDVQTYNVMISGFCGKSAISDANVLFHKMKDNGHEPDNSTYNTLIRGCLKAGEIDKSIE 623
Query: 267 LLHKMILENINPRMYTFSILVD 288
L+ +M + +T ++ D
Sbjct: 624 LISEMRSNGFSGDAFTIKMVAD 645
Score = 64.3 bits (155), Expect = 9e-09
Identities = 49/203 (24%), Positives = 90/203 (44%), Gaps = 4/203 (1%)
Query: 35 SSISLYSQLHHHQDEENNLISS---FNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALS 91
++ + Y+ L H E +NL ++ F ++ H P I N +L + + AL
Sbjct: 460 ANTTTYNTLIHGFCEVDNLNAAQDLFQEMISH-GVCPDTITCNILLYGFCENEKLEEALE 518
Query: 92 LHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLC 151
L + ++++ I D +NI+I+ + ++ +F + G +P+ T+N +I G C
Sbjct: 519 LFEVIQMSKIDLDTVAYNIIIHGMCKGSKVDEAWDLFCSLPIHGVEPDVQTYNVMISGFC 578
Query: 152 LKGHIHQALNFHDKVVAQGFHLDQVSYGTLINGLCKVGRITAALQLLKRVDGKLVQPNAV 211
K I A K+ G D +Y TLI G K G I +++L+ + +A
Sbjct: 579 GKSAISDANVLFHKMKDNGHEPDNSTYNTLIRGCLKAGEIDKSIELISEMRSNGFSGDAF 638
Query: 212 MYNMIIDNMCKAKLVNDAFDLYS 234
M+ D + +L D+ S
Sbjct: 639 TIKMVADLITDGRLDKSFSDMLS 661
>UniRef100_Q8L8A3 Fertility restorer-like protein [Petunia hybrida]
Length = 591
Score = 321 bits (822), Expect = 4e-86
Identities = 181/545 (33%), Positives = 302/545 (55%), Gaps = 7/545 (1%)
Query: 16 PNFSHFQQSPNPNFIGLFPSSISLYSQLHHHQDEEN-----NLISSFNSLLHHKNPTPPI 70
P FS F S P SIS+ + +N + S F ++ K P P +
Sbjct: 14 PFFSFFAYSIAPRHYSTNTCSISVKGNFGVSNEFQNVKCLDDAFSLFRQMVRTK-PLPSV 72
Query: 71 IQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLGLNSLSFSVFGK 130
F+K+L ++V KH+S+ +SL +++ I F +I++N + L FSV
Sbjct: 73 ASFSKLLKAMVHMKHYSSVVSLFREIHKLRIPVHEFILSIVVNSCCLMHRTDLGFSVLAI 132
Query: 131 ILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGF-HLDQVSYGTLINGLCKVG 189
KKG N +TF TLI+GL + + A++ K+V + ++V YGT++NGLCK G
Sbjct: 133 HFKKGIPYNEVTFTTLIRGLFAENKVKDAVHLFKKLVRENICEPNEVMYGTVMNGLCKKG 192
Query: 190 RITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAKRISPDDFTCN 249
A LL+ ++ +PN Y ++ID CK +++ A L ++M K I PD FT +
Sbjct: 193 HTQKAFDLLRLMEQGSTKPNTRTYTIVIDAFCKDGMLDGATSLLNEMKQKSIPPDIFTYS 252
Query: 250 SLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEAKMMLGVTMKK 309
+LI C + Q + L +MI NI P + TF+ ++D CKEGKV++A+ ++ ++K
Sbjct: 253 TLIDALCKLSQWENVRTLFLEMIHLNIYPNVCTFNSVIDGLCKEGKVEDAEEIMRYMIEK 312
Query: 310 DIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMINGLCKIKMVDEA 369
+ DV+TYN ++DGY L ++++A++IFDSM ++ + ++ SY +ING + K +DEA
Sbjct: 313 GVDPDVITYNMIIDGYGLRGQVDRAREIFDSMINKSIEPDIISYNILINGYARQKKIDEA 372
Query: 370 VNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPNIITYNSILDA 429
+ + E+ + + P++VT N L+ GL +LG+ DEM G P++ T+ ++L
Sbjct: 373 MQVCREISQKGLKPSIVTCNVLLHGLFELGRTKSAQNFFDEMLSAGHIPDLYTHCTLLGG 432
Query: 430 LCKNHHVDKAIALLTNLKDQGIRPDMYTYTVLIKGLCQSGKLEDAQKVFEDLLVKGYNLD 489
KN V++A++ L+ + ++ YT +I GLC++GKL+ A FE L + G + D
Sbjct: 433 YFKNGLVEEAMSHFHKLERRREDTNIQIYTAVIDGLCKNGKLDKAHATFEKLPLIGLHPD 492
Query: 490 VYTYTVMIQGFCVKGLFDAALALLSKMEDNGCIPNAKTYEIVILSLFEKDENDMAEKLLR 549
V TYT MI G+C +GL D A +L KMEDNGC+ + +TY +++ ++ + L
Sbjct: 493 VITYTAMISGYCQEGLLDEAKDMLRKMEDNGCLADNRTYNVIVRGFLRSNKVSEMKAFLE 552
Query: 550 EMIAR 554
E+ +
Sbjct: 553 EIAGK 557
Score = 151 bits (382), Expect = 4e-35
Identities = 88/379 (23%), Positives = 186/379 (48%)
Query: 60 LLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILINCFSQLG 119
L+ + P + ++ + K A SL +M+ I D FT++ LI+ +L
Sbjct: 203 LMEQGSTKPNTRTYTIVIDAFCKDGMLDGATSLLNEMKQKSIPPDIFTYSTLIDALCKLS 262
Query: 120 LNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHLDQVSYG 179
++F +++ PN TFN++I GLC +G + A ++ +G D ++Y
Sbjct: 263 QWENVRTLFLEMIHLNIYPNVCTFNSVIDGLCKEGKVEDAEEIMRYMIEKGVDPDVITYN 322
Query: 180 TLINGLCKVGRITAALQLLKRVDGKLVQPNAVMYNMIIDNMCKAKLVNDAFDLYSQMVAK 239
+I+G G++ A ++ + K ++P+ + YN++I+ + K +++A + ++ K
Sbjct: 323 MIIDGYGLRGQVDRAREIFDSMINKSIEPDIISYNILINGYARQKKIDEAMQVCREISQK 382
Query: 240 RISPDDFTCNSLIYGFCIMGQLKEAVGLLHKMILENINPRMYTFSILVDAFCKEGKVKEA 299
+ P TCN L++G +G+ K A +M+ P +YT L+ + K G V+EA
Sbjct: 383 GLKPSIVTCNVLLHGLFELGRTKSAQNFFDEMLSAGHIPDLYTHCTLLGGYFKNGLVEEA 442
Query: 300 KMMLGVTMKKDIILDVVTYNSLMDGYCLVKEINKAKDIFDSMASRGVIANVQSYTTMING 359
++ ++ Y +++DG C +++KA F+ + G+ +V +YT MI+G
Sbjct: 443 MSHFHKLERRREDTNIQIYTAVIDGLCKNGKLDKAHATFEKLPLIGLHPDVITYTAMISG 502
Query: 360 LCKIKMVDEAVNLFEEMRCRKIIPNVVTYNSLIDGLGKLGKISCVLKLVDEMHDRGQPPN 419
C+ ++DEA ++ +M + + TYN ++ G + K+S + ++E+ +
Sbjct: 503 YCQEGLLDEAKDMLRKMEDNGCLADNRTYNVIVRGFLRSNKVSEMKAFLEEIAGKSFSFE 562
Query: 420 IITYNSILDALCKNHHVDK 438
T ++D + ++ + +
Sbjct: 563 AATVELLMDIIAEDPSITR 581
Score = 36.6 bits (83), Expect = 2.0
Identities = 27/146 (18%), Positives = 64/146 (43%), Gaps = 1/146 (0%)
Query: 54 ISSFNSLLHHKNPTPPIIQFNKILSSLVKAKHHSTALSLHQQMELNGIVSDFFTFNILIN 113
+S F+ L + T I + ++ L K A + +++ L G+ D T+ +I+
Sbjct: 443 MSHFHKLERRREDTNIQI-YTAVIDGLCKNGKLDKAHATFEKLPLIGLHPDVITYTAMIS 501
Query: 114 CFSQLGLNSLSFSVFGKILKKGFDPNAITFNTLIKGLCLKGHIHQALNFHDKVVAQGFHL 173
+ Q GL + + K+ G + T+N +++G + + F +++ + F
Sbjct: 502 GYCQEGLLDEAKDMLRKMEDNGCLADNRTYNVIVRGFLRSNKVSEMKAFLEEIAGKSFSF 561
Query: 174 DQVSYGTLINGLCKVGRITAALQLLK 199
+ + L++ + + IT + +K
Sbjct: 562 EAATVELLMDIIAEDPSITRKMHWIK 587
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.324 0.140 0.406
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 904,148,057
Number of Sequences: 2790947
Number of extensions: 37827756
Number of successful extensions: 114800
Number of sequences better than 10.0: 1189
Number of HSP's better than 10.0 without gapping: 975
Number of HSP's successfully gapped in prelim test: 215
Number of HSP's that attempted gapping in prelim test: 87650
Number of HSP's gapped (non-prelim): 7764
length of query: 557
length of database: 848,049,833
effective HSP length: 133
effective length of query: 424
effective length of database: 476,853,882
effective search space: 202186045968
effective search space used: 202186045968
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 77 (34.3 bits)
Medicago: description of AC138580.2


