

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.4 + phase: 0
(835 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
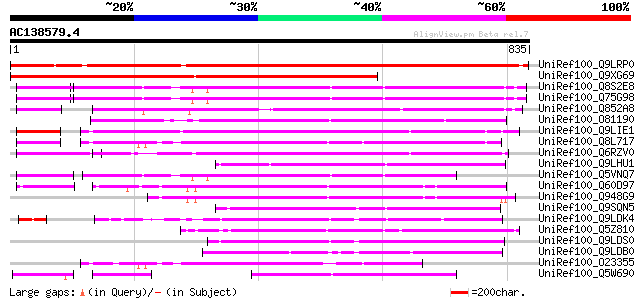
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LRP0 Emb|CAB39405.1 [Arabidopsis thaliana] 947 0.0
UniRef100_Q9XG69 Hypothetical protein [Nicotiana tabacum] 853 0.0
UniRef100_Q8S2E8 P0022F10.3 protein [Oryza sativa] 361 4e-98
UniRef100_Q75G98 Hypothetical protein OSJNBa0042F15.17 [Oryza sa... 351 6e-95
UniRef100_Q852A8 Hypothetical protein OSJNBb0081B07.17 [Oryza sa... 348 4e-94
UniRef100_O81190 Putative transposase [Arabidopsis thaliana] 347 6e-94
UniRef100_Q9LIE1 Transposase-like protein [Arabidopsis thaliana] 343 2e-92
UniRef100_Q8L717 Hypothetical protein At4g15020 [Arabidopsis tha... 340 1e-91
UniRef100_Q6RZV0 Transposase-like protein [Musa acuminata] 330 1e-88
UniRef100_Q9LHU1 Similar to Arabidopsis thaliana chromosome 1 [O... 288 5e-76
UniRef100_Q5VNQ7 HAT dimerisation domain-containing protein-like... 285 3e-75
UniRef100_Q60D97 Putative polyprotein [Oryza sativa] 276 1e-72
UniRef100_Q948G9 Putative transposable element [Oryza sativa] 254 1e-65
UniRef100_Q9SQN5 Hypothetical protein F19K16.28 [Arabidopsis tha... 251 6e-65
UniRef100_Q9LDK4 Transposase-like protein [Arabidopsis thaliana] 246 3e-63
UniRef100_Q5Z810 HAT dimerisation domain-containing protein-like... 232 4e-59
UniRef100_Q9LDS0 Transposase-like protein [Arabidopsis thaliana] 211 6e-53
UniRef100_Q9LDB0 Transposase-like protein [Arabidopsis thaliana] 201 7e-50
UniRef100_O23355 Hypothetical protein AT4g15020 [Arabidopsis tha... 200 2e-49
UniRef100_Q5W690 Hypothetical protein OSJNBa0076A09.5 [Oryza sat... 199 3e-49
>UniRef100_Q9LRP0 Emb|CAB39405.1 [Arabidopsis thaliana]
Length = 883
Score = 947 bits (2449), Expect = 0.0
Identities = 478/850 (56%), Positives = 631/850 (74%), Gaps = 37/850 (4%)
Query: 1 MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKA 60
MAP S G VDPGW+HG+AQD+RKKKV+CNYCGK+VSGGIYRLKQHLARVSGEVTYC+K+
Sbjct: 1 MAPPGSIGVVDPGWEHGVAQDQRKKKVKCNYCGKIVSGGIYRLKQHLARVSGEVTYCDKS 60
Query: 61 PEEVYLKMKENLEGCRSNKKQKQVD---AQAYMNFQ--SNDDEDDEEQVGC---RSKGKQ 112
PEEV ++MKENL RS KK +Q + Q+ +F +NDDE DEE+ C RSKGK
Sbjct: 61 PEEVCMRMKENL--VRSTKKLRQSEDNSGQSCSSFHQSNNDDEADEEERRCWSIRSKGKL 118
Query: 113 -LMDGRNVSVNLTPLRSLGYVDPGWEHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLAR 171
L DG + LRS GY+DPGWEHG+AQDERKKKVKC+YC K+VSGGINRFKQHLAR
Sbjct: 119 GLSDG-------SLLRSSGYIDPGWEHGIAQDERKKKVKCNYCNKIVSGGINRFKQHLAR 171
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAK-DLMPFYPKSDNEDDEYEQQED 230
IPGEVAPCK+APEEVY+KIKENMKWHR GKR +P+ + + F S + D E ++++
Sbjct: 172 IPGEVAPCKTAPEEVYVKIKENMKWHRAGKRQNRPDDEMGALTFRTVSQDPDQEEDREDH 231
Query: 231 TLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCK 290
+ +++ L+ + R+SKD K+F + + E +R+R+ F Q+ + K
Sbjct: 232 DFYPTSQDRLMLGNGRFSKDKRKSFDSTNMRSVSEAKTKRARMIPF-------QSPSSSK 284
Query: 291 QLKVKTGPTKKL--RKEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSS 348
Q K+ + + ++ RK+V SSI KF H G+P +AA+S+YF KM+EL G YG+G PSS
Sbjct: 285 QRKLYSSCSNRVVSRKDVTSSISKFLHHVGVPTEAANSLYFQKMIELIGMYGEGFVVPSS 344
Query: 349 QLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFV 408
QL SGR LQEE+++IK+YL EY++SW +TGCSIMAD+W + +G+ +I+FLVS P GVYF
Sbjct: 345 QLFSGRLLQEEMSTIKSYLREYRSSWVVTGCSIMADTWTNTEGKKMISFLVSCPRGVYFH 404
Query: 409 SSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTP 468
SS+DAT++VEDA LFK LDK+V+++GEENVVQVIT+NT +++AGK+LEE+R+NL+WTP
Sbjct: 405 SSIDATDIVEDALSLFKCLDKLVDDIGEENVVQVITQNTAIFRSAGKLLEEKRKNLYWTP 464
Query: 469 CAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQ 528
CAI+C VLEDF K+ V EC+EK Q+IT+ IYNQ WLLNLMK+EFT G +LL+PA +
Sbjct: 465 CAIHCTELVLEDFSKLEFVSECLEKAQRITRFIYNQTWLLNLMKNEFTQGLDLLRPAVMR 524
Query: 529 CASSFATLQNLLDHRVSLRRMFLSNKW-MSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRN 587
AS F TLQ+L+DH+ SLR +F S+ W +S + S +G+EV+K+VL+ FWKK+Q V
Sbjct: 525 HASGFTTLQSLMDHKASLRGLFQSDGWILSQTAAKSEEGREVEKMVLSAVFWKKVQYVLK 584
Query: 588 SVYPILQVFQKVS-SGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSL 646
SV P++QV ++ G+ LSMPY Y + AK+AIKSIH DDARKY PFW+VI+ N L
Sbjct: 585 SVDPVMQVIHMINDGGDRLSMPYAYGYMCCAKMAIKSIHSDDARKYGPFWRVIEYRWNPL 644
Query: 647 FCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQ 706
F HPLY+AAYF NP+Y+YR DF++ S+VVRG+NECIVRLE DN RRI+A MQIP Y A+
Sbjct: 645 FHHPLYVAAYFFNPAYKYRPDFMAQSEVVRGVNECIVRLEPDNTRRITALMQIPDYTCAK 704
Query: 707 DDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYDQIYS 766
DFGT++AI TRT L+P+AWWQQHGISCLELQR+AVRILS TCSS CE S+YDQ+ S
Sbjct: 705 ADFGTDIAIGTRTELDPSAWWQQHGISCLELQRVAVRILSHTCSSVGCEPKWSVYDQVNS 764
Query: 767 KRKNRLSQKKLNDIMYVHYNLRLRECQVRKR--SRESKSTSAENVLQEHLLGDWIVDTTA 824
+ +++ +K D+ YVHYNLRLRE Q+++R + + + L + LL DW+V
Sbjct: 765 QCQSQFGKKSTKDLTYVHYNLRLREKQLKQRLHYEDEPPPTLNHALLDRLLPDWLV---- 820
Query: 825 QSSDSDKVIS 834
+S+ ++V++
Sbjct: 821 -TSEKEEVLN 829
>UniRef100_Q9XG69 Hypothetical protein [Nicotiana tabacum]
Length = 593
Score = 853 bits (2204), Expect = 0.0
Identities = 410/594 (69%), Positives = 494/594 (83%), Gaps = 5/594 (0%)
Query: 1 MAPIRSTGFVDPGWDHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKA 60
MA +RSTG+VDPGW+HG+AQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYC+KA
Sbjct: 1 MASLRSTGYVDPGWEHGVAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCDKA 60
Query: 61 PEEVYLKMKENLEGCRSNKKQKQV--DAQAYMNFQSNDDEDDEEQVGCRSKGKQLMDGRN 118
PE+V LKM+ENLEGCR +KK + V D QAY+NF ++DD ++E+ +G R+KGKQLM+ +
Sbjct: 61 PEDVCLKMRENLEGCRLSKKPRHVEYDEQAYLNFHASDDAEEEDHIGYRNKGKQLMNDKG 120
Query: 119 VSVNLTPLRSLGYVDPGWEHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLARIPGEVAP 178
+ +NLTPLRSLGYVDPGWEHGV QDERKKKVKC+YCEK++SGGINRFKQHLARIPGEVAP
Sbjct: 121 LVINLTPLRSLGYVDPGWEHGVPQDERKKKVKCNYCEKIISGGINRFKQHLARIPGEVAP 180
Query: 179 CKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHH-MNK 237
CKSAPEEVYL+IKENMKWHRTG+RHR+P K+L FY SDNE+++ +Q+E+ LHH M+
Sbjct: 181 CKSAPEEVYLRIKENMKWHRTGRRHRRPHTKELSSFYMNSDNEEEDEDQEEEALHHHMSN 240
Query: 238 EALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQLKVKTG 297
E L+ D+R +D ++FKGMS E L+R + ++ K P + + K +KV +
Sbjct: 241 EKLLIGDKRLDRDCRRSFKGMSPGIGSESLLKRPKYETLGTKEPKSLFQASGKHVKVCSN 300
Query: 298 PTKKLRKEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQ 357
KK RKEV SSICKFF HAGI AA S YF KMLEL GQYG+GL PSS+++SGR LQ
Sbjct: 301 --KKSRKEVISSICKFFYHAGISPHAASSPYFQKMLELVGQYGEGLVGPSSRVLSGRLLQ 358
Query: 358 EEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVV 417
+EI SI+NYL+EYKASWA+TG SI+ADSW+D QGRT+IN LVS PHG+YFV SVDAT VV
Sbjct: 359 DEIVSIRNYLSEYKASWAVTGYSILADSWQDTQGRTLINVLVSCPHGMYFVCSVDATGVV 418
Query: 418 EDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQV 477
EDATY+FKLLD+VVE++GEENVVQVIT+NTPNY+AAGKMLEE+RRNLFWTPCA YCI+++
Sbjct: 419 EDATYIFKLLDRVVEDMGEENVVQVITQNTPNYQAAGKMLEEKRRNLFWTPCAAYCIDRI 478
Query: 478 LEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQ 537
LEDF+KI+ V ECMEK QKITK IYN WLL+LMK EFT G ELLKP+ T+ +S+FAT+Q
Sbjct: 479 LEDFVKIKWVRECMEKAQKITKFIYNSFWLLSLMKKEFTAGQELLKPSFTRYSSTFATVQ 538
Query: 538 NLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYP 591
+LLDHR L+RMF SNKW+SSR+S GKEV+KIVLN TFW+KMQ VR SV P
Sbjct: 539 SLLDHRNGLKRMFQSNKWLSSRYSKLEDGKEVEKIVLNATFWRKMQYVRKSVDP 592
>UniRef100_Q8S2E8 P0022F10.3 protein [Oryza sativa]
Length = 965
Score = 361 bits (927), Expect = 4e-98
Identities = 223/756 (29%), Positives = 392/756 (51%), Gaps = 44/756 (5%)
Query: 98 EDDEEQVGCR---SKGKQLMDGRNVSVNLTPLRS-LGYVDPGWEHGVAQDERKKK-VKCS 152
+ DEE C + +Q +VSV P ++ + DP W H D KK ++C
Sbjct: 151 KSDEEVRQCNVSEALKQQHQSAASVSVEAGPKQAGINSDDPAWAHCFCPDITKKHHLRCK 210
Query: 153 YCEKVVSGGINRFKQHLARIPG-EVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDL 211
YC+KV + GI R K HLA I G C+ P V+ ++ + + +T ++ ++ + K++
Sbjct: 211 YCDKVCTAGITRIKYHLAGIKGFNTTKCQKVPSPVHQEMFDLLT-KKTSEKEQKNKEKEV 269
Query: 212 MPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRS 271
+ D E+ + E + H N ++ + ++ G +
Sbjct: 270 AR--AQVDIENSDCESGSEGSDHGNNVLVVKPKETTGSSSSRSVAGGHT----------- 316
Query: 272 RLDSFYLKHPTNQNLQTCKQL-----KVKTGPTKKLRKEVFSSICKFFCH----AGIPLQ 322
+D +Y ++ +++ KV+T T + R+E + C++ C A IP
Sbjct: 317 -IDKYYKPPSIEESASMTQRVIKLSNKVQTALTTQKREERRNRTCEYICQWFYEASIPHN 375
Query: 323 AADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIM 382
F MLE GQ+G+ L PS +SG FLQ+ + + E+K SW +TGCSIM
Sbjct: 376 TVTLPSFAHMLEAIGQFGRSLKGPSPYEMSGSFLQKRKEKVMDGFKEHKESWELTGCSIM 435
Query: 383 ADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQV 442
D+W D +GR ++N +V S HGV F+ SV+ + +D Y+FKL+D+ +EE+GE++VVQV
Sbjct: 436 TDAWTDRKGRGVMNLVVHSAHGVLFLDSVECSGDRKDGKYIFKLVDRYIEEIGEQHVVQV 495
Query: 443 ITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIY 502
+T+N A +L +R ++FW CA +C++ +LED K+ VEE + +++T +Y
Sbjct: 496 VTDNASVNTIAASLLTAKRPSIFWNGCAAHCLDLMLEDIGKLGPVEETIANARQVTVFLY 555
Query: 503 NQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSS 562
+L+LM+ +L++ T+ A+++ L++LLD++ L R+F S++ +
Sbjct: 556 AHTRVLDLMRKFL--NRDLVRSGVTRFATAYLNLKSLLDNKKELVRLFKSDEMEQLGYLK 613
Query: 563 SSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIK 622
++GK+ K++ + TFWK + N P+ V +++ S + SM + + + AK I
Sbjct: 614 QTKGKKASKVIRSETFWKNVDIAVNYFEPLANVLRRMDS-DVPSMGFFHGLMLEAKKEIS 672
Query: 623 SIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRY---RQDFVSHSDVVRGLN 679
+D ++ W +ID+ ++ PL+LA Y+LNP Y Y +Q+ S G+
Sbjct: 673 QRFDNDKSRFIEVWDIIDKRWDNKLKTPLHLAGYYLNPYYYYYPNKQEIESDGSFRAGVI 732
Query: 680 ECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTR--TGLEPAAWWQQHGISCLEL 737
CI +L D + ++ Y FG E+A+ R PA WW HG S L L
Sbjct: 733 SCIDKLVDDEDIQDKIIEELNLYQDQHGSFGHEIAVRQRKNKNFNPAKWWLNHGTSTLNL 792
Query: 738 QRIAVRILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKR 797
+++A RILS TCSS ACE + S+++Q+++K++NRL +++ D++++ +N +LR + K
Sbjct: 793 RKLAARILSLTCSSSACERNWSVFEQVHTKKRNRLLHERMRDLVFIKFNSKLRNKRENK- 851
Query: 798 SRESKSTSAENVLQEHLLGD--WIVDTTAQSSDSDK 831
R+ ++V+ + GD +I SS+ D+
Sbjct: 852 GRDPIEKEVDDVVAD---GDNEFITGVVPSSSEMDQ 884
Score = 63.9 bits (154), Expect = 2e-08
Identities = 37/97 (38%), Positives = 53/97 (54%), Gaps = 5/97 (5%)
Query: 11 DPGWDHGIAQDERKKK-VRCNYCGKVVSGGIYRLKQHLARVSG-EVTYCEKAPEEVYLKM 68
DP W H D KK +RC YC KV + GI R+K HLA + G T C+K P V+ +M
Sbjct: 190 DPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKYHLAGIKGFNTTKCQKVPSPVHQEM 249
Query: 69 KENLEGCRSNKKQKQVD---AQAYMNFQSNDDEDDEE 102
+ L S K+QK + A+A ++ +++D E E
Sbjct: 250 FDLLTKKTSEKEQKNKEKEVARAQVDIENSDCESGSE 286
>UniRef100_Q75G98 Hypothetical protein OSJNBa0042F15.17 [Oryza sativa]
Length = 1005
Score = 351 bits (900), Expect = 6e-95
Identities = 220/755 (29%), Positives = 389/755 (51%), Gaps = 43/755 (5%)
Query: 98 EDDEEQVGCR---SKGKQLMDGRNVSVNLTPLRS-LGYVDPGWEHGVAQDERKKK-VKCS 152
+ DEE C + +Q +VSV P ++ + DP W H D KK ++C
Sbjct: 196 KSDEEVRQCNVSEALKQQHQSAASVSVEAGPKQAGINSDDPAWAHCFCPDITKKHHLRCK 255
Query: 153 YCEKVVSGGINRFKQHLARIPG-EVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDL 211
YC+KV + GI R K HLA I G C+ P V ++ + + +T ++ ++ + K++
Sbjct: 256 YCDKVCTAGITRIKYHLAGIKGFNTTKCQKVPSPVQQEMFDLLT-KKTSEKEQKNKEKEV 314
Query: 212 MPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRS 271
+ D E+ + E + H N ++ + ++ G +
Sbjct: 315 AR--AEVDIENSDCESGSEGSDHGNNVLVVKPKETTGSSSSRSVAGGHT----------- 361
Query: 272 RLDSFYLKHPTNQNLQTCKQL-----KVKTGPTKKLRKEVFSSICKFFCH----AGIPLQ 322
+D +Y ++ +++ KV+T T + R+E + C++ C A IP
Sbjct: 362 -IDKYYKPPSIEESASMTQRVIKLSNKVQTALTTQKREERRNRTCEYICQWFYEASIPHN 420
Query: 323 AADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIM 382
F MLE GQ+G+ L PS +SG FLQ+ + + E+K SW +TGCSIM
Sbjct: 421 TVTLPSFAHMLEAIGQFGRSLKGPSPYEMSGSFLQKRKEKVMDGFKEHKESWELTGCSIM 480
Query: 383 ADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQV 442
D+W D +GR ++N +V S HGV F+ SV+ + +D Y+F+L+D+ +EE+GE++VVQV
Sbjct: 481 TDAWTDRKGRGVMNLVVHSAHGVLFLDSVECSGDRKDGKYIFELVDRYIEEIGEQHVVQV 540
Query: 443 ITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIY 502
+T+N A +L +R ++FW A +C++ +LED K+ VEE + +++T +Y
Sbjct: 541 VTDNASVNTTAASLLTAKRPSIFWNGYAAHCLDLMLEDIGKLGPVEETIANARQVTVFLY 600
Query: 503 NQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSS 562
+L+LM+ +L++ T+ A+++ L++LLD++ L R+F S++ +
Sbjct: 601 AHTRVLDLMRKFL--NRDLVRSGVTRFATAYLNLKSLLDNKKELVRLFKSDEMEQLGYLK 658
Query: 563 SSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIK 622
++GK+ K++ + TFWK + N P+ V +++ S + SM + + + AK I
Sbjct: 659 QAKGKKASKVIRSETFWKNVDIAVNYFEPLANVLRRMDS-DVPSMGFFHGLMLEAKKEIS 717
Query: 623 SIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRY--RQDFVSHSDVVRGLNE 680
+D ++ W +ID+ ++ PL+LA Y+LNP Y + +Q+ S G+
Sbjct: 718 QRFDNDKSRFIEVWDIIDKRWDNKIKAPLHLAGYYLNPYYYFPNKQEIESDGSFRAGVIS 777
Query: 681 CIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTR--TGLEPAAWWQQHGISCLELQ 738
CI +L D + ++ Y FG E+A+ R PA WW HG S L+
Sbjct: 778 CIDKLIDDEDIQDKIIEELNLYQDQHGSFGHEIAVRQRKNKNFNPAKWWLNHGTSTPNLR 837
Query: 739 RIAVRILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRS 798
++A RILS TCSS ACE + S+++Q+++K++NRL +++ D+++V +N +LR + K
Sbjct: 838 KLAARILSLTCSSSACERNWSVFEQVHTKKRNRLLHERMRDLVFVKFNSKLRNKRENK-G 896
Query: 799 RESKSTSAENVLQEHLLGD--WIVDTTAQSSDSDK 831
R+ ++V+ + GD +I SS+ D+
Sbjct: 897 RDPIEKEVDDVVAD---GDNEFITGVVPSSSEMDQ 928
Score = 62.4 bits (150), Expect = 5e-08
Identities = 37/97 (38%), Positives = 52/97 (53%), Gaps = 5/97 (5%)
Query: 11 DPGWDHGIAQDERKKK-VRCNYCGKVVSGGIYRLKQHLARVSG-EVTYCEKAPEEVYLKM 68
DP W H D KK +RC YC KV + GI R+K HLA + G T C+K P V +M
Sbjct: 235 DPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKYHLAGIKGFNTTKCQKVPSPVQQEM 294
Query: 69 KENLEGCRSNKKQKQVD---AQAYMNFQSNDDEDDEE 102
+ L S K+QK + A+A ++ +++D E E
Sbjct: 295 FDLLTKKTSEKEQKNKEKEVARAEVDIENSDCESGSE 331
>UniRef100_Q852A8 Hypothetical protein OSJNBb0081B07.17 [Oryza sativa]
Length = 779
Score = 348 bits (893), Expect = 4e-94
Identities = 230/725 (31%), Positives = 359/725 (48%), Gaps = 59/725 (8%)
Query: 133 DPGWEH-GVAQDERKKKVKCSYCEK-VVSGGINRFKQHLARIPGEVAPCKSAPEEVYLKI 190
DP W+H + + + ++KC YC K + GGI+RFK+HLA PG C P EV +
Sbjct: 25 DPAWKHCQMVRSAGRVRLKCVYCHKHFLGGGIHRFKEHLANRPGNACCCPKVPREVQETM 84
Query: 191 KENMKWHRTGKRHRQPEAKDLMPF--------YPKSDNEDDEYEQQEDTLHHMNKEALID 242
++ K+ +Q A+ + S + + E +H + ++D
Sbjct: 85 LHSLDAVAAKKKRKQSLAEGIRRITHSAPAAAASASPPAPADAAEMESPIHMIPLNEVLD 144
Query: 243 IDRRYSKDTGKTFKGMSSNTSPE-PALRRSRLDSFYLKHPTNQNLQT------------- 288
+ ++T + M + S + L + + L H Q LQ+
Sbjct: 145 LGSVPLEETPPETREMKGSISKKRKKLAARQASTAPLAHQNQQPLQSTPAGLTQPFHQMV 204
Query: 289 ------CKQLKVKTGPTKKLRKEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQG 342
QL P +++V+ +I +F AG+ L+A +SVYF MLE G
Sbjct: 205 VAFDSAASQLMHFDQPGSN-KEQVYMAIGRFLYDAGVSLEAVNSVYFQPMLEAVASAGGK 263
Query: 343 LACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSP 402
S G L++ ++ + L YK SW TGC+++AD W +GRT+INF V P
Sbjct: 264 PEAFSYHDFRGSILKKSLDEVTAQLEFYKGSWTRTGCTLLADEWTTDRGRTLINFSVYCP 323
Query: 403 HGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRR 462
ED L++LL VVEE+GE+NVVQVIT N+ + AGK L E
Sbjct: 324 ---------------EDP--LYELLKNVVEEVGEKNVVQVITNNSEIHAVAGKRLCETFP 366
Query: 463 NLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELL 522
LFW+ C+ CI+ +LEDF K+ + E + + IT IYN + NLMK HG +LL
Sbjct: 367 TLFWSQCSFQCIDGMLEDFSKVGAINEIICNAKVITGFIYNSAFAFNLMKRHL-HGKDLL 425
Query: 523 KPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKM 582
PA T+ A +F TL+N+ + + SL M S++W+ G EV ++ N+ FW
Sbjct: 426 VPAETRAAMNFVTLKNMYNLKDSLEAMISSDEWIHYLLPKKPGGVEVTNLIGNLQFWSSC 485
Query: 583 QSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKS--IHGDDARKYEPFWKVID 640
+V P++ + + V S + SM Y+Y LY+AK AIK + +D Y +W +ID
Sbjct: 486 AAVVRITEPLVHLLKLVGSNKRPSMGYVYAGLYQAKAAIKKELVRKND---YMAYWDIID 542
Query: 641 RHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIP 700
N PL+LA +FLNP + + S++ G+ +C+ RL D + ++
Sbjct: 543 WRWNKDAPRPLHLAGFFLNPLFFDGVRGGTSSEIFSGMLDCVERLVSDVKIQDKIQKELN 602
Query: 701 HYNS-AQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGS 759
Y S A DF ++AI R L PA WW +G +C L R+AVRILSQTCS+ C+
Sbjct: 603 VYRSEAAGDFRRQMAIRARHTLPPAEWWYTYGGACPNLTRLAVRILSQTCSAKGCDRRHI 662
Query: 760 MYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAENVLQEHLLGDWI 819
++QI+ +R N +++++ + +V YNLRL+ Q K ++ S +N+ ++ DW+
Sbjct: 663 SFEQIHDQRMNLFERQRMHHLTFVQYNLRLQHRQQHK-TKAFDPVSVDNI---DIVDDWV 718
Query: 820 VDTTA 824
VD +A
Sbjct: 719 VDRSA 723
Score = 55.8 bits (133), Expect = 5e-06
Identities = 28/75 (37%), Positives = 41/75 (54%), Gaps = 2/75 (2%)
Query: 11 DPGWDH-GIAQDERKKKVRCNYCGK-VVSGGIYRLKQHLARVSGEVTYCEKAPEEVYLKM 68
DP W H + + + +++C YC K + GGI+R K+HLA G C K P EV M
Sbjct: 25 DPAWKHCQMVRSAGRVRLKCVYCHKHFLGGGIHRFKEHLANRPGNACCCPKVPREVQETM 84
Query: 69 KENLEGCRSNKKQKQ 83
+L+ + KK+KQ
Sbjct: 85 LHSLDAVAAKKKRKQ 99
>UniRef100_O81190 Putative transposase [Arabidopsis thaliana]
Length = 729
Score = 347 bits (891), Expect = 6e-94
Identities = 201/673 (29%), Positives = 350/673 (51%), Gaps = 42/673 (6%)
Query: 130 GYVDPGWEHGV-AQDERKKKV-KCSYCEKVV-SGGINRFKQHLARIPGEVAPCKSAPEEV 186
G D W + + ++DE+ K V +C++C K GGINR K HLA + G C ++
Sbjct: 21 GKSDIAWSYVIQSKDEKGKTVLECAFCHKKKRGGGINRMKHHLAGVKGNTDACLKISADI 80
Query: 187 YLKIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRR 246
KI +K K+ + D P+ ++ D + + + K+ ID+
Sbjct: 81 RFKIVNALK-EAENKKKQNIVLDDTNLDGPEIEDVDGDDIRVDVRPSQKRKKQGIDLHDY 139
Query: 247 YSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQLKVKTGPTKKLRKEV 306
+ + G+ T P +++ C Q K + V
Sbjct: 140 FKR-------GVHDQTQP--------------------SIKACMQSKERI-------HAV 165
Query: 307 FSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNY 366
S+ +F A IP+ A +S F M+ + G G PS + L++ +
Sbjct: 166 DMSVALWFYDACIPMNAVNSPLFQPMMSMVASMGHGYVGPSYHALRVGLLRDAKLQVSLI 225
Query: 367 LAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKL 426
+ ++K+SWA TGC++MAD W+D + R +INFLV P G+ F+ SVDA+++ A L L
Sbjct: 226 IDKFKSSWASTGCTLMADGWKDTRQRPLINFLVYCPKGITFLKSVDASDIYASAENLCNL 285
Query: 427 LDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRC 486
++V +G EN+V +T++ PNYKAAGK+L E+ + W+PC+ +CIN +LED K+
Sbjct: 286 FAELVGIIGSENIVHFVTDSAPNYKAAGKLLVEKFPTIAWSPCSAHCINLILEDVAKLPH 345
Query: 487 VEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSL 546
V + + K+T +YN LN ++ + E+++P T+ A++F LQ+L H+ L
Sbjct: 346 VHHIVRRMSKVTIFVYNHKPALNWVRKR-SGWREIIRPGETRFATTFIALQSLYQHKEDL 404
Query: 547 RRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLS 606
+ + S + +S+ K + ++L+ W + + PI+++ + + E S
Sbjct: 405 QALVTSADPELKQLFKTSKAKVAKSVILDERMWNDCLIIVKVMTPIIRLLRICDADEKPS 464
Query: 607 MPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQ 666
+PY+Y +YRA+L IK+I + Y+P+ +IDR + + H L+ AAY+LNP++ Y Q
Sbjct: 465 LPYVYEGMYRARLGIKNIFQEKETLYKPYTNIIDRRWDRMLRHDLHAAAYYLNPAFMYDQ 524
Query: 667 -DFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAA 725
F +V+ GL + + D+ ++ +++ Y + F ++A++ +P
Sbjct: 525 PTFCEKPEVMSGLMNLFEKQKNDSKTKLFQELRV--YREREGSFSLDMALTCSKTSQPDE 582
Query: 726 WWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHY 785
WW+ G LQ++A+RILSQT SS CE + ++++I++K++NRL ++LND+++VHY
Sbjct: 583 WWRYFGHDAPNLQKMAIRILSQTASSSGCERNWCVFERIHTKKRNRLEHQRLNDLVFVHY 642
Query: 786 NLRLRECQVRKRS 798
NLRL+ RKRS
Sbjct: 643 NLRLQHRSKRKRS 655
Score = 46.2 bits (108), Expect = 0.004
Identities = 36/113 (31%), Positives = 53/113 (46%), Gaps = 4/113 (3%)
Query: 8 GFVDPGWDHGI-AQDERKKKV-RCNYCGKVV-SGGIYRLKQHLARVSGEVTYCEKAPEEV 64
G D W + I ++DE+ K V C +C K GGI R+K HLA V G C K ++
Sbjct: 21 GKSDIAWSYVIQSKDEKGKTVLECAFCHKKKRGGGINRMKHHLAGVKGNTDACLKISADI 80
Query: 65 YLKMKENLEGCRSNKKQKQVDAQAYMNFQSNDDED-DEEQVGCRSKGKQLMDG 116
K+ L+ + KKQ V ++ +D D D+ +V R K+ G
Sbjct: 81 RFKIVNALKEAENKKKQNIVLDDTNLDGPEIEDVDGDDIRVDVRPSQKRKKQG 133
>UniRef100_Q9LIE1 Transposase-like protein [Arabidopsis thaliana]
Length = 761
Score = 343 bits (879), Expect = 2e-92
Identities = 220/712 (30%), Positives = 367/712 (50%), Gaps = 22/712 (3%)
Query: 114 MDGRNVSVNLTPLRSLGYVDPGWEH-GVAQDERKKKVKCSYCEKVVSGG-INRFKQHLAR 171
MD V LTP + D W+H V + + +++C YC K+ GG I R K+HLA
Sbjct: 1 MDSDLEPVALTPQKQ----DSAWKHCEVYKYGDRVQMRCLYCRKMFKGGGITRVKEHLAG 56
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDLMP--FYPKSDNEDDEYEQQE 229
G+ C P+EV L +++ + +R R+ + + +P ++P + E +
Sbjct: 57 KKGQGTICDQVPDEVRLFLQQCIDGTVRRQRKRRKSSPEPLPIAYFPPCEVETQVAASSD 116
Query: 230 DTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTC 289
+ + + + + + +T++ +N L +D + N
Sbjct: 117 VNNGFKSPSSDVVVGQSTGRTKQRTYRSRKNNAFERNDLANVEVD----RDMDNLIPVAI 172
Query: 290 KQLKVKTGPTKKLR-KEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSS 348
+K PT K R K V ++ +F G AA+SV ++ G G++ P+
Sbjct: 173 SSVKNIVHPTSKEREKTVHMAMGRFLFDIGADFDAANSVNVQPFIDAIVSGGFGVSIPTH 232
Query: 349 QLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFV 408
+ + G L+ + +K + E K W TGCS++ +G I+ FLV P V F+
Sbjct: 233 EDLRGWILKSCVEEVKKEIDECKTLWKRTGCSVLVQELNSNEGPLILKFLVYCPEKVVFL 292
Query: 409 SSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTP 468
SVDA+ +++ L++LL +VVEE+G+ NVVQVIT+ +Y AAGK L + +L+W P
Sbjct: 293 KSVDASEILDSEDKLYELLKEVVEEIGDTNVVQVITKCEDHYAAAGKKLMDVYPSLYWVP 352
Query: 469 CAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQ 528
CA +CI+++LE+F K+ + E +E+ + +T++IYN +LNLM+ +FT GN++++P T
Sbjct: 353 CAAHCIDKMLEEFGKMDWIREIIEQARTVTRIIYNHSGVLNLMR-KFTFGNDIVQPVCTS 411
Query: 529 CASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNS 588
A++F T+ + D + L+ M S++W +S + G + + + + FWK + +
Sbjct: 412 SATNFTTMGRIADLKPYLQAMVTSSEWNDCSYSKEAGGLAMTETINDEDFWKALTLANHI 471
Query: 589 VYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFC 648
PIL+V + V S +M Y+Y +YRAK AIK+ + +Y +WK+IDR
Sbjct: 472 TAPILRVLRIVCSERKPAMGYVYAAMYRAKEAIKT-NLAHREEYIVYWKIIDRW---WLQ 527
Query: 649 HPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDD 708
PLY A ++LNP + Y D S++ + +CI +L D + I Y +A
Sbjct: 528 QPLYAAGFYLNPKFFYSIDEEMRSEIHLAVVDCIEKLVPDVNIQDIVIKDINSYKNAVGI 587
Query: 709 FGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTC-SSFACEHDGSMYDQIYSK 767
FG LAI R + PA WW +G SCL L R A+RILSQTC SS + + QIY +
Sbjct: 588 FGRNLAIRARDTMLPAEWWSTYGESCLNLSRFAIRILSQTCSSSIGSVRNLTSISQIY-E 646
Query: 768 RKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAENVLQEHLLGDWI 819
KN + +++LND+++V YN+RLR ++ S + + +L DW+
Sbjct: 647 SKNSIERQRLNDLVFVQYNMRLR--RIGSESSGDDTVDPLSHSNMEVLEDWV 696
Score = 49.7 bits (117), Expect = 4e-04
Identities = 27/76 (35%), Positives = 46/76 (60%), Gaps = 4/76 (5%)
Query: 11 DPGWDH-GIAQDERKKKVRCNYCGKVVSGG-IYRLKQHLARVSGEVTYCEKAPEEVYLKM 68
D W H + + + ++RC YC K+ GG I R+K+HLA G+ T C++ P+EV L +
Sbjct: 16 DSAWKHCEVYKYGDRVQMRCLYCRKMFKGGGITRVKEHLAGKKGQGTICDQVPDEVRLFL 75
Query: 69 KENLEGC--RSNKKQK 82
++ ++G R K++K
Sbjct: 76 QQCIDGTVRRQRKRRK 91
>UniRef100_Q8L717 Hypothetical protein At4g15020 [Arabidopsis thaliana]
Length = 768
Score = 340 bits (871), Expect = 1e-91
Identities = 232/701 (33%), Positives = 351/701 (49%), Gaps = 48/701 (6%)
Query: 114 MDGRNVSVNLTPLRSLGYVDPGWEH-GVAQDERKKKVKCSYCEKVVSGG-INRFKQHLAR 171
MD V LTP + D W+H + + + +++C YC K+ GG I R K+HLA
Sbjct: 1 MDAELEPVALTPQKQ----DNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVKEHLAG 56
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKW--HRTGKRHRQ-----------PEAKDLMPFYP-- 216
G+ C PE+V L +++ + R KRH+ P D+M P
Sbjct: 57 KKGQGTICDQVPEDVRLFLQQCIDGTVRRQRKRHKSSSEPLSVASLPPIEGDMMVVQPDV 116
Query: 217 ----KSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSR 272
KS D Q E L K+ R Y G +SN
Sbjct: 117 NDGFKSPGSSDVVVQNESLLSGRTKQ------RTYRSKKNAFENGSASNNV--------- 161
Query: 273 LDSFYLKHPTNQNLQTCKQLKVKTGPTKKLRKE-VFSSICKFFCHAGIPLQAADSVYFHK 331
+ N +K P+ + R+ + +I +F G A +SV F
Sbjct: 162 --DLIGRDMDNLIPVAISSVKNIVHPSFRDRENTIHMAIGRFLFGIGADFDAVNSVNFQP 219
Query: 332 MLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQG 391
M++ G G++ P+ + G L+ + + + E KA W TGCSI+ + +G
Sbjct: 220 MIDAIASGGFGVSAPTHDDLRGWILKNCVEEMAKEIDECKAMWKRTGCSILVEELNSDKG 279
Query: 392 RTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYK 451
++NFLV P V F+ SVDA+ V+ A LF+LL ++VEE+G NVVQVIT+ Y
Sbjct: 280 FKVLNFLVYCPEKVVFLKSVDASEVLSSADKLFELLSELVEEVGSTNVVQVITKCDDYYV 339
Query: 452 AAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLM 511
AGK L +L+W PCA +CI+Q+LE+F K+ + E +E+ Q IT+ +YN +LNLM
Sbjct: 340 DAGKRLMLVYPSLYWVPCAAHCIDQMLEEFGKLGWISETIEQAQAITRFVYNHSGVLNLM 399
Query: 512 KSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQK 571
+FT GN++L PA + A++FATL + + + +L+ M S +W +S G V
Sbjct: 400 -WKFTSGNDILLPAFSSSATNFATLGRIAELKSNLQAMVTSAEWNECSYSEEPSGL-VMN 457
Query: 572 IVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARK 631
+ + FWK + V + P+L+ + V S + +M Y+Y LYRAK AIK+ H +
Sbjct: 458 ALTDEAFWKAVALVNHLTSPLLRALRIVCSEKRPAMGYVYAALYRAKDAIKT-HLVNRED 516
Query: 632 YEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMR 691
Y +WK+IDR PL A +FLNP Y + S+++ + +CI RL D+
Sbjct: 517 YIIYWKIIDRWWEQQQHIPLLAAGFFLNPKLFYNTNEEMRSELILSVLDCIERLVPDDKI 576
Query: 692 RISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTC-S 750
+ ++ Y +A FG LAI R + PA WW +G SCL L R A+RILSQTC S
Sbjct: 577 QDKIIKELTSYKTAGGVFGRNLAIRARDTMLPAEWWSTYGESCLNLSRFAIRILSQTCSS 636
Query: 751 SFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRE 791
S +C + + IY + KN + QK+L+D+++V YN+RLR+
Sbjct: 637 SVSCRRNQIPVEHIY-QSKNSIEQKRLSDLVFVQYNMRLRQ 676
Score = 49.3 bits (116), Expect = 5e-04
Identities = 28/76 (36%), Positives = 45/76 (58%), Gaps = 4/76 (5%)
Query: 11 DPGWDH-GIAQDERKKKVRCNYCGKVVSGG-IYRLKQHLARVSGEVTYCEKAPEEVYLKM 68
D W H I + + ++RC YC K+ GG I R+K+HLA G+ T C++ PE+V L +
Sbjct: 16 DNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVKEHLAGKKGQGTICDQVPEDVRLFL 75
Query: 69 KENLEGC--RSNKKQK 82
++ ++G R K+ K
Sbjct: 76 QQCIDGTVRRQRKRHK 91
>UniRef100_Q6RZV0 Transposase-like protein [Musa acuminata]
Length = 670
Score = 330 bits (845), Expect = 1e-88
Identities = 208/672 (30%), Positives = 334/672 (48%), Gaps = 47/672 (6%)
Query: 133 DPGWEHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLARIPG-EVAPCKSAPEEVYLKIK 191
D WEH V D ++KV+C+YC + SGG+ R K HLA+I ++ PC P++V I
Sbjct: 6 DACWEHCVLVDATRQKVRCNYCHREFSGGVYRMKFHLAQIKNKDIVPCSEVPDDVRNLIH 65
Query: 192 ENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDT 251
+ T ++ + P K+ ID + +
Sbjct: 66 SILT---TPRKQKAP------------------------------KKLKIDHTANGPQHS 92
Query: 252 GKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQLKVKTGPTKKLRKEVFSSIC 311
+ G ++ + S S L P+ T + K+ + I
Sbjct: 93 SSSASGYNAKNAGSSGQHGSTCPSLLLPLPSPGAQPTANDAQ------KQKYDNADNKIA 146
Query: 312 KFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYK 371
FF H IP A+ S+Y+ M++ G G P+ + + L++ I + K
Sbjct: 147 LFFFHNSIPFSASKSIYYQAMIDAIADCGAGYKPPTYEGLRSTLLEKVKEEINENHRKLK 206
Query: 372 ASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVV 431
W TGC+I++D+W D + ++++ V+SP G F+ VD ++ +DA YLF+LLD V+
Sbjct: 207 DEWKDTGCTILSDNWSDGRSKSLLVLSVASPKGTQFLKLVDISSRADDAYYLFELLDSVI 266
Query: 432 EELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECM 491
E+G ENVVQVIT++ +Y A +L ++ +LFW PCA Y I ++LED K+ V +
Sbjct: 267 MEVGAENVVQVITDSATSYTYAAGLLLKKYPSLFWFPCASYSIEKMLEDISKLEWVSTTL 326
Query: 492 EKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFL 551
E+ + I + I + W+L+LMK + T G EL++P + + F TL+++++ L+ F
Sbjct: 327 EETRTIARFICSDGWILSLMK-KLTGGRELVRPKVARFMTHFLTLRSIVNQEDDLKHFFS 385
Query: 552 SNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIY 611
W+SS S ++ ++ FWK + P+L++ + V G+ +M YIY
Sbjct: 386 HADWLSSVHSRRPDALAIKSLLYLERFWKSAHEIIGMSEPLLKLLRLV-DGDMPAMGYIY 444
Query: 612 NDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHP-LYLAAYFLNPSYRYRQDFVS 670
+ RAK+AIK+ + KY ++I+R S+ CH L+ AA FLNPS Y F
Sbjct: 445 EGIERAKMAIKAFYKGCEEKYMSVLEIIERRW-SMHCHSHLHAAAAFLNPSIFYDPSFKF 503
Query: 671 HSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQH 730
++ G + + ++ + RI P Y AQ G++ AI RT P WW +
Sbjct: 504 DVNMRNGFHAAMWKMFPEENDRIELIKDQPVYIKAQGALGSKFAIMGRTLNSPGDWWATY 563
Query: 731 GISCLELQRIAVRILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLR 790
G LQR AVRILSQ CSS+ + + S ++ IY+K R+ +KLND+++VH NLRL+
Sbjct: 564 GYEIPVLQRAAVRILSQPCSSYWFKWNWSAFENIYTKNHTRMELEKLNDLVFVHCNLRLQ 623
Query: 791 ECQVRKRSRESK 802
E RSR +K
Sbjct: 624 EI---SRSRGAK 632
Score = 59.7 bits (143), Expect = 4e-07
Identities = 39/139 (28%), Positives = 61/139 (43%), Gaps = 8/139 (5%)
Query: 11 DPGWDHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSG-EVTYCEKAPEEVYLKMK 69
D W+H + D ++KVRCNYC + SGG+YR+K HLA++ ++ C + P++V +
Sbjct: 6 DACWEHCVLVDATRQKVRCNYCHREFSGGVYRMKFHLAQIKNKDIVPCSEVPDDVRNLIH 65
Query: 70 ENLEGCRSNKKQKQVDAQAYMNFQSNDDEDDEEQVGCRSKGKQLMDGRNVSVNLTPLRSL 129
L R K K++ N + ++ G G L PL S
Sbjct: 66 SILTTPRKQKAPKKLKIDHTANGPQH-SSSSASGYNAKNAGSSGQHGSTCPSLLLPLPSP 124
Query: 130 GYVDPGWEHGVAQDERKKK 148
G A D +K+K
Sbjct: 125 G------AQPTANDAQKQK 137
>UniRef100_Q9LHU1 Similar to Arabidopsis thaliana chromosome 1 [Oryza sativa]
Length = 521
Score = 288 bits (737), Expect = 5e-76
Identities = 154/466 (33%), Positives = 267/466 (57%), Gaps = 5/466 (1%)
Query: 332 MLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQG 391
M+E G GQG PS++++ ++L + + + E + WA TGC+I+ADSW D +
Sbjct: 1 MMEALG--GQGFRGPSAEVLKTKWLHKLKSEVLQKTKEIEKDWATTGCTILADSWTDNKS 58
Query: 392 RTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYK 451
+ +INF VSSP G +F+ +VDA+ ++ + L++L D V+ E+G +NVVQ+IT+ NY
Sbjct: 59 KALINFSVSSPLGTFFLKTVDASPHIK-SHQLYELFDDVIREVGPDNVVQIITDRNINYG 117
Query: 452 AAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLM 511
+ K++ + +FW+PCA C+N +L+DF KI V C+ + Q IT+ +YN W+L+LM
Sbjct: 118 SVDKLIMQNYNTIFWSPCASSCVNSMLDDFSKIDWVNRCICQAQTITRFVYNNKWVLDLM 177
Query: 512 KSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQK 571
+ + G EL+ T+C S F TLQ+LL HR L++MF S+ + SS +++ S +
Sbjct: 178 R-KCIAGQELVCSGITKCVSDFLTLQSLLRHRPKLKQMFHSSDYASSSYANRSLSSSCVE 236
Query: 572 IVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARK 631
I+ + FW+ ++ + P+L+V + V G++ ++ YIY + + +I++ + D K
Sbjct: 237 ILDDDEFWRAVEEIAAVSEPLLRVMRDVLGGKA-AIGYIYESMTKVMDSIRTYYIMDEGK 295
Query: 632 YEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMR 691
+ F ++++ PL+ AA FLNPS +Y + + + + ++ +
Sbjct: 296 CKSFLDIVEQKWQVELHSPLHSAAAFLNPSIQYNPEVKFFTSIKEEFYHVLDKVLTVPDQ 355
Query: 692 RISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSS 751
R ++++ + AQ FG+ +A R P WW+Q+G S LQ AVRI+SQ CS+
Sbjct: 356 RQGITVELHAFRKAQGMFGSNIAKEARNNTSPGMWWEQYGDSAPSLQHAAVRIVSQVCST 415
Query: 752 FACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKR 797
+ D S+ + +S+++N+L ++ L D YVHYN L K+
Sbjct: 416 LTFQRDWSIIVRNHSEKRNKLDKEALADQAYVHYNFMLHSDSKMKK 461
>UniRef100_Q5VNQ7 HAT dimerisation domain-containing protein-like [Oryza sativa]
Length = 622
Score = 285 bits (730), Expect = 3e-75
Identities = 177/615 (28%), Positives = 313/615 (50%), Gaps = 32/615 (5%)
Query: 118 NVSVNLTPLRS-LGYVDPGWEHGVAQDERKKK-VKCSYCEKVVSGGINRFKQHLARIPG- 174
+VSV P ++ + DP W H D KK ++C YC+KV + GI R K HLA I G
Sbjct: 13 SVSVEAGPKQAGINSDDPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKYHLAGIKGF 72
Query: 175 EVAPCKSAPEEVYLKIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHH 234
C+ P V ++ + + +T ++ ++ + K++ + D E+ + E + H
Sbjct: 73 NTTKCQKVPSPVQQEMFDLLT-KKTSEKEQKNKEKEVAR--AEVDIENSDCESGSEGSDH 129
Query: 235 MNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQL-- 292
N ++ + ++ G + +D +Y ++ +++
Sbjct: 130 GNNVLVVKPKETTGSSSSRSVAGGHT------------IDKYYKPPSIEESASMTQRVIK 177
Query: 293 ---KVKTGPTKKLRKEVFSSICKFFCH----AGIPLQAADSVYFHKMLELAGQYGQGLAC 345
KV+T T + R+E + C++ C A IP F MLE GQ+G+ L
Sbjct: 178 LSNKVQTTLTTQKREERRNRTCEYICQWFYEASIPHNTVTLPSFAHMLEAIGQFGRSLKG 237
Query: 346 PSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGV 405
PS +SG FLQ+ + + E+K SW +TGCSIM D+W D +GR ++N +V S HGV
Sbjct: 238 PSPYEMSGSFLQKRKEKVMDGFKEHKESWELTGCSIMTDAWTDRKGRGVMNLVVHSAHGV 297
Query: 406 YFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLF 465
F+ SV+ + +D Y+F+L+D+ +EE+GE++VVQV+T+N A +L +R ++F
Sbjct: 298 LFLDSVECSGDRKDGKYIFELVDRYIEEIGEQHVVQVVTDNASVNTTAASLLTAKRPSIF 357
Query: 466 WTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPA 525
W CA +C++ +LED K+ VEE + +++T +Y +L+LM+ +L++
Sbjct: 358 WNGCAAHCLDLMLEDIGKLGPVEETIANARQVTVFLYAHTRVLDLMRKFL--NRDLVRSG 415
Query: 526 GTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSV 585
T+ A+++ L++LLD++ L R+F SN+ + ++GK+ K++++ TFWK +
Sbjct: 416 VTRFATAYLNLKSLLDNKKELVRLFKSNEMEQLGYLKQAKGKKASKVIISETFWKNVDIA 475
Query: 586 RNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNS 645
N P+ V +++ S + SM + + + AK I +D ++ W +ID+ ++
Sbjct: 476 VNYFEPLANVLRRMDS-DVPSMGFFHGLMLEAKKEISQRFDNDKSRFIEVWDIIDKRWDN 534
Query: 646 LFCHPLYLAAYFLNPSYRY--RQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYN 703
L+LA Y+LNP Y Y +Q+ S G+ CI +L D + ++ Y
Sbjct: 535 KLKTLLHLAGYYLNPYYYYPNKQEIESDGSFRAGVISCIDKLVDDEDIQDKIIEELNLYQ 594
Query: 704 SAQDDFGTELAISTR 718
FG E+A+ R
Sbjct: 595 DQHGSFGHEIAVRQR 609
Score = 62.4 bits (150), Expect = 5e-08
Identities = 37/97 (38%), Positives = 52/97 (53%), Gaps = 5/97 (5%)
Query: 11 DPGWDHGIAQDERKKK-VRCNYCGKVVSGGIYRLKQHLARVSG-EVTYCEKAPEEVYLKM 68
DP W H D KK +RC YC KV + GI R+K HLA + G T C+K P V +M
Sbjct: 29 DPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKYHLAGIKGFNTTKCQKVPSPVQQEM 88
Query: 69 KENLEGCRSNKKQKQVD---AQAYMNFQSNDDEDDEE 102
+ L S K+QK + A+A ++ +++D E E
Sbjct: 89 FDLLTKKTSEKEQKNKEKEVARAEVDIENSDCESGSE 125
>UniRef100_Q60D97 Putative polyprotein [Oryza sativa]
Length = 1086
Score = 276 bits (707), Expect = 1e-72
Identities = 195/695 (28%), Positives = 338/695 (48%), Gaps = 42/695 (6%)
Query: 133 DPGWEHGVAQDERKKKVKCSYCE-KVVSGGINRFKQHLARIPGEVAPCKSAPEEVYL--- 188
DP WEHG + +C YC K GG R KQHLA V C S P +V
Sbjct: 129 DPVWEHG---ENIPPGWRCKYCHTKRGGGGATRLKQHLAARGKGVTYCNSVPPDVREFFC 185
Query: 189 ----KIKENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDID 244
+IK+ ++ R A+ + +Y + + D+E EQ E + ++
Sbjct: 186 RELDRIKDAGDQRKSDSGRRVEAAR--VNYYDLTGDADEE-EQMEAAIAASRQDENF--- 239
Query: 245 RRYSKDTGKTFK--GMSSNTSPEPALRRSRLDSFYLKHPTNQ---------NLQTCK--- 290
RR ++ G T++ G S + PE RS + L+ T+ NL + K
Sbjct: 240 RRDVEERGGTYEHGGGSGSAQPEARKGRSNPITNMLRRATSHRESPAVRDYNLASAKAPV 299
Query: 291 QLKVKTG----PTKKLRKEVFSSICKFFCHAGIPLQAADSVYFHKMLELAGQYGQGLACP 346
Q ++ TG K+ R+ + S +FF AGIP + AD+ YF + ++G+ + P
Sbjct: 300 QPRIDTGFFTKKGKQARQAIGESWARFFFTAGIPGRNADNPYFVSAVRETQKWGESVPSP 359
Query: 347 SSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVY 406
+ I G++L +K +K W G +IM DSW ++INFL+ ++
Sbjct: 360 TGNEIDGKYLDSTEKDVKKQFDRFKKDWDEYGVTIMCDSWTGPTSMSVINFLIYCNGIMF 419
Query: 407 FVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFW 466
F S+DAT +DA ++ K + KVV E+G E+VVQ+IT+N NYK A ++L + + + W
Sbjct: 420 FHKSIDATGQSQDANFVLKEIRKVVREIGSEHVVQIITDNGSNYKKACRLLRQEYKTIVW 479
Query: 467 TPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAG 526
PC + +N +L++ K+ E +E +KI + +YN L +M G EL+K
Sbjct: 480 QPCVAHTVNLMLKEVGKMPDHEMVIESARKICRWLYNHNKLHAMM--VLAIGGELVKWNA 537
Query: 527 TQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVR 586
T+ +++ LQ+ L R + S+ +M S+FS + +G+ + ++++W+ +++V
Sbjct: 538 TRFGTNYMFLQSFLRKRDLFMQWMASSVFMQSKFSGTLEGRYAHACLSSLSWWENLEAVV 597
Query: 587 NSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSL 646
NSV P+ + ++ ++ + K+ ++ + K+E + ++++R + L
Sbjct: 598 NSVQPMYSFLRFADEDKNPNLSEVLLRYQLLKMEYDTLFANQRDKFEAYMEIVNRRMHDL 657
Query: 647 FCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQ 706
L AA LNP Y + + V + L + + D +A +++ Y
Sbjct: 658 TNETLINAAAALNPRTHYA--YSPSATVFQDLRQAFEWM-TDIDTAAAALLEVEMYRRKT 714
Query: 707 DDFGTELA--ISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSSFACEHDGSMYDQI 764
+FG LA ++ PA WW L+++A+R++ Q CSS CE + S + +
Sbjct: 715 GEFGRALARRMAIDGKTSPAQWWSMFASDTPNLKKLALRLVGQCCSSSGCERNWSTFAFV 774
Query: 765 YSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSR 799
++K +NRL+ KKLN ++YV+YNLRLR Q + R
Sbjct: 775 HTKVRNRLTHKKLNKLVYVNYNLRLRIQQANAQIR 809
Score = 50.4 bits (119), Expect = 2e-04
Identities = 33/101 (32%), Positives = 48/101 (46%), Gaps = 10/101 (9%)
Query: 11 DPGWDHGIAQDERKKKVRCNYCG-KVVSGGIYRLKQHLARVSGEVTYCEKAPEEV---YL 66
DP W+HG + RC YC K GG RLKQHLA VTYC P +V +
Sbjct: 129 DPVWEHG---ENIPPGWRCKYCHTKRGGGGATRLKQHLAARGKGVTYCNSVPPDVREFFC 185
Query: 67 KMKENLEGCRSNKKQ---KQVDAQAYMNFQSNDDEDDEEQV 104
+ + ++ +K ++V+A + D D+EEQ+
Sbjct: 186 RELDRIKDAGDQRKSDSGRRVEAARVNYYDLTGDADEEEQM 226
>UniRef100_Q948G9 Putative transposable element [Oryza sativa]
Length = 891
Score = 254 bits (648), Expect = 1e-65
Identities = 173/626 (27%), Positives = 310/626 (48%), Gaps = 42/626 (6%)
Query: 223 DEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFK--GMSSNTSPEPALRRSRLDSFYLKH 280
DE EQ E + ++ RR ++ G T++ G S + PE RS + L+
Sbjct: 109 DEEEQMEAAIAASRQDENF---RRDVEERGGTYEHGGGSGSAQPEARKGRSNPITNMLRR 165
Query: 281 PTNQ---------NLQTCK---QLKVKTG----PTKKLRKEVFSSICKFFCHAGIPLQAA 324
T+ NL + K Q ++ TG K+ R+ + S +FF AGIP + A
Sbjct: 166 ATSHRESPAVRDYNLASAKAPVQPRIDTGFFTKKGKQARQAIGESWARFFFTAGIPGRNA 225
Query: 325 DSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMAD 384
D+ YF + ++G+ + P+ I G++L +K +K W G +IM D
Sbjct: 226 DNPYFVSAVRETQKWGESVPSPTGNEIDGKYLDSTEKDVKKQFDRFKKDWDEYGVTIMCD 285
Query: 385 SWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVIT 444
SW ++INFL+ ++F S+DAT +DA ++ K + KVV E+G E+VVQ+IT
Sbjct: 286 SWTGPTSMSVINFLIYCNGIMFFHKSIDATGQSQDANFVLKEIRKVVREIGSEHVVQIIT 345
Query: 445 ENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQ 504
+N NYK A ++L + + + W PC + +N +L++ K+ E +E +KI + +YN
Sbjct: 346 DNGSNYKKACRLLRQEYKTIVWQPCVAHTVNLMLKEVGKMPDHEMVIESARKICRWLYNH 405
Query: 505 IWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSS 564
L +M G EL+K T+ +++ LQ+ L R + S+ +M S+FS +
Sbjct: 406 NKLHAMM--VLAIGGELVKWNATRFGTNYMFLQSFLRKRDLFMQWMASSVFMQSKFSGTL 463
Query: 565 QGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSI 624
+G+ + ++++W+ +++V +SV P+ + ++ ++ + K+ S+
Sbjct: 464 EGRYAHACLSSLSWWENLEAVVSSVQPMYSFLRFADEDKNPNLGEVLLRYQLLKMEYDSL 523
Query: 625 HGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVR 684
+ K+E + ++++R + L L AA LNP Y + + V + L +
Sbjct: 524 FANQRDKFEAYMEIVNRRMHDLTNETLINAAAALNPRTHYA--YSPSATVFQDLRQAFEW 581
Query: 685 LELDNMRRISASMQIPHYNSAQDDFGTELA--ISTRTGLEPAAWWQQHGISCLELQRIAV 742
+ D +A +++ Y +FG LA ++ PA WW L+++A+
Sbjct: 582 M-TDIDTAAAALLKVEMYRRKTGEFGRALARRMAIDGKTSPAQWWSMFASDTPNLKKLAL 640
Query: 743 RILSQTCSSFACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLR----ECQVR--- 795
R++ Q CSS CE + S + +++K +NRL+ KKLN ++YV+YNLRLR Q+R
Sbjct: 641 RLVGQCCSSSGCERNWSTFAFVHTKVRNRLTHKKLNKLVYVNYNLRLRIQQANAQIRCLD 700
Query: 796 -------KRSRESKSTSAENVLQEHL 814
KR +E++ST + ++ + L
Sbjct: 701 GLVLIAMKRPKEAQSTKSPMIVAQEL 726
>UniRef100_Q9SQN5 Hypothetical protein F19K16.28 [Arabidopsis thaliana]
Length = 518
Score = 251 bits (641), Expect = 6e-65
Identities = 140/458 (30%), Positives = 258/458 (55%), Gaps = 7/458 (1%)
Query: 332 MLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQG 391
ML+ + G G PS + +L + I L + + W TGC+I+A++W D +
Sbjct: 1 MLDAVAKCGPGFVAPSPKT---EWLDRVKSDISLQLKDTEKEWVTTGCTIIAEAWTDNKS 57
Query: 392 RTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYK 451
R +INF VSSP ++F SVDA++ +++ L L D V++++G+E++VQ+I +N+ Y
Sbjct: 58 RALINFSVSSPSRIFFHKSVDASSYFKNSKCLADLFDSVIQDIGQEHIVQIIMDNSFCYT 117
Query: 452 AAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLM 511
L + +F +PCA C+N +LE+F K+ V +C+ + Q I+K +YN +L+L+
Sbjct: 118 GISNHLLQNYATIFVSPCASQCLNIILEEFSKVDWVNQCISQAQVISKFVYNNSPVLDLL 177
Query: 512 KSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQK 571
+ + T G ++++ T+ S+F +LQ+++ + L+ MF ++ ++ ++ Q
Sbjct: 178 R-KLTGGQDIIRSGVTRSVSNFLSLQSMMKQKARLKHMFNCPEYTTN--TNKPQSISCVN 234
Query: 572 IVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARK 631
I+ + FW+ ++ PIL+V ++VS+G+ ++ IY + +AK +I++ + D K
Sbjct: 235 ILEDNDFWRAVEESVAISEPILKVLREVSTGKP-AVGSIYELMSKAKESIRTYYIMDENK 293
Query: 632 YEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMR 691
++ F ++D + PL+ AA FLNPS +Y + + + + + +L +
Sbjct: 294 HKVFSDIVDTNWCEHLHSPLHAAAAFLNPSIQYNPEIKFLTSLKEDFFKVLEKLLPTSDL 353
Query: 692 RISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSS 751
R + QI + A+ FG LA+ R + P WW+Q G S LQR+A+RILSQ CS
Sbjct: 354 RRDITNQIFTFTRAKGMFGCNLAMEARDSVSPGLWWEQFGDSAPVLQRVAIRILSQVCSG 413
Query: 752 FACEHDGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRL 789
+ E S + Q++ +R+N++ ++ LN + YV+ NL+L
Sbjct: 414 YNLERQWSTFQQMHWERRNKIDREILNKLAYVNQNLKL 451
>UniRef100_Q9LDK4 Transposase-like protein [Arabidopsis thaliana]
Length = 605
Score = 246 bits (627), Expect = 3e-63
Identities = 178/666 (26%), Positives = 308/666 (45%), Gaps = 80/666 (12%)
Query: 137 EHGVAQDERKKKVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSAPEEVYLKIKENMKW 196
EHG+ D++K +VKC+YC K ++ +R K HL + +V C ++V L ++E +
Sbjct: 10 EHGICVDKKKSRVKCNYCGKEMNS-FHRLKHHLGAVGTDVTHC----DQVSLTLRETFRT 64
Query: 197 HRTGKR--HRQPEAKDLMPFYPKSDNEDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKT 254
+ + P+ K + F Q D S+ KT
Sbjct: 65 MLMEDKSGYTTPKTKRVGKF------------QMAD-----------------SRKRRKT 95
Query: 255 FKGMSSNTSPEPALRRSRLDSFYLKHPTNQNLQTCKQLKVKTGPTKKLRKEVFSSICKFF 314
S + SPE +D NQ+L + K K I +FF
Sbjct: 96 EDSSSKSVSPEQGNVAVEVD--------NQDLLSSKAQKC---------------IGRFF 132
Query: 315 CHAGIPLQAADSVYFHKMLELAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASW 374
+ L A DS F +M+ G G P S ++GR LQE + +++Y+ K SW
Sbjct: 133 YEHCVDLSAVDSPCFKEMMMALGV---GQKIPDSHDLNGRLLQEAMKEVQDYVKNIKDSW 189
Query: 375 AITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEEL 434
ITGCSI+ D+W D +G +++F+ P G ++ S+D + V D T L L++ +VEE+
Sbjct: 190 KITGCSILLDAWIDPKGHDLVSFVADCPAGPVYLKSIDVSVVKNDVTALLSLVNGLVEEV 249
Query: 435 GEENVVQVITENTPNYKA-AGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEK 493
G NV Q+I +T + GK+ R +FW+ +C +L K+R + ++K
Sbjct: 250 GVHNVTQIIACSTSGWVGELGKLFSGHDREVFWSVSLSHCFELMLVKIGKMRSFGDILDK 309
Query: 494 GQKITKLIYNQIWLLNLMKSEFTHGNEL-LKPAGTQCASSFATLQNLLDHRVSLRRMFLS 552
I + I N L + + + +HG ++ + + + + L+++ + +L MF S
Sbjct: 310 VNTIWEFINNNPSALKIYRDQ-SHGKDITVSSSEFEFVKPYLILKSVFKAKKNLAAMFAS 368
Query: 553 NKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPI---LQVFQKVSSGESLSMPY 609
+ W +GK V +V + +FW+ ++ + P+ L++F + + + Y
Sbjct: 369 SVW------KKEEGKSVSNLVNDSSFWEAVEEILKCTSPLTDGLRLFSNADNNQHVG--Y 420
Query: 610 IYNDLYRAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFV 669
IY+ L KL+IK D+ + Y W VID N +PL+ A Y+LNP+ Y DF
Sbjct: 421 IYDTLDGIKLSIKKEFNDEKKHYLTLWDVIDDVWNKHLHNPLHAAGYYLNPTSFYSTDFH 480
Query: 670 SHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQ 729
+V GL +V + + +I++ Q+ Y +D F +G+ P WW +
Sbjct: 481 LDPEVSSGLTHSLVHVAKEGQIKIAS--QLDRYRLGKDCFNEASQPDQISGISPIDWWTE 538
Query: 730 HGISCLELQRIAVRILSQTCSSFA-CEHDGSMYDQ-IYSKRKNRLSQKKLNDIMYVHYNL 787
ELQ A++ILSQTC + + S+ ++ + ++ + +K L ++ +VHYNL
Sbjct: 539 KASQHPELQSFAIKILSQTCEGASRYKLKRSLAEKLLLTEGMSHCERKHLEELAFVHYNL 598
Query: 788 RLRECQ 793
L+ C+
Sbjct: 599 HLQSCK 604
Score = 52.4 bits (124), Expect = 6e-05
Identities = 21/45 (46%), Positives = 33/45 (72%), Gaps = 1/45 (2%)
Query: 15 DHGIAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEK 59
+HGI D++K +V+CNYCGK ++ +RLK HL V +VT+C++
Sbjct: 10 EHGICVDKKKSRVKCNYCGKEMN-SFHRLKHHLGAVGTDVTHCDQ 53
>UniRef100_Q5Z810 HAT dimerisation domain-containing protein-like [Oryza sativa]
Length = 657
Score = 232 bits (591), Expect = 4e-59
Identities = 155/549 (28%), Positives = 274/549 (49%), Gaps = 21/549 (3%)
Query: 275 SFYLKHPTNQNLQTCKQLKVKTGPTKKLRKEVFSSICKFFCHAGIPLQAADSVYFHKMLE 334
SF +++ ++Q +Q + K T L + SI K AG+ F M++
Sbjct: 116 SFSMENSSSQ-MQDSESSKEPTNDY--LNSQARKSIGKLIFEAGLEPGILHLPSFKDMVD 172
Query: 335 LAGQYGQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTI 394
+ + Q +A P+ + I ++E++ I+ + + K W + GCS++ D+W G++
Sbjct: 173 VLA-WAQ-VAIPTYESI----MEEQLREIQCHARDLKKHWEMNGCSVILDTWESRCGKSF 226
Query: 395 INFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAG 454
I+ LV G+ F+ S+D +++++D L +L +VVEE+G N+VQVIT + Y A
Sbjct: 227 ISVLVHCSKGMLFIKSMDVSDIIDDVDELAVMLFRVVEEVGVLNIVQVITNDESPYMQAA 286
Query: 455 K--MLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMK 512
+ +L+ + F+T CA +CIN +LE+ + V E + K ++IT+ IY+ + L K
Sbjct: 287 EHAVLKRYGYSFFFTLCADHCINLLLENIAALDHVNEVLIKAREITRFIYSHAVPMEL-K 345
Query: 513 SEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKI 572
++ G E+L + + + F TL L+ R++L MF S +W SS +S S + V ++
Sbjct: 346 GKYIQGGEILSSSNLKFVAMFITLGKLVSERINLVEMFSSPEWASSDLASRSSFRHVYEV 405
Query: 573 V-LNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARK 631
V + FW + P++ V K+ + ++ + +Y+ + AK IK D K
Sbjct: 406 VKTDNAFWSAAADILKLTDPLITVLYKLEA-DNCPIGILYDAMDCAKEDIKCNLRD---K 461
Query: 632 YEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVVRGLNECIVRLELDNMR 691
+ +W ++D + P++ A Y LNP Y + F +++ G N C+ RL ++
Sbjct: 462 HGDYWPMVDEIWDHYLHTPVHAAGYILNPRIFYTERFSYDTEIKSGTNACVTRLAKNHYD 521
Query: 692 RISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCLELQRIAVRILSQTCSS 751
++Q+ Y F ++ AI + WW HG ELQ A+RILSQTC
Sbjct: 522 PKKVAIQMDRYRRKSAPFDSDSAIQQTMEIPQVRWWSAHGTDTPELQTFAIRILSQTCFG 581
Query: 752 FACEH-DGSMYDQIYSKRKNRLSQKKLNDIMYVHYNLRLRECQVRKRSRESKSTSAENVL 810
+ + D S+ +Q++ ++ Q++ + Y+HYNLRL C+ R +
Sbjct: 582 ASIYNIDRSISEQLHVVKRTYPEQERFRTMEYLHYNLRLAHCEPCVRGASGAQQHSRLTS 641
Query: 811 QEHLLGDWI 819
Q LGDWI
Sbjct: 642 Q---LGDWI 647
>UniRef100_Q9LDS0 Transposase-like protein [Arabidopsis thaliana]
Length = 544
Score = 211 bits (538), Expect = 6e-53
Identities = 133/482 (27%), Positives = 243/482 (49%), Gaps = 14/482 (2%)
Query: 319 IPLQAADSVYFHKMLELAGQYGQ-GLACPSSQLISGRFLQEEINSIKNYLAEYKASWAIT 377
+ L A D+ F +M+ + G GQ GL ++G LQ+ + +++ + + K SWAIT
Sbjct: 62 VNLSAVDAPCFKEMMTVDG--GQMGLESSDCHDLNGWRLQDALEEVQDRVEKIKESWAIT 119
Query: 378 GCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEE 437
GCSI+ D+W D +GR ++ F+ P G+ ++ S D ++ +D T L L++ +VEE+G
Sbjct: 120 GCSILLDAWVDQKGRDLVTFVADCPAGLVYLISFDVSDFKDDVTALLSLVNGLVEEVGVR 179
Query: 438 NVVQVITENTPNYKAA-GKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQK 496
NV Q+I +T + G++ R +FW+ +C +L KIR + +K
Sbjct: 180 NVTQIIACSTSGWVGELGELFAGHDREVFWSVSVSHCFELMLVKISKIRSFGDIFDKVNN 239
Query: 497 ITKLIYNQIWLLNLMKSEFTHGNEL-LKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKW 555
I I N +LN+ + + HG ++ + + + + + L+++ + +L MF S+ W
Sbjct: 240 IWLFINNNPSVLNIFRDQ-CHGIDITVSSSEFEFVTPYLILESIFKAKKNLTAMFASSNW 298
Query: 556 MSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLY 615
++ Q + +V + +FW+ ++SV P++ S+ + + Y+Y+ +
Sbjct: 299 ------NNEQCIAISNLVSDSSFWETVESVLKCTSPLIHGLLLFSTANNQHLGYVYDTMD 352
Query: 616 RAKLAIKSIHGDDARKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSDVV 675
K +I + Y+P W VID N +PL+ A YFLNP+ Y +F +VV
Sbjct: 353 SIKESIAREFNHKPQFYKPLWDVIDDVWNKHLHNPLHAAGYFLNPTAFYSTNFHLDIEVV 412
Query: 676 RGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEPAAWWQQHGISCL 735
GL ++ + D + S QI Y +D F TG+ PA WW
Sbjct: 413 TGLISSLIHMVEDCHVQFKISTQIDMYRLGKDCFNEASQADQITGISPAEWWAHKASQYP 472
Query: 736 ELQRIAVRILSQTCSSFA-CEHDGSMYDQ-IYSKRKNRLSQKKLNDIMYVHYNLRLRECQ 793
ELQ +A++ILSQTC + + S+ ++ + S+ + ++ L+++++V YNL L+ +
Sbjct: 473 ELQSLAIKILSQTCEGASKYKLKRSLAEKLLLSEGMSNRERQHLDELVFVQYNLHLQSYK 532
Query: 794 VR 795
+
Sbjct: 533 AK 534
>UniRef100_Q9LDB0 Transposase-like protein [Arabidopsis thaliana]
Length = 572
Score = 201 bits (511), Expect = 7e-50
Identities = 140/492 (28%), Positives = 233/492 (46%), Gaps = 27/492 (5%)
Query: 310 ICKFFCHAGIPLQAADSVYFHKMLEL--AGQYGQGLACPSSQLISGRFLQEEINSIKNYL 367
+ +FF G+ A DS F KM+ + G G G P S+ ++G QE + +++ +
Sbjct: 99 VAQFFYEHGVDFSAVDSTSFKKMMMIKTVGGEGGGQMIPDSRDLNGWMFQEALKKVQDRV 158
Query: 368 AEYKASWAITGCSIMADSWRDAQGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLL 427
E KASW ITGCSI+ D+W +GR ++ F+ P G ++ S D +++ D T L L+
Sbjct: 159 KEIKASWEITGCSILFDAWIGPKGRDLVTFVADCPAGAVYLKSADVSDIKTDVTALTSLV 218
Query: 428 DKVVEELGEENVVQVITENTPNYKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCV 487
+ +VEE+G NV Q+I +T + G + ++ +FW+ YC+ +L + K+
Sbjct: 219 NGIVEEVGVRNVTQIIACSTSGW--VGDLGKQLAGQVFWSVSLSYCLKLMLVEIGKMYSF 276
Query: 488 EECMEKGQKITKLIYNQIWLLNLMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLR 547
E+ EK + + LI N L + + N ++C F L+H +R
Sbjct: 277 EDIFEKVKLLLDLINNNPSFLYVFRE-----NSHKVDVSSEC--EFVMPYLTLEHIYWVR 329
Query: 548 R--MFLSNKWMSSRFSSSSQGKEVQKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESL 605
R +F S +W QG + V + TFW+ + + S ++ + S G S
Sbjct: 330 RAGLFASPEW------KKEQGIAISSFVNDSTFWESLDKIVGSTSSLVHGWLWFSRG-SK 382
Query: 606 SMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVIDR--HCNSLFCHPLYLAAYFLNPSYR 663
+ Y Y+ + K + + + YEP W VID H N +PL+ A YFLNP
Sbjct: 383 HVAYAYHFIESIKKNVAWTFKYERQFYEPTWNVIDDVWHNNH---NPLHAAGYFLNPMAY 439
Query: 664 YRQDFVSHSDVVRGLNECIVRLELDNMRRISASMQIPHYNSAQDDFGTELAISTRTGLEP 723
Y DF ++ V GL +V L + ++ Q+ Y + F G+ P
Sbjct: 440 YSDDFHTYQHVYTGLAFSLVHLVKEPHLQVKIGTQLDVYRYGRGCFMKASQAGQLNGVSP 499
Query: 724 AAWWQQHGISCLELQRIAVRILSQTCSSFA-CEHDGSMYDQ-IYSKRKNRLSQKKLNDIM 781
WW Q ELQ +AV+ILSQT + + S+ ++ + ++ + +K L ++
Sbjct: 500 VNWWTQKANQYPELQNLAVKILSQTSEGASRYKLKRSVAEKLLLTEGMSHCERKHLEELA 559
Query: 782 YVHYNLRLRECQ 793
VHYNL+L+ C+
Sbjct: 560 VVHYNLQLQSCK 571
>UniRef100_O23355 Hypothetical protein AT4g15020 [Arabidopsis thaliana]
Length = 522
Score = 200 bits (508), Expect = 2e-49
Identities = 160/565 (28%), Positives = 251/565 (44%), Gaps = 64/565 (11%)
Query: 114 MDGRNVSVNLTPLRSLGYVDPGWEH-GVAQDERKKKVKCSYCEKVVSGG-INRFKQHLAR 171
MD V LTP + D W+H + + + +++C YC K+ GG I R K
Sbjct: 1 MDAELEPVALTPQKQ----DNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVK----- 51
Query: 172 IPGEVAPCKSAPEEVYLKIKENMKWHRTGKRHRQ-----PEAKDLMPFYP------KSDN 220
++ K + + ++ K H++ P D+M P KS
Sbjct: 52 ---DILLVKRDKQCIDGTVRRQRKRHKSSSEPLSVASLPPIEGDMMVVQPDVNDGFKSPG 108
Query: 221 EDDEYEQQEDTLHHMNKEALIDIDRRYSKDTGKTFKGMSSNTSPEPALRRSRLDSFYLKH 280
D Q E L K+ R Y G +SN +
Sbjct: 109 SSDVVVQNESLLSGRTKQ------RTYRSKKNAFENGSASNNV-----------DLIGRD 151
Query: 281 PTNQNLQTCKQLKVKTGPTKKLRKE-VFSSICKFFCHAGIPLQAADSVYFHKMLELAGQY 339
N +K P+ + R+ + +I +F G A +SV F M++
Sbjct: 152 MDNLIPVAISSVKNIVHPSFRDRENTIHMAIGRFLFGIGADFDAVNSVNFQPMIDAIASG 211
Query: 340 GQGLACPSSQLISGRFLQEEINSIKNYLAEYKASWAITGCSIMADSWRDAQGRTIINFLV 399
G G++ P+ + G L+ + + + E KA W TGCSI+ + +G ++NFLV
Sbjct: 212 GFGVSAPTHDDLRGWILKNCVEEMAKEIDECKAMWKRTGCSILVEELNSDKGFKVLNFLV 271
Query: 400 SSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPNYKAAGKMLEE 459
P V F+ SVDA+ V+ A LF+LL ++VEE+G NVVQVIT+ Y AGK L
Sbjct: 272 YCPEKVVFLKSVDASEVLSSADKLFELLSELVEEVGSTNVVQVITKCDDYYVDAGKRLML 331
Query: 460 RRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLNLMKSEFTHGN 519
+L+W PCA +CI+Q+LE+F K+ + E +E+ Q IT+ +YN
Sbjct: 332 VYPSLYWVPCAAHCIDQMLEEFGKLGWISETIEQAQAITRFVYNH--------------- 376
Query: 520 ELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEVQKIVLNVTFW 579
A + A++FATL + + + +L+ M S +W +S G V + + FW
Sbjct: 377 ----SAFSSSATNFATLGRIAELKSNLQAMVTSAEWNECSYSEEPSGL-VMNALTDEAFW 431
Query: 580 KKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDARKYEPFWKVI 639
K + V + P+L+ + V S + +M Y+Y LYRAK AIK+ H + Y +WK+I
Sbjct: 432 KAVALVNHLTSPLLRALRIVCSEKRPAMGYVYAALYRAKDAIKT-HLVNREDYIIYWKII 490
Query: 640 DRHCNSLFCHPLYLAAYFLNPSYRY 664
D ++ L +FLNP Y
Sbjct: 491 DHGGSNNSIFLCLLLGFFLNPKLFY 515
>UniRef100_Q5W690 Hypothetical protein OSJNBa0076A09.5 [Oryza sativa]
Length = 1030
Score = 199 bits (506), Expect = 3e-49
Identities = 110/331 (33%), Positives = 186/331 (55%), Gaps = 5/331 (1%)
Query: 390 QGRTIINFLVSSPHGVYFVSSVDATNVVEDATYLFKLLDKVVEELGEENVVQVITENTPN 449
+GR+++N + G+ F+ ++DA+ V D Y++ L+ ++E+G + VVQV+T+N N
Sbjct: 522 RGRSLMNLVAHCARGMCFIDAIDASLEVHDGKYIYSLVSSCIDEIGPKKVVQVVTDNASN 581
Query: 450 YKAAGKMLEERRRNLFWTPCAIYCINQVLEDFLKIRCVEECMEKGQKITKLIYNQIWLLN 509
+A KML+ + ++FWT CA + I+ +LED KI V + G+ IT L+Y Q+ LL
Sbjct: 582 NMSASKMLQVKHPHIFWTSCAAHYIDLMLEDIGKITMVHNIIRDGKSITNLLYAQVRLLA 641
Query: 510 LMKSEFTHGNELLKPAGTQCASSFATLQNLLDHRVSLRRMFLSNKWMSSRFSSSSQGKEV 569
+M+ +FT G +L++ T+ A+S+ L++L D R L+++F S W S ++ +G+
Sbjct: 642 IMR-QFTKG-DLVRAGTTRFATSYLNLKSLYDKRNELKQLFASQDWAKSSWAKKIKGQNA 699
Query: 570 QKIVLNVTFWKKMQSVRNSVYPILQVFQKVSSGESLSMPYIYNDLYRAKLAIKSIHGDDA 629
+V+N FW +M V N P+ V ++V G+ +M YIY DL +AK + + +
Sbjct: 700 HNLVMNNKFWSQMLEVINYFEPLAYVLRRV-DGDVPAMGYIYGDLIKAKKDVAACLNGNE 758
Query: 630 RKYEPFWKVIDRHCNSLFCHPLYLAAYFLNPSYRYRQDFVSHSD-VVRGLNECIVRLELD 688
+KY WK+ID +S L+ A YFLNP + Y D +++ + EC + D
Sbjct: 759 KKYSHIWKIIDARWDSKLKTTLHKAGYFLNPCFFYENKREIKEDFLMQAVVECATCMYRD 818
Query: 689 NMR-RISASMQIPHYNSAQDDFGTELAISTR 718
++ + Q+ Y A D FGT +AI R
Sbjct: 819 DITVQDICVAQLSLYTEAMDSFGTTMAIRQR 849
Score = 71.2 bits (173), Expect = 1e-10
Identities = 40/104 (38%), Positives = 57/104 (54%), Gaps = 7/104 (6%)
Query: 5 RSTGFVDPGWDHGI--AQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCEKAPE 62
+ST DP W+ + +E K +V C YC GGI RLK+HLA V G+ YC + P+
Sbjct: 365 QSTSRKDPAWEFALPLTGNETKNQVGCLYCKNYFKGGITRLKRHLAGVRGQSVYCTQVPD 424
Query: 63 EVYLKMKENLEGCRSNKKQKQVDAQ----AYMNFQSNDDEDDEE 102
+V K+K L+ KK ++D+Q +N NDDE+ EE
Sbjct: 425 DVKEKVKAMLD-AHGEKKSAKLDSQLRLRQQVNINGNDDEEMEE 467
Score = 58.2 bits (139), Expect = 1e-06
Identities = 31/97 (31%), Positives = 47/97 (47%), Gaps = 2/97 (2%)
Query: 133 DPGWEHGV--AQDERKKKVKCSYCEKVVSGGINRFKQHLARIPGEVAPCKSAPEEVYLKI 190
DP WE + +E K +V C YC+ GGI R K+HLA + G+ C P++V K+
Sbjct: 371 DPAWEFALPLTGNETKNQVGCLYCKNYFKGGITRLKRHLAGVRGQSVYCTQVPDDVKEKV 430
Query: 191 KENMKWHRTGKRHRQPEAKDLMPFYPKSDNEDDEYEQ 227
K + H K + L + N+D+E E+
Sbjct: 431 KAMLDAHGEKKSAKLDSQLRLRQQVNINGNDDEEMEE 467
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.133 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,395,171,592
Number of Sequences: 2790947
Number of extensions: 59482087
Number of successful extensions: 170908
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 48
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 170667
Number of HSP's gapped (non-prelim): 163
length of query: 835
length of database: 848,049,833
effective HSP length: 136
effective length of query: 699
effective length of database: 468,481,041
effective search space: 327468247659
effective search space used: 327468247659
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 79 (35.0 bits)
Medicago: description of AC138579.4


