

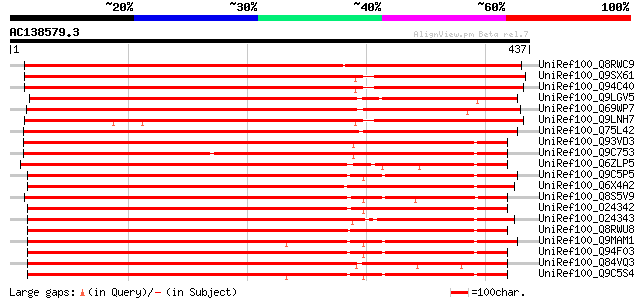
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.3 + phase: 0 /pseudo
(437 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8RWC9 CBL-interacting serine/threonine-protein kinase... 590 e-167
UniRef100_Q9SX61 F11A17.18 protein [Arabidopsis thaliana] 553 e-156
UniRef100_Q94C40 CBL-interacting protein kinase 17 [Arabidopsis ... 553 e-156
UniRef100_Q9LGV5 ESTs D41739 [Oryza sativa] 548 e-154
UniRef100_Q69WP7 Putative serine/threonine kinase [Oryza sativa] 543 e-153
UniRef100_Q9LNH7 F21D18.2 [Arabidopsis thaliana] 536 e-151
UniRef100_Q75L42 Hypothetical protein OJ1127_B08.5 [Oryza sativa] 527 e-148
UniRef100_Q93VD3 At1g30270/F12P21_6 [Arabidopsis thaliana] 450 e-125
UniRef100_Q9C753 Serine/threonine kinase, putative [Arabidopsis ... 444 e-123
UniRef100_Q6ZLP5 Putative CBL-interacting protein kinase 23 [Ory... 443 e-123
UniRef100_Q9C5P5 SOS2-like protein kinase PKS6 [Arabidopsis thal... 433 e-120
UniRef100_Q6X4A2 CIPK-like protein [Oryza sativa] 432 e-119
UniRef100_Q8S5V9 Putative serine/threonine kinase [Oryza sativa] 430 e-119
UniRef100_O24342 Serine/threonine kinase [Sorghum bicolor] 430 e-119
UniRef100_O24343 Serine/threonine kinase [Sorghum bicolor] 430 e-119
UniRef100_Q8RWU8 Hypothetical protein At2g26980 [Arabidopsis tha... 429 e-119
UniRef100_Q9MAM1 T25K16.13 [Arabidopsis thaliana] 428 e-118
UniRef100_Q94F03 Similar to wpk4 protein kinase [Arabidopsis tha... 427 e-118
UniRef100_Q84VQ3 NAF specific protein kinase family [Arabidopsis... 424 e-117
UniRef100_Q9C5S4 CBL-interacting protein kinase 9 [Arabidopsis t... 423 e-117
>UniRef100_Q8RWC9 CBL-interacting serine/threonine-protein kinase 1 [Arabidopsis
thaliana]
Length = 444
Score = 590 bits (1521), Expect = e-167
Identities = 301/419 (71%), Positives = 347/419 (81%), Gaps = 1/419 (0%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K +RLGKYELG+TLGEGNFGKVK A+DT G FAVKI++K++I DLN + QIKREI TL
Sbjct: 13 KGMRLGKYELGRTLGEGNFGKVKFAKDTVSGHSFAVKIIDKSRIADLNFSLQIKREIRTL 72
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
K+LKHP++VRL+EVLASKTKI MV+E V GGELFD+I S GKLTE GRKMFQQLIDG+S
Sbjct: 73 KMLKHPHIVRLHEVLASKTKINMVMELVTGGELFDRIVSNGKLTETDGRKMFQQLIDGIS 132
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
YCH+KGVFHRDLKLENVL+DAKG+IKITDF LSALPQH R DGLLHTTCGSPNYVAPE+L
Sbjct: 133 YCHSKGVFHRDLKLENVLLDAKGHIKITDFGLSALPQHFRDDGLLHTTCGSPNYVAPEVL 192
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
ANRGYDGA SDIWSCGVILYVILTG LPFDDRNLAVLYQKI K D IPRWLSPGA+ +I
Sbjct: 193 ANRGYDGAASDIWSCGVILYVILTGCLPFDDRNLAVLYQKICKGDPPIPRWLSPGARTMI 252
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQS 312
K++LDPNP TR+T+ IK EWFK Y P ++ D+++EE+V DD+AFSI E+ + +
Sbjct: 253 KRMLDPNPVTRITVVGIKASEWFKLEYIP-SIPDDDDEEEVDTDDDAFSIQELGSEEGKG 311
Query: 313 PKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
SP +INAFQLIGMSS LDLSGFFEQE+VSER+IRFTSN S KDL EKIE V EMGF
Sbjct: 312 SDSPTIINAFQLIGMSSFLDLSGFFEQENVSERRIRFTSNSSAKDLLEKIETAVTEMGFS 371
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCILLL 431
V KK+ KL+V QE++ K LSV AEVFE+ PSL VVELRKSYGD+ +YRQ+ LL
Sbjct: 372 VQKKHAKLRVKQEERNQKGQVGLSVTAEVFEIKPSLNVVELRKSYGDSCLYRQLYERLL 430
>UniRef100_Q9SX61 F11A17.18 protein [Arabidopsis thaliana]
Length = 421
Score = 553 bits (1426), Expect = e-156
Identities = 286/426 (67%), Positives = 337/426 (78%), Gaps = 12/426 (2%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K +R+GKYELG+TLGEGN KVK A DT G+ FA+KI+EK+ I LN + QIKREI TL
Sbjct: 4 KGMRVGKYELGRTLGEGNSAKVKFAIDTLTGESFAIKIIEKSCITRLNVSFQIKREIRTL 63
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
K+LKHPN+VRL+EVLASKTKIYMVLE V GG+LFD+I SKGKL+E GRKMFQQLIDGVS
Sbjct: 64 KVLKHPNIVRLHEVLASKTKIYMVLECVTGGDLFDRIVSKGKLSETQGRKMFQQLIDGVS 123
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
YCHNKGVFHRDLKLENVL+DAKG+IKITDF LSAL QH R DGLLHTTCGSPNYVAPE+L
Sbjct: 124 YCHNKGVFHRDLKLENVLLDAKGHIKITDFGLSALSQHYREDGLLHTTCGSPNYVAPEVL 183
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
AN GYDGA SDIWSCGVILYVILTG LPFDD NLAV+ +KIFK D IPRW+S GA+ +I
Sbjct: 184 ANEGYDGAASDIWSCGVILYVILTGCLPFDDANLAVICRKIFKGDPPIPRWISLGAKTMI 243
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEE----EEDVFIDDEAFSIHEVSLD 308
K++LDPNP TRVT+ IK +WFK Y P+N +D+++ +EDVF+ E +
Sbjct: 244 KRMLDPNPVTRVTIAGIKAHDWFKHDYTPSNYDDDDDVYLIQEDVFMMKE--------YE 295
Query: 309 GDQSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIE 368
++SP SP +INAFQLIGMSS LDLSGFFE E +SER+IRFTSN KDL E IE I E
Sbjct: 296 EEKSPDSPTIINAFQLIGMSSFLDLSGFFETEKLSERQIRFTSNSLAKDLLENIETIFTE 355
Query: 369 MGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCI 428
MGF + KK+ KLK I+E+ K LSV AEVFE+ PSL VVELRKS+GD+S+Y+QV +
Sbjct: 356 MGFCLQKKHAKLKAIKEESTQKRQCGLSVTAEVFEISPSLNVVELRKSHGDSSLYKQVMM 415
Query: 429 LLLSLS 434
++S
Sbjct: 416 HSCNIS 421
>UniRef100_Q94C40 CBL-interacting protein kinase 17 [Arabidopsis thaliana]
Length = 432
Score = 553 bits (1424), Expect = e-156
Identities = 286/424 (67%), Positives = 336/424 (78%), Gaps = 12/424 (2%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K +R+GKYELG+TLGEGN KVK A DT G+ FA+KI+EK+ I LN + QIKREI TL
Sbjct: 4 KGMRVGKYELGRTLGEGNSAKVKFAIDTLTGESFAIKIIEKSCITRLNVSFQIKREIRTL 63
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
K+LKHPN+VRL+EVLASKTKIYMVLE V GG+LFD+I SKGKL+E GRKMFQQLIDGVS
Sbjct: 64 KVLKHPNIVRLHEVLASKTKIYMVLECVTGGDLFDRIVSKGKLSETQGRKMFQQLIDGVS 123
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
YCHNKGVFHRDLKLENVL+DAKG+IKITDF LSAL QH R DGLLHTTCGSPNYVAPE+L
Sbjct: 124 YCHNKGVFHRDLKLENVLLDAKGHIKITDFGLSALSQHYREDGLLHTTCGSPNYVAPEVL 183
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
AN GYDGA SDIWSCGVILYVILTG LPFDD NLAV+ +KIFK D IPRW+S GA+ +I
Sbjct: 184 ANEGYDGAASDIWSCGVILYVILTGCLPFDDANLAVICRKIFKGDPPIPRWISLGAKTMI 243
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEE----EEDVFIDDEAFSIHEVSLD 308
K++LDPNP TRVT+ IK +WFK Y P+N +D+++ +EDVF+ E +
Sbjct: 244 KRMLDPNPVTRVTIAGIKAHDWFKHDYTPSNYDDDDDVYLIQEDVFMMKE--------YE 295
Query: 309 GDQSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIE 368
++SP SP +INAFQLIGMSS LDLSGFFE E +SER+IRFTSN KDL E IE I E
Sbjct: 296 EEKSPDSPTIINAFQLIGMSSFLDLSGFFETEKLSERQIRFTSNSLAKDLLENIETIFTE 355
Query: 369 MGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCI 428
MGF + KK+ KLK I+E+ K LSV AEVFE+ PSL VVELRKS+GD+S+Y+Q+
Sbjct: 356 MGFCLQKKHAKLKAIKEESTQKRQCGLSVTAEVFEISPSLNVVELRKSHGDSSLYKQLYE 415
Query: 429 LLLS 432
LL+
Sbjct: 416 RLLN 419
>UniRef100_Q9LGV5 ESTs D41739 [Oryza sativa]
Length = 461
Score = 548 bits (1412), Expect = e-154
Identities = 272/414 (65%), Positives = 329/414 (78%), Gaps = 8/414 (1%)
Query: 17 LGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLLK 76
LG+YE+G+TLGEGNFGKVK AR G FA+KIL++NKI+ L DQI+REI TLKLLK
Sbjct: 16 LGRYEIGRTLGEGNFGKVKYARHLATGAHFAIKILDRNKILSLRFDDQIRREIGTLKLLK 75
Query: 77 HPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCHN 136
HPNVVRL+EV ASKTKIYMVLEYVNGGELFDKI+ KGKL+E GR++FQQLID VSYCH+
Sbjct: 76 HPNVVRLHEVAASKTKIYMVLEYVNGGELFDKIAVKGKLSEHEGRRLFQQLIDAVSYCHD 135
Query: 137 KGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANRG 196
KGV+HRDLK ENVLVD +GNIKI+DF LSALPQH DGLLHTTCGSPNY+APE+L NRG
Sbjct: 136 KGVYHRDLKPENVLVDRRGNIKISDFGLSALPQHLGNDGLLHTTCGSPNYIAPEVLQNRG 195
Query: 197 YDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKIL 256
YDG+ SDIWSCGVILYV+L GYLPFDDRNL VLYQKIFK D QIP+WLSP A++++++IL
Sbjct: 196 YDGSLSDIWSCGVILYVMLVGYLPFDDRNLVVLYQKIFKGDTQIPKWLSPSARDLLRRIL 255
Query: 257 DPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPKSP 316
+PNP R+ + IKE EWF++ Y P D++++ + D I E + Q + P
Sbjct: 256 EPNPMKRINIAGIKEHEWFQKDYTPVVPYDDDDDNYL---DSVLPIKEQIDEAKQ--EKP 310
Query: 317 ALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKVHKK 376
INAFQLIGM+S LDLSGFFE+ED S+RKIRFTS HSPKDL +KIE++V EMGF+V +
Sbjct: 311 THINAFQLIGMASALDLSGFFEEEDASQRKIRFTSTHSPKDLFDKIENVVTEMGFQVQRG 370
Query: 377 NGKLKVIQEDKAHKSL---DSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVC 427
N KLKV++ + K+L S V EV E+GPSLYVVEL+KS+GD +YRQ+C
Sbjct: 371 NSKLKVMKNGRGSKNLRNPSSFLVCTEVVELGPSLYVVELKKSHGDPILYRQLC 424
>UniRef100_Q69WP7 Putative serine/threonine kinase [Oryza sativa]
Length = 444
Score = 543 bits (1400), Expect = e-153
Identities = 271/419 (64%), Positives = 333/419 (78%), Gaps = 7/419 (1%)
Query: 15 VRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKL 74
+R+GKYE+G+ LGEG+FGKVKLAR D G FA+KIL++ +I+ + +QIKREI+TLKL
Sbjct: 1 MRMGKYEMGRALGEGHFGKVKLARHADTGAAFAIKILDRQRILAMKIDEQIKREIATLKL 60
Query: 75 LKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYC 134
LKHPNVVRL+EV ASKTKIYMVLEYVNGGELFDKI+ KGKL+E GRK+FQQL+D VSYC
Sbjct: 61 LKHPNVVRLHEVSASKTKIYMVLEYVNGGELFDKIALKGKLSEKEGRKLFQQLMDAVSYC 120
Query: 135 HNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILAN 194
H KGV+HRDLK ENVLVDAKGNIK++DF LSALPQ+ R DGLLHTTCGSPNY+APE+L N
Sbjct: 121 HEKGVYHRDLKPENVLVDAKGNIKVSDFGLSALPQNQRKDGLLHTTCGSPNYIAPEVLLN 180
Query: 195 RGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKK 254
RGYDG+ SDIWSCGVILYV+LTG LPFDD+N VLYQKI K D +IP+WLSPGAQ+I++K
Sbjct: 181 RGYDGSLSDIWSCGVILYVMLTGNLPFDDQNTVVLYQKILKGDARIPKWLSPGAQDILRK 240
Query: 255 ILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPK 314
ILDPNP TR+ + I+ EWF++ Y PA D++++ ++ ++H ++
Sbjct: 241 ILDPNPITRLDITGIRAHEWFRQDYTPAMPFDDDDDNNI----SDGNLHMTENQDIETSP 296
Query: 315 SPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKVH 374
+ + INAFQLIGMSSCLDLSGFFE+EDVSERKIRF SN+SP L EKIE V E GF+V
Sbjct: 297 AISQINAFQLIGMSSCLDLSGFFEKEDVSERKIRFVSNYSPTSLFEKIESTVTEKGFQVQ 356
Query: 375 KKNGKLKVIQ---EDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVCILL 430
K +GKLKVIQ E + +L ++AEVFE+ SLYVVEL++S GD S+YRQ+C L
Sbjct: 357 KNSGKLKVIQVCKEPANPRGHGNLLISAEVFEINESLYVVELKRSSGDCSLYRQLCASL 415
>UniRef100_Q9LNH7 F21D18.2 [Arabidopsis thaliana]
Length = 454
Score = 536 bits (1380), Expect = e-151
Identities = 286/446 (64%), Positives = 336/446 (75%), Gaps = 34/446 (7%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
K +R+GKYELG+TLGEGN KVK A DT G+ FA+KI+EK+ I LN + QIKREI TL
Sbjct: 4 KGMRVGKYELGRTLGEGNSAKVKFAIDTLTGESFAIKIIEKSCITRLNVSFQIKREIRTL 63
Query: 73 KLLKHPNVVRLYEV-----LASKTKIYMVLEYVNGGELFDKIS----------------- 110
K+LKHPN+VRL+EV LASKTKIYMVLE V GG+LFD+I
Sbjct: 64 KVLKHPNIVRLHEVIILLVLASKTKIYMVLECVTGGDLFDRIVRFLLAPVTVLLVSKRLV 123
Query: 111 SKGKLTEAHGRKMFQQLIDGVSYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQH 170
SKGKL+E GRKMFQQLIDGVSYCHNKGVFHRDLKLENVL+DAKG+IKITDF LSAL QH
Sbjct: 124 SKGKLSETQGRKMFQQLIDGVSYCHNKGVFHRDLKLENVLLDAKGHIKITDFGLSALSQH 183
Query: 171 CRADGLLHTTCGSPNYVAPEILANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLY 230
R DGLLHTTCGSPNYVAPE+LAN GYDGA SDIWSCGVILYVILTG LPFDD NLAV+
Sbjct: 184 YREDGLLHTTCGSPNYVAPEVLANEGYDGAASDIWSCGVILYVILTGCLPFDDANLAVIC 243
Query: 231 QKIFKADVQIPRWLSPGAQNIIKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEE- 289
+KIFK D IPRW+S GA+ +IK++LDPNP TRVT+ IK +WFK Y P+N +D+++
Sbjct: 244 RKIFKGDPPIPRWISLGAKTMIKRMLDPNPVTRVTIAGIKAHDWFKHDYTPSNYDDDDDV 303
Query: 290 ---EEDVFIDDEAFSIHEVSLDGDQSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERK 346
+EDVF+ E + ++SP SP +INAFQLIGMSS LDLSGFFE E +SER+
Sbjct: 304 YLIQEDVFMMKE--------YEEEKSPDSPTIINAFQLIGMSSFLDLSGFFETEKLSERQ 355
Query: 347 IRFTSNHSPKDLTEKIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGP 406
IRFTSN KDL E IE I EMGF + KK+ KLK I+E+ K LSV AEVFE+ P
Sbjct: 356 IRFTSNSLAKDLLENIETIFTEMGFCLQKKHAKLKAIKEESTQKRQCGLSVTAEVFEISP 415
Query: 407 SLYVVELRKSYGDASVYRQVCILLLS 432
SL VVELRKS+GD+S+Y+Q+ LL+
Sbjct: 416 SLNVVELRKSHGDSSLYKQLYERLLN 441
>UniRef100_Q75L42 Hypothetical protein OJ1127_B08.5 [Oryza sativa]
Length = 454
Score = 527 bits (1357), Expect = e-148
Identities = 260/416 (62%), Positives = 327/416 (78%), Gaps = 3/416 (0%)
Query: 12 GKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREIST 71
G+ LG YE+G+TLGEGNFGKVK AR G FAVKIL++ ++V L DQI+REI+T
Sbjct: 5 GREALLGGYEMGRTLGEGNFGKVKYARHLATGGHFAVKILDRGRVVSLRAGDQIRREIAT 64
Query: 72 LKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGV 131
LKLL+HP+VVRL+EV ASKTKIYMVLE+VNGGELF++I+ KGKL+E GR++FQQLIDGV
Sbjct: 65 LKLLRHPHVVRLHEVAASKTKIYMVLEFVNGGELFERIAVKGKLSEKEGRRLFQQLIDGV 124
Query: 132 SYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEI 191
SYCH++GV+HRDLK ENVLVD KGNIKI+DF LSALPQH DGLLHTTCGSPNY+APE+
Sbjct: 125 SYCHDRGVYHRDLKPENVLVDQKGNIKISDFGLSALPQHLGNDGLLHTTCGSPNYIAPEV 184
Query: 192 LANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNI 251
L N+GYDG+ SDIWSCGVILYV+L GYLPFDDRN+ VLYQKIFK D QIP+WLS AQN+
Sbjct: 185 LQNKGYDGSLSDIWSCGVILYVMLIGYLPFDDRNIVVLYQKIFKGDTQIPKWLSHSAQNL 244
Query: 252 IKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQ 311
+++IL+PNP R+ M IK EWF++ Y P D+++E+ F A + ++ +
Sbjct: 245 LRRILEPNPMKRIDMAGIKSHEWFQKDYIPVLPYDDDDEDVQF---GARLPAKEQINDEP 301
Query: 312 SPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGF 371
K+ INAFQLIGM+S LDLSGFFE E+VS+R+IRFTS H PKD +KIE E+GF
Sbjct: 302 GDKNSHQINAFQLIGMASSLDLSGFFEDEEVSQRRIRFTSTHPPKDAFDKIESSATELGF 361
Query: 372 KVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVC 427
+V + + KLK+++ K K+ +S V+AEVFE+GPS+ VVELRKS GD ++YRQ+C
Sbjct: 362 QVQRGHSKLKLMRNCKGSKNPESFMVSAEVFELGPSVNVVELRKSNGDPALYRQLC 417
>UniRef100_Q93VD3 At1g30270/F12P21_6 [Arabidopsis thaliana]
Length = 482
Score = 450 bits (1158), Expect = e-125
Identities = 225/412 (54%), Positives = 292/412 (70%), Gaps = 6/412 (1%)
Query: 12 GKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREIST 71
G R+GKYELG+TLGEG F KVK AR+ + G A+K+++K K++ QIKREIST
Sbjct: 23 GGRTRVGKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMIAQIKREIST 82
Query: 72 LKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGV 131
+KL+KHPNV+R++EV+ASKTKIY VLE+V GGELFDKISS G+L E RK FQQLI+ V
Sbjct: 83 MKLIKHPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARKYFQQLINAV 142
Query: 132 SYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEI 191
YCH++GV+HRDLK EN+L+DA G +K++DF LSALPQ R DGLLHTTCG+PNYVAPE+
Sbjct: 143 DYCHSRGVYHRDLKPENLLLDANGALKVSDFGLSALPQQVREDGLLHTTCGTPNYVAPEV 202
Query: 192 LANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNI 251
+ N+GYDGAK+D+WSCGVIL+V++ GYLPF+D NL LY+KIFKA+ P W S A+ +
Sbjct: 203 INNKGYDGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEFTCPPWFSASAKKL 262
Query: 252 IKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEE---EEEDVFIDDEAFSIHEVSLD 308
IK+ILDPNP TR+T + E+EWFK+GY E+ + ++ D DD S + V
Sbjct: 263 IKRILDPNPATRITFAEVIENEWFKKGYKAPKFENADVSLDDVDAIFDDSGESKNLVVER 322
Query: 309 GDQSPKSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVI 367
++ K+P +NAF+LI S L+L FE Q + +RK RFTS S ++ KIE
Sbjct: 323 REEGLKTPVTMNAFELISTSQGLNLGSLFEKQMGLVKRKTRFTSKSSANEIVTKIEAAAA 382
Query: 368 EMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
MGF V N K+K+ E K L+VA EVF+V PSLY+VE+RKS GD
Sbjct: 383 PMGFDVKTNNYKMKLTGEKSGRKG--QLAVATEVFQVAPSLYMVEMRKSGGD 432
>UniRef100_Q9C753 Serine/threonine kinase, putative [Arabidopsis thaliana]
Length = 480
Score = 444 bits (1142), Expect = e-123
Identities = 224/412 (54%), Positives = 292/412 (70%), Gaps = 8/412 (1%)
Query: 12 GKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREIST 71
G R+GKYELG+TLGEG F KVK AR+ + G A+K+++K K++ QIKREIST
Sbjct: 23 GGRTRVGKYELGRTLGEGTFAKVKFARNVENGDNVAIKVIDKEKVLKNKMIAQIKREIST 82
Query: 72 LKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGV 131
+KL+KHPNV+R++EV+ASKTKIY VLE+V GGELFDKISS G+L E RK FQQLI+ V
Sbjct: 83 MKLIKHPNVIRMFEVMASKTKIYFVLEFVTGGELFDKISSNGRLKEDEARKYFQQLINAV 142
Query: 132 SYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEI 191
YCH++GV+HRDLK EN+L+DA G +K++DF LSALPQ + DGLLHTTCG+PNYVAPE+
Sbjct: 143 DYCHSRGVYHRDLKPENLLLDANGALKVSDFGLSALPQ--QEDGLLHTTCGTPNYVAPEV 200
Query: 192 LANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNI 251
+ N+GYDGAK+D+WSCGVIL+V++ GYLPF+D NL LY+KIFKA+ P W S A+ +
Sbjct: 201 INNKGYDGAKADLWSCGVILFVLMAGYLPFEDSNLTSLYKKIFKAEFTCPPWFSASAKKL 260
Query: 252 IKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEE---EEEDVFIDDEAFSIHEVSLD 308
IK+ILDPNP TR+T + E+EWFK+GY E+ + ++ D DD S + V
Sbjct: 261 IKRILDPNPATRITFAEVIENEWFKKGYKAPKFENADVSLDDVDAIFDDSGESKNLVVER 320
Query: 309 GDQSPKSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVI 367
++ K+P +NAF+LI S L+L FE Q + +RK RFTS S ++ KIE
Sbjct: 321 REEGLKTPVTMNAFELISTSQGLNLGSLFEKQMGLVKRKTRFTSKSSANEIVTKIEAAAA 380
Query: 368 EMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
MGF V N K+K+ E K L+VA EVF+V PSLY+VE+RKS GD
Sbjct: 381 PMGFDVKTNNYKMKLTGEKSGRKG--QLAVATEVFQVAPSLYMVEMRKSGGD 430
>UniRef100_Q6ZLP5 Putative CBL-interacting protein kinase 23 [Oryza sativa]
Length = 450
Score = 443 bits (1140), Expect = e-123
Identities = 226/419 (53%), Positives = 297/419 (69%), Gaps = 18/419 (4%)
Query: 10 IEGKIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREI 69
+ G R+G+YELG+TLGEG F KVK AR+ D G+ A+KIL+K+K++ QIKREI
Sbjct: 3 VSGGRTRVGRYELGRTLGEGTFAKVKFARNADSGENVAIKILDKDKVLKHKMIAQIKREI 62
Query: 70 STLKLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLID 129
ST+KL++HPNV+R++EV+ASKTKIY+V+E V GGELFDKI+S+G+L E RK FQQLI+
Sbjct: 63 STMKLIRHPNVIRMHEVMASKTKIYIVMELVTGGELFDKIASRGRLKEDDARKYFQQLIN 122
Query: 130 GVSYCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAP 189
V YCH++GV+HRDLK EN+L+DA G +K++DF LSAL Q R DGLLHTTCG+PNYVAP
Sbjct: 123 AVDYCHSRGVYHRDLKPENLLLDASGTLKVSDFGLSALSQQVREDGLLHTTCGTPNYVAP 182
Query: 190 EILANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQ 249
E++ N+GYDGAK+D+WSCGVIL+V++ GYLPF+D NL LY+KIFKAD P W S A+
Sbjct: 183 EVINNKGYDGAKADLWSCGVILFVLMAGYLPFEDSNLMSLYKKIFKADFSCPSWFSTSAK 242
Query: 250 NIIKKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDG 309
+IKKILDPNP TR+T+ + +EWFK+GY P E DV +DD +E G
Sbjct: 243 KLIKKILDPNPSTRITIAELINNEWFKKGYQPPRF----ETADVNLDDINSIFNE---SG 295
Query: 310 DQS-------PKSPALINAFQLIGMSSCLDLSGFFEQEDVS--ERKIRFTSNHSPKDLTE 360
DQ+ + P+++NAF+LI S L+L FE++ +R+ RF S ++
Sbjct: 296 DQTQLVVERREERPSVMNAFELISTSQGLNLGTLFEKQSQGSVKRETRFASRLPANEILS 355
Query: 361 KIEDIVIEMGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
KIE MGF V K+N KLK+ E+ K L++A EVFEV PSLY+VELRKS GD
Sbjct: 356 KIEAAAGPMGFNVQKRNYKLKLQGENPGRKG--QLAIATEVFEVTPSLYMVELRKSNGD 412
>UniRef100_Q9C5P5 SOS2-like protein kinase PKS6 [Arabidopsis thaliana]
Length = 451
Score = 433 bits (1114), Expect = e-120
Identities = 216/415 (52%), Positives = 298/415 (71%), Gaps = 8/415 (1%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+G YE+G+TLGEG+F KVK A++T G A+KIL++ K+ +Q+KREIST+KL+
Sbjct: 15 RVGNYEMGRTLGEGSFAKVKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREISTMKLI 74
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV + EV+ASKTKIY+VLE VNGGELFDKI+ +G+L E R+ FQQLI+ V YCH
Sbjct: 75 KHPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINAVDYCH 134
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+++DA G +K++DF LSA + R DGLLHT CG+PNYVAPE+L+++
Sbjct: 135 SRGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPEVLSDK 194
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGVIL+V++ GYLPFD+ NL LY++I KA+ P W S GA+ +IK+I
Sbjct: 195 GYDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRICKAEFSCPPWFSQGAKRVIKRI 254
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGDQSP 313
L+PNP TR+++ + EDEWFK+GY P + ++++ED+ ID D AFS + L ++
Sbjct: 255 LEPNPITRISIAELLEDEWFKKGYKPPSF--DQDDEDITIDDVDAAFSNSKECLVTEKKE 312
Query: 314 KSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
K P +NAF+LI SS L FE Q + +++ RFTS S ++ K+E+ +GF
Sbjct: 313 K-PVSMNAFELISSSSEFSLENLFEKQAQLVKKETRFTSQRSASEIMSKMEETAKPLGFN 371
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVC 427
V K N K+K+ + K LSVA EVFEV PSL+VVELRK+ GD + +VC
Sbjct: 372 VRKDNYKIKMKGDKSGRKG--QLSVATEVFEVAPSLHVVELRKTGGDTLEFHKVC 424
>UniRef100_Q6X4A2 CIPK-like protein [Oryza sativa]
Length = 449
Score = 432 bits (1110), Expect = e-119
Identities = 211/410 (51%), Positives = 293/410 (71%), Gaps = 3/410 (0%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+GKYELG+T+GEG F KV+ A++T+ + A+KIL+K K+ +QI+REI T+KL+
Sbjct: 16 RVGKYELGRTIGEGTFAKVRFAKNTENDEPVAIKILDKEKVQKHRLVEQIRREICTMKLV 75
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVVRL+EV+ SK +I++VLEYV GGELF+ I++ G+L E RK FQQLI+ V YCH
Sbjct: 76 KHPNVVRLFEVMGSKARIFIVLEYVTGGELFEIIATNGRLKEEEARKYFQQLINAVDYCH 135
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLKLEN+L+DA GN+K++DF LSAL + +ADGLLHTTCG+PNYVAPE++ +R
Sbjct: 136 SRGVYHRDLKLENLLLDASGNLKVSDFGLSALTEQVKADGLLHTTCGTPNYVAPEVIEDR 195
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +DIWSCGVILYV+L G+LPF+D N+ LY+KI +A P W S GA+ +I +I
Sbjct: 196 GYDGAAADIWSCGVILYVLLAGFLPFEDDNIIALYKKISEAQFTCPSWFSTGAKKLITRI 255
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPKS 315
LDPNP TR+T+ I ED WFK+GY P + DE+ E D AF E +++
Sbjct: 256 LDPNPTTRITISQILEDPWFKKGYKPP-VFDEKYETSFDDVDAAFGDSEDRHVKEETEDQ 314
Query: 316 PALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKVHK 375
P +NAF+LI ++ L+L FE + +R+ RFTS PK++ KIE+ +GF + K
Sbjct: 315 PTSMNAFELISLNQALNLDNLFEAKKEYKRETRFTSQCPPKEIITKIEEAAKPLGFDIQK 374
Query: 376 KNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQ 425
KN K+++ K +L+VA EVF+V PSL+VVEL+K+ GD +++
Sbjct: 375 KNYKMRMENLKAGRKG--NLNVATEVFQVAPSLHVVELKKAKGDTLEFQK 422
>UniRef100_Q8S5V9 Putative serine/threonine kinase [Oryza sativa]
Length = 542
Score = 430 bits (1106), Expect = e-119
Identities = 218/411 (53%), Positives = 293/411 (71%), Gaps = 10/411 (2%)
Query: 13 KIVRLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTL 72
+ R+G+YELGKT+GEG+F KVK+ARDT G A+K+L++N ++ +QIKREIST+
Sbjct: 20 RTTRVGRYELGKTIGEGSFAKVKVARDTRTGDTLAIKVLDRNHVLRHKMVEQIKREISTM 79
Query: 73 KLLKHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVS 132
KL+KHPNVV+L+EV+ASK+KIYMVLEYV+GGELFDKI + G+L E R+ F QLI+ V
Sbjct: 80 KLIKHPNVVQLHEVMASKSKIYMVLEYVDGGELFDKIVNSGRLGEDEARRYFHQLINAVD 139
Query: 133 YCHNKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEIL 192
YCH++GV+HRDLK EN+L+D+ G +K++DF LSA + DGLLHT CG+PNYVAPE+L
Sbjct: 140 YCHSRGVYHRDLKPENLLLDSHGALKVSDFGLSAFAPQTKEDGLLHTACGTPNYVAPEVL 199
Query: 193 ANRGYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNII 252
A++GYDG +D+WSCG+IL+V++ GYLPFDD NL LY+ I KA V P W S GA+ I
Sbjct: 200 ADKGYDGMAADVWSCGIILFVLMAGYLPFDDPNLMTLYKLICKAKVSCPHWFSSGAKKFI 259
Query: 253 KKILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGD 310
K+ILDPNP TR+T+ I ED+WFK+ Y P E+ EDV +D D AF E +L +
Sbjct: 260 KRILDPNPCTRITIAQILEDDWFKKDYKPPLF---EQGEDVSLDDVDAAFDCSEENLVAE 316
Query: 311 QSPKSPALINAFQLIGMSSCLDLSGFFEQE--DVSERKIRFTSNHSPKDLTEKIEDIVIE 368
+ K P +NAF LI S +L FE+E + +R+ FTS +P+++ KIE+
Sbjct: 317 KREK-PESMNAFALISRSQGFNLGNLFEKEMMGMVKRETSFTSQCTPQEIMSKIEEACGP 375
Query: 369 MGFKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
+GF V K+N K+K+ + K LSVA EVFEV PSL++VELRK+ GD
Sbjct: 376 LGFNVRKQNYKMKLKGDKTGRKGY--LSVATEVFEVAPSLHMVELRKTGGD 424
>UniRef100_O24342 Serine/threonine kinase [Sorghum bicolor]
Length = 440
Score = 430 bits (1106), Expect = e-119
Identities = 211/406 (51%), Positives = 291/406 (70%), Gaps = 7/406 (1%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+GKYELG+T+GEG F KV+ ARDT G+ A+KIL+K+K++ +QIKREIST+KL+
Sbjct: 9 RVGKYELGRTIGEGTFAKVRFARDTVTGEAVAIKILDKDKVLKHKMVEQIKREISTMKLI 68
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVVR+YEV+ SKTKIY+VLE+ GGELF +I + G++ E R+ FQQLI+ V YCH
Sbjct: 69 KHPNVVRIYEVMGSKTKIYIVLEFATGGELFQRIVNHGRMREDEARRYFQQLINAVDYCH 128
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+L+D+ GN+K++DF LSAL Q + DGLLHTTCG+PNYVAPE+L ++
Sbjct: 129 SRGVYHRDLKPENLLLDSYGNLKVSDFGLSALSQQMKDDGLLHTTCGTPNYVAPEVLEDQ 188
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGVIL+V+L GYLPF+D NL LY+KI A+ P W S A+ ++ +I
Sbjct: 189 GYDGAMADLWSCGVILFVLLAGYLPFEDSNLMTLYKKISNAEFTFPPWTSFPAKRLLTRI 248
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGDQSP 313
LDPNP TR+T+ I EDEWFK+GY +E+ D +D D F+ E ++
Sbjct: 249 LDPNPMTRITIPEILEDEWFKKGYKRPEF---DEKYDTTLDDVDAVFNDSEEHHVTEKKE 305
Query: 314 KSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKV 373
+ P +NAF+LI MS+ L+L F+ E +R+ RFTS PK++ KIE+ +GF V
Sbjct: 306 EEPVALNAFELISMSAGLNLGNLFDSEQEFKRETRFTSKCPPKEIVRKIEEAAKPLGFDV 365
Query: 374 HKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
KKN KL++ + K +L+VA E+ +V PSL++VE+RK+ GD
Sbjct: 366 QKKNYKLRLEKVKAGRKG--NLNVATEILQVAPSLHMVEVRKAKGD 409
>UniRef100_O24343 Serine/threonine kinase [Sorghum bicolor]
Length = 440
Score = 430 bits (1105), Expect = e-119
Identities = 210/415 (50%), Positives = 298/415 (71%), Gaps = 13/415 (3%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+GKYELG+T+GEG F KV+ A++T+ G+ A+KIL+K K++ +QIKREIST+KL+
Sbjct: 9 RVGKYELGRTIGEGTFAKVRFAKNTETGEPVAIKILDKEKVLRHKMVEQIKREISTMKLI 68
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVVR+YEV+ SKTKIY+VLEYV GGELFD I++ G++ E R+ FQQLI+ V YCH
Sbjct: 69 KHPNVVRIYEVMGSKTKIYIVLEYVTGGELFDTIANHGRMREDEARRYFQQLINAVDYCH 128
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+L+D+ GN+K++DF LSAL Q + DGLLHTTCG+PNYVAPE+L ++
Sbjct: 129 SRGVYHRDLKPENLLLDSYGNLKVSDFGLSALSQQIKDDGLLHTTCGTPNYVAPEVLEDQ 188
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGVIL+V+L GYLPF+D NL LY+KI A+ P W S A+ ++ +
Sbjct: 189 GYDGAMADLWSCGVILFVLLAGYLPFEDSNLMTLYKKISNAEYTFPPWTSFPAKRLLTRF 248
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDE-----EEEEDVFIDDEAFSIHEVSLDGD 310
LDPNP TR+T+ I EDEWFK+GY +++ ++ + VF D E H V+ +
Sbjct: 249 LDPNPMTRITIPEILEDEWFKKGYKRPEFDEKYDTPLDDVDAVFNDSEE---HHVT---E 302
Query: 311 QSPKSPALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMG 370
+ + P ++NAF+LI S+ L+L F+ E +R+ RFTS PK++ KIE+ +G
Sbjct: 303 KKEEEPVVLNAFELISRSAGLNLGNLFDSEQEFKRETRFTSKCPPKEIVRKIEEAAKPLG 362
Query: 371 FKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQ 425
F V KKN KL++ + K +L+VA E+ +V PSL++VE+RK+ GD +++
Sbjct: 363 FGVQKKNYKLRLEKVKAGRKG--NLNVATEILQVAPSLHMVEVRKAKGDTLEFQK 415
>UniRef100_Q8RWU8 Hypothetical protein At2g26980 [Arabidopsis thaliana]
Length = 441
Score = 429 bits (1104), Expect = e-119
Identities = 212/404 (52%), Positives = 288/404 (70%), Gaps = 3/404 (0%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+GKYE+G+T+GEG F KVK AR+++ G+ A+KIL+K K++ +QI+REI+T+KL+
Sbjct: 10 RVGKYEVGRTIGEGTFAKVKFARNSETGEPVALKILDKEKVLKHKMAEQIRREIATMKLI 69
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV+LYEV+ASKTKI+++LEYV GGELFDKI + G++ E R+ FQQLI V YCH
Sbjct: 70 KHPNVVQLYEVMASKTKIFIILEYVTGGELFDKIVNDGRMKEDEARRYFQQLIHAVDYCH 129
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+L+D+ GN+KI+DF LSAL Q R DGLLHT+CG+PNYVAPE+L +R
Sbjct: 130 SRGVYHRDLKPENLLLDSYGNLKISDFGLSALSQQVRDDGLLHTSCGTPNYVAPEVLNDR 189
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGV+LYV+L GYLPFDD NL LY+KI + P WLS GA +I +I
Sbjct: 190 GYDGATADMWSCGVVLYVLLAGYLPFDDSNLMNLYKKISSGEFNCPPWLSLGAMKLITRI 249
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFIDDEAFSIHEVSLDGDQSPKS 315
LDPNP TRVT + EDEWFK+ Y P E E ++ ++ D F E L ++ +
Sbjct: 250 LDPNPMTRVTPQEVFEDEWFKKDYKPPVFE-ERDDSNMDDIDAVFKDSEEHLVTEKREEQ 308
Query: 316 PALINAFQLIGMSSCLDLSGFFEQEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFKVHK 375
PA INAF++I MS L+L F+ E +R+ R T ++ EKIE+ +GF V K
Sbjct: 309 PAAINAFEIISMSRGLNLENLFDPEQEFKRETRITLRGGANEIIEKIEEAAKPLGFDVQK 368
Query: 376 KNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
KN K+++ K +L+VA E+F+V PSL++V++ KS GD
Sbjct: 369 KNYKMRLENVKAGRKG--NLNVATEIFQVAPSLHMVQVSKSKGD 410
>UniRef100_Q9MAM1 T25K16.13 [Arabidopsis thaliana]
Length = 453
Score = 428 bits (1101), Expect = e-118
Identities = 216/417 (51%), Positives = 298/417 (70%), Gaps = 10/417 (2%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+G YE+G+TLGEG+F KVK A++T G A+KIL++ K+ +Q+KREIST+KL+
Sbjct: 15 RVGNYEMGRTLGEGSFAKVKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREISTMKLI 74
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV + EV+ASKTKIY+VLE VNGGELFDKI+ +G+L E R+ FQQLI+ V YCH
Sbjct: 75 KHPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINAVDYCH 134
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+++DA G +K++DF LSA + R DGLLHT CG+PNYVAPE+L+++
Sbjct: 135 SRGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPEVLSDK 194
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQ--KIFKADVQIPRWLSPGAQNIIK 253
GYDGA +D+WSCGVIL+V++ GYLPFD+ NL LY+ +I KA+ P W S GA+ +IK
Sbjct: 195 GYDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRVRICKAEFSCPPWFSQGAKRVIK 254
Query: 254 KILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGDQ 311
+IL+PNP TR+++ + EDEWFK+GY P + ++++ED+ ID D AFS + L ++
Sbjct: 255 RILEPNPITRISIAELLEDEWFKKGYKPPSF--DQDDEDITIDDVDAAFSNSKECLVTEK 312
Query: 312 SPKSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVIEMG 370
K P +NAF+LI SS L FE Q + +++ RFTS S ++ K+E+ +G
Sbjct: 313 KEK-PVSMNAFELISSSSEFSLENLFEKQAQLVKKETRFTSQRSASEIMSKMEETAKPLG 371
Query: 371 FKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGDASVYRQVC 427
F V K N K+K+ + K LSVA EVFEV PSL+VVELRK+ GD + +VC
Sbjct: 372 FNVRKDNYKIKMKGDKSGRKG--QLSVATEVFEVAPSLHVVELRKTGGDTLEFHKVC 426
>UniRef100_Q94F03 Similar to wpk4 protein kinase [Arabidopsis thaliana]
Length = 447
Score = 427 bits (1097), Expect = e-118
Identities = 213/407 (52%), Positives = 293/407 (71%), Gaps = 8/407 (1%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+G YE+G+TLGEG+F K K A++T G A+KIL++ K+ +Q+KREIST+KL+
Sbjct: 15 RVGNYEMGRTLGEGSFAKAKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREISTMKLI 74
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV + EV+ASKTKIY+VLE VNGGELFDKI+ +G+L E R+ FQQLI+ V YCH
Sbjct: 75 KHPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINAVDYCH 134
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+++DA G +K++DF LSA + R DGLLHT CG+PNYVAPE+L+++
Sbjct: 135 SRGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPEVLSDK 194
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGVIL+V++ GYLPFD+ NL LY++I KA+ P W S GA+ +IK+I
Sbjct: 195 GYDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRICKAEFSCPPWFSQGAKRVIKRI 254
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGDQSP 313
L+PNP TR+++ + EDEWFK+GY P + ++++ED+ ID D AFS + L ++
Sbjct: 255 LEPNPITRISIAELLEDEWFKKGYKPPSF--DQDDEDITIDDVDAAFSNSKECLVTEKKE 312
Query: 314 KSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVIEMGFK 372
K P +NAF+LI SS L FE Q + +++ RFTS S ++ K+E+ +GF
Sbjct: 313 K-PVSMNAFELISSSSEFSLENLFEKQAQLVKKETRFTSQRSASEIMSKMEETAKPLGFN 371
Query: 373 VHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
V K N K+K+ + K LSVA EVFEV PSL+VVELRK+ GD
Sbjct: 372 VRKDNYKIKMKGDKSGRKG--QLSVATEVFEVAPSLHVVELRKTGGD 416
>UniRef100_Q84VQ3 NAF specific protein kinase family [Arabidopsis thaliana]
Length = 452
Score = 424 bits (1091), Expect = e-117
Identities = 214/417 (51%), Positives = 291/417 (69%), Gaps = 16/417 (3%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+GKYE+GKTLG+G F KV+ A +T+ G+ A+KIL+K K++ +QI+REI T+KL+
Sbjct: 9 RVGKYEVGKTLGQGTFAKVRCAVNTETGERVALKILDKEKVLKHKMAEQIRREICTMKLI 68
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
HPNVVRLYEVLASKTKIY+VLE+ GGELFDKI G+L E + RK FQQLI+ V YCH
Sbjct: 69 NHPNVVRLYEVLASKTKIYIVLEFGTGGELFDKIVHDGRLKEENARKYFQQLINAVDYCH 128
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+L+DA+GN+K++DF LSAL + R DGLLHT CG+PNY APE+L ++
Sbjct: 129 SRGVYHRDLKPENLLLDAQGNLKVSDFGLSALSRQVRGDGLLHTACGTPNYAAPEVLNDQ 188
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQKIFKADVQIPRWLSPGAQNIIKKI 255
GYDGA +D+WSCGVIL+V+L GYLPF+D NL LY+KI + P WLSPGA+N+I +I
Sbjct: 189 GYDGATADLWSCGVILFVLLAGYLPFEDSNLMTLYKKIIAGEYHCPPWLSPGAKNLIVRI 248
Query: 256 LDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEE--EDVFIDDEAFSIHEVSLDGDQSP 313
LDPNP TR+T+ + D WFK+ Y PA E++EE +DV D F E ++
Sbjct: 249 LDPNPMTRITIPEVLGDAWFKKNYKPAVFEEKEEANLDDV---DAVFKDSEEHHVTEKKE 305
Query: 314 KSPALINAFQLIGMSSCLDLSGFFEQEDV-------SERKIRFTSNHSPKDLTEKIEDIV 366
+ P +NAF+LI MS LDL FE+E+ +R+ RF + + DL +KIE+
Sbjct: 306 EQPTSMNAFELISMSRALDLGNLFEEEESIPFNLQGFKRETRFAAKGAANDLVQKIEEAS 365
Query: 367 IEMGFKVHKKNGK----LKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
+GF + KKN K ++ E+ +L VA E+F+V PSL+++E+RK+ GD
Sbjct: 366 KPLGFDIQKKNYKRYLCTQMRLENVTAGRKGNLRVATEIFQVSPSLHMIEVRKTKGD 422
>UniRef100_Q9C5S4 CBL-interacting protein kinase 9 [Arabidopsis thaliana]
Length = 449
Score = 423 bits (1088), Expect = e-117
Identities = 214/409 (52%), Positives = 294/409 (71%), Gaps = 10/409 (2%)
Query: 16 RLGKYELGKTLGEGNFGKVKLARDTDCGQFFAVKILEKNKIVDLNNTDQIKREISTLKLL 75
R+G YE+G+TLGEG+F KVK A++T G A+KIL++ K+ +Q+KREIST+KL+
Sbjct: 15 RVGNYEMGRTLGEGSFAKVKYAKNTVTGDQAAIKILDREKVFRHKMVEQLKREISTMKLI 74
Query: 76 KHPNVVRLYEVLASKTKIYMVLEYVNGGELFDKISSKGKLTEAHGRKMFQQLIDGVSYCH 135
KHPNVV + EV+ASKTKIY+VLE VNGGELFDKI+ +G+L E R+ FQQLI+ V YCH
Sbjct: 75 KHPNVVEIIEVMASKTKIYIVLELVNGGELFDKIAQQGRLKEDEARRYFQQLINAVDYCH 134
Query: 136 NKGVFHRDLKLENVLVDAKGNIKITDFNLSALPQHCRADGLLHTTCGSPNYVAPEILANR 195
++GV+HRDLK EN+++DA G +K++DF LSA + R DGLLHT CG+PNYVAPE+L+++
Sbjct: 135 SRGVYHRDLKPENLILDANGVLKVSDFGLSAFSRQVREDGLLHTACGTPNYVAPEVLSDK 194
Query: 196 GYDGAKSDIWSCGVILYVILTGYLPFDDRNLAVLYQ--KIFKADVQIPRWLSPGAQNIIK 253
GYDGA +D+WSCGVIL+V++ GYLPFD+ NL LY+ +I KA+ P W S GA+ +IK
Sbjct: 195 GYDGAAADVWSCGVILFVLMAGYLPFDEPNLMTLYKRVRICKAEFSCPPWFSQGAKRVIK 254
Query: 254 KILDPNPKTRVTMDMIKEDEWFKEGYNPANLEDEEEEEDVFID--DEAFSIHEVSLDGDQ 311
+IL+PNP TR+++ + EDEWFK+GY P + ++++ED+ ID D AFS + L ++
Sbjct: 255 RILEPNPITRISIAELLEDEWFKKGYKPPSF--DQDDEDITIDDVDAAFSNSKECLVTEK 312
Query: 312 SPKSPALINAFQLIGMSSCLDLSGFFE-QEDVSERKIRFTSNHSPKDLTEKIEDIVIEMG 370
K P +NAF+LI SS L FE Q + +++ RFTS S ++ K+E+ +G
Sbjct: 313 KEK-PVSMNAFELISSSSEFSLENLFEKQAQLVKKETRFTSQRSASEIMSKMEETAKPLG 371
Query: 371 FKVHKKNGKLKVIQEDKAHKSLDSLSVAAEVFEVGPSLYVVELRKSYGD 419
F V K N K+K+ + K LSVA EVFEV PSL+VVELRK+ GD
Sbjct: 372 FNVRKDNYKIKMKGDKSGRKG--QLSVATEVFEVAPSLHVVELRKTGGD 418
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.138 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 748,398,699
Number of Sequences: 2790947
Number of extensions: 33311274
Number of successful extensions: 161215
Number of sequences better than 10.0: 19311
Number of HSP's better than 10.0 without gapping: 12692
Number of HSP's successfully gapped in prelim test: 6621
Number of HSP's that attempted gapping in prelim test: 109551
Number of HSP's gapped (non-prelim): 22991
length of query: 437
length of database: 848,049,833
effective HSP length: 130
effective length of query: 307
effective length of database: 485,226,723
effective search space: 148964603961
effective search space used: 148964603961
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC138579.3


