

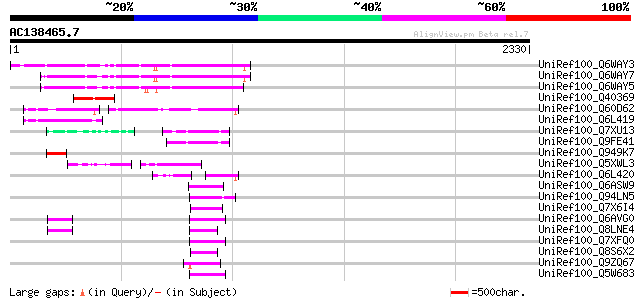
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138465.7 + phase: 0 /pseudo
(2330 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum] 505 e-141
UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum] 503 e-140
UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum] 496 e-138
UniRef100_Q40369 RPE15 protein [Medicago sativa] 256 6e-66
UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solan... 228 2e-57
UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum] 141 3e-31
UniRef100_Q7XU13 OSJNBa0091D06.8 protein [Oryza sativa] 106 1e-20
UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic ... 105 1e-20
UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum] 99 2e-18
UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum] 97 5e-18
UniRef100_Q6L420 Putative polyprotein [Solanum demissum] 87 8e-15
UniRef100_Q6ASW9 Reverse transcriptase (RNA-dependent DNA polyme... 83 1e-13
UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sa... 79 2e-12
UniRef100_Q7X6I4 OSJNBa0039G19.2 protein [Oryza sativa] 78 3e-12
UniRef100_Q6AVG0 Putative retrotransposon gag protein [Oryza sat... 77 6e-12
UniRef100_Q8LNE4 Putative retroelement [Oryza sativa] 77 8e-12
UniRef100_Q7XFQ0 Putative retroelement [Oryza sativa] 77 8e-12
UniRef100_Q8S6X2 Putative pol polyprotein [Oryza sativa] 77 8e-12
UniRef100_Q9ZQ67 Putative retroelement pol polyprotein [Arabidop... 76 1e-11
UniRef100_Q5W683 Hypothetical protein OSJNBa0076A09.14 [Oryza sa... 76 1e-11
>UniRef100_Q6WAY3 Gag/pol polyprotein [Pisum sativum]
Length = 2262
Score = 505 bits (1300), Expect = e-141
Identities = 386/1160 (33%), Positives = 563/1160 (48%), Gaps = 142/1160 (12%)
Query: 3 QLRTELASLREELAKANDTMTALLAAQEQSATAIPVATAIPIATSVIPTASTDARFAMPT 62
Q++TELA +R +A+ M + QE+ + A+ +
Sbjct: 3 QVQTELAEMRANMAQFMHMMQGVAQGQEELRALVQRQEAVTPPPN--------------Q 48
Query: 63 GFPYGLPPFFTPSTAAGTSGTANNGAVPGTNTI-------SINTTLPQTTATVTEPLVHA 115
P G P TP+ A + A + G + N + A P+V
Sbjct: 49 ALPEGNPVHDTPAAAIPVNNYAVGEELMGIRVDGQPIAPDAANARVIHAPARNRIPIVDR 108
Query: 116 ISQGVNINAHHGSIPVIKTMEERMEELAKELRREIKANRGNGDSVKTHDLCLVPKVDVPK 175
+ + IP +R + E R ++ G V ++ LV + +P
Sbjct: 109 QEDLFTMFSEDEDIPGRNDARDRKVDALAEKIRAMECQNSLGFDVT--NMGLVEGLRIPY 166
Query: 176 KFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDD 235
KFK P FD+YNG +CP+ H+ Y RK+ Y+D++ + ++ FQDSL + +WY L D
Sbjct: 167 KFKAPSFDKYNGTSCPRTHVQAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKSDS 226
Query: 236 VHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEE 295
+ + +L AF Y N + P+R L SL QK +ESF+EYAQ WR AAR+ P + E
Sbjct: 227 IRCWRDLGEAFLRQYKHNMDMAPSRTQLQSLCQKSNESFKEYAQRWRELAARVQPPMLER 286
Query: 296 EMTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNASKRY 355
E+T F+ TL+ +++RM +F ++V G R E ++ G I + V SS +SK+
Sbjct: 287 ELTDMFIGTLQGVFMDRMGSCPFGSFSDVVICGERTESLIKTGKI--QDVGSS--SSKKP 342
Query: 356 GNDHHKKKETEVGMVSAGAGQSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQYPLPS 415
+++E E V Q+ A +P P Q P
Sbjct: 343 FAGAPRRREGETNAVQHRRDQNRIEYRQAAAVTIPAPQPRQQQQQRVQQP---------- 392
Query: 416 GQPQVPVNAVVQQMQQQPPVQQQQHQQARPTFPPIPMLYAELLPTLLLRGH---CTTRHG 472
QQ QQQ P Q +Q R F +PM YAELLP LL G CT
Sbjct: 393 -----------QQQQQQRPYQPRQRMPDR-RFDSLPMSYAELLPELLRLGLVELCTMA-- 438
Query: 473 KPPPDPLLLRFRSDLKCDFHQGALGHDVEGCYTLKYIVKKLIDQGKLTFENNVPYVLDNP 532
PP L + ++++CDFH GA GH E C L++ V+ LID + F VP V++NP
Sbjct: 439 --PPTVLPPGYDANVRCDFHSGAPGHHTEKCRALQHKVQDLIDANAINFAP-VPNVVNNP 495
Query: 533 LPNHAA--VNMIEVYEEAPRLD-VHNIATPLVPLHIKLCQASLF---DHDHASCPE*FHN 586
+P H VN IE E +D V ++ T L+ + +L ++ D D C E +
Sbjct: 496 MPQHGGHRVNNIEGKEAEDLVDNVDDVQTSLLVVKSRLLNEGVYSGCDEDCLGCAE---S 552
Query: 587 TLGCYVVQNEIQSLMNDNYLTVS-------DVCVIVPVF-----HDPPVKSIPSKENVEP 634
GC ++ IQ +M++ L S V I F H V S P+ N P
Sbjct: 553 ENGCDQLRAGIQGMMDEGCLQFSRAVKDRGTVSTITIYFKPSEGHGQRVVSAPAT-NGTP 611
Query: 635 LVIRLPGPV--PYTL----------EKAIPYKY-----------------NATMIENGV- 664
+ I +P + P T+ +A+P+KY N T + G+
Sbjct: 612 VTIPVPVTISAPTTIVASGRRAVENSRAVPWKYDNAYRSNRRVESQTKPVNQTPVTIGLA 671
Query: 665 -EVPLASLAMVSNIAEGTTAALRSGRV--RPPLFQKKADTPTTPPIDKATLTDVSPVTKD 721
VP V N+ G RSGR+ PL A+ K + + PV K+
Sbjct: 672 NRVPATVGPAVDNVG-GPGGFTRSGRLFAPQPLRDNNAEALAKAK-GKQAVVEEEPVQKE 729
Query: 722 VSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRSTLLKVLEQ 781
P S E +++E +++IK+SDYKIVDQL QTPSKIS+LSLLL SEAHR+ LLK+L
Sbjct: 730 A--PEGSFE-KDVEEFMKMIKKSDYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNL 786
Query: 782 AYVDHEVTVDRFGGIVGNITACNNLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVD 841
AYV E++V++ G++ N++ + + F+ ++P + HN ALHI++ K +LS+VLVD
Sbjct: 787 AYVPQEISVNQLEGVMANVSTRHGVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVD 846
Query: 842 TGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQ 901
TGSSLNV+PK L ++ L S +V+AFD S+++V GE+ LP+ IG E F I F
Sbjct: 847 TGSSLNVLPKQILKKIDVEGFVLTPSDLIVRAFDRSKRSVCGEVTLPVKIGPEVFDIIFY 906
Query: 902 VMDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIE 961
VMDI +YSCLLGRPWIH AGAV+STLH+KLK+V NG++VTV GEE LVS LSSF +E
Sbjct: 907 VMDIQPAYSCLLGRPWIHAAGAVSSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVE 966
Query: 962 A-GSAEGT---AFQGLTIEG---TELKKTGTAMASLKDAQKAVQEGQAAGWGKLIQLREN 1014
G T F+ + +E E +K G ++ S K A++ V G+A GWGK++ L
Sbjct: 967 VDGEIHETLCQVFETVALEKVAYAEQRKPGVSITSYKQAKEVVDSGKAEGWGKMVDLPVK 1026
Query: 1015 KHKEGLGFSPTSG-----VST*TFHSAGFVNAITEEAAGFGP-----------RPVFVIP 1058
+ K G+G+ P TF SAG +N A G RP P
Sbjct: 1027 EDKFGVGYEPLQAEQNGQAGPSTFTSAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAP 1084
Query: 1059 GGIARDWDAIDIPTIMHVSE 1078
GG +W A ++ + ++E
Sbjct: 1085 GGSINNWTAEEVVQVTLLTE 1104
>UniRef100_Q6WAY7 Gag/pol polyprotein [Pisum sativum]
Length = 1814
Score = 503 bits (1294), Expect = e-140
Identities = 357/1017 (35%), Positives = 528/1017 (51%), Gaps = 115/1017 (11%)
Query: 136 EERMEELAKELRREIKANRGNGDSVKTHDLCLVPKVDVPKKFKIPEFDRYNGLTCPQNHI 195
+ +++ LA+++R ++ G V ++ LV + +P KFK P FD+YNG +CP+ H+
Sbjct: 131 DRKVDALAEKIRA-MECQNSLGFDVT--NMGLVEGLRIPYKFKAPSFDKYNGTSCPRTHV 187
Query: 196 IKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTR 255
Y RK+ Y+D++ + ++ FQDSL + +WY L +D + + +L AF Y N
Sbjct: 188 QAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKRDSIRCWKDLGEAFLRQYKHNMD 247
Query: 256 LKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEEMTQTFLKTLKKDYVERMII 315
+ P+R L SL QK ESF+EYAQ WR AAR+ P + E E+T F+ TL+ +++RM
Sbjct: 248 MAPSRTQLQSLCQKSGESFKEYAQRWRELAARVQPPMLERELTDMFIGTLQGVFMDRMGS 307
Query: 316 AAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNASKRYGNDHHKKKETEVGMVSAGAG 375
+F ++V G R E ++ G I ++ ++SK+ +++E E V
Sbjct: 308 CPFVSFSDVVICGERTESLIKTGKIQ----DAGSSSSKKPFAGAPRRREGETNAVQYRRD 363
Query: 376 QSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPV 435
Q+ + + A +P P P Q Q V QQ QQQ P
Sbjct: 364 QNRSQRCQVAAVTIPA--------------------PQPRQQQQQRVQQPQQQQQQQRPY 403
Query: 436 QQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPPPDPLLLRFRSDLKCDFHQGA 495
Q +Q R F +PM YAELLP LL G R PP L + ++++CDFH GA
Sbjct: 404 QPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT-MAPPTVLPPGYDANVRCDFHSGA 461
Query: 496 LGHDVEGCYTLKYIVKKLIDQGKLTFENNVPYVLDNPLPNHAA--VNMIEVYE-EAPRLD 552
GH E C L++ V+ LID + F VP V++NP+P H VN IE E E ++
Sbjct: 462 PGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQHGGHRVNNIEGKEAEDLVVN 520
Query: 553 VHNIATPLVPLHIKLCQASLF---DHDHASCPE*FHNTLGCYVVQNEIQSLMNDNYLTVS 609
V ++ T L+ + +L ++ D D C E + GC ++ IQ +M++ L S
Sbjct: 521 VDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAE---SENGCDQLRTGIQGMMDEGCLQFS 577
Query: 610 ------DVCVIVPVFHDPP------VKSIPSKENVEPLVIRLPGPV--PYTL-------- 647
+ ++ P V S P+ N P+ I +P + P T+
Sbjct: 578 RAVKDRGTVSTITIYFKPSEGRGQRVVSAPAT-NGTPVTISVPVTISAPTTIAASGRRAV 636
Query: 648 --EKAIPYKY-----------------NATMIENGV--EVPLASLAMVSNIAEGTTAALR 686
+A+P+KY N + G+ VP V N+ G R
Sbjct: 637 ENSRAVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVG-GPGGFTR 695
Query: 687 SGRV--RPPLFQKKADTPTTPPIDKATLTDVSPVTKDVSRPSQSIEDSNLDEILRIIKRS 744
SGR+ PL A+ K + + PV K+ P S E +++E ++IIK+S
Sbjct: 696 SGRLFAPQPLRDNNAEALAKAK-GKQAVVEEEPVQKEA--PEGSFE-KDVEEFMKIIKKS 751
Query: 745 DYKIVDQLLQTPSKISVLSLLLSSEAHRSTLLKVLEQAYVDHEVTVDRFGGIVGNITACN 804
DYKIVDQL QTPSKIS+LSLLL SEAHR+ LLK+L AYV E++V++ G++ N++ +
Sbjct: 752 DYKIVDQLNQTPSKISILSLLLCSEAHRNALLKMLNLAYVPQEISVNQLEGVMANVSTRH 811
Query: 805 NLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPL 864
+ F+ ++ + HN ALHI++ K +LS+VLVDTGSSLNV+PK L ++ L
Sbjct: 812 GVGFTNLDLTPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPKQILKKIDVEGFVL 871
Query: 865 RRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAV 924
S +V+AFDGS+++V GE+ LP+ IG E F I F VMDI +YSCLLGRPWIH AGAV
Sbjct: 872 TPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAV 931
Query: 925 TSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEA-GSAEGT---AFQGLTIEG--- 977
+STLH+KLK+V NG++VTV GEE LVS LSSF +E G T AF+ + +E
Sbjct: 932 SSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVALEKVAY 991
Query: 978 TELKKTGTAMASLKDAQKAVQEGQAAGWGKLIQLRENKHKEGLGFSPTSG-----VST*T 1032
E +K G ++ S K A++ V G+A GWGK++ L + K G+G+ P T
Sbjct: 992 AEQRKPGASITSYKQAKEVVDSGKAEGWGKMVYLPVKEDKFGVGYEPLQAEQNGQAGPST 1051
Query: 1033 FHSAGFVNAITEEAAGFGP-----------RPVFVIPGGIARDWDAIDIPTIMHVSE 1078
F SAG +N A G RP PGG +W A ++ + ++E
Sbjct: 1052 FTSAGLMNHGDVSATGSEDCDSDCDLDNWVRP--CAPGGSINNWTAEEVVQVTLLTE 1106
>UniRef100_Q6WAY5 Gag/pol polyprotein [Pisum sativum]
Length = 1105
Score = 496 bits (1278), Expect = e-138
Identities = 349/976 (35%), Positives = 508/976 (51%), Gaps = 102/976 (10%)
Query: 136 EERMEELAKELRREIKANRGNGDSVKTHDLCLVPKVDVPKKFKIPEFDRYNGLTCPQNHI 195
+ +++ LA+++R N S ++ LV + +P KFK P FD+YN +CP+ H+
Sbjct: 131 DRKVDALAEKIRA---MECQNSLSFDVTNMGLVEGLRIPYKFKAPSFDKYNDTSCPRTHV 187
Query: 196 IKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTR 255
Y RK+ Y+D++ + ++ FQDSL + +WY L +D + + +L AF Y N
Sbjct: 188 QAYYRKISAYTDDEKMWMYFFQDSLSGASLDWYMELKRDSIRCWRDLGEAFLRQYKHNMD 247
Query: 256 LKPNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEEMTQTFLKTLKKDYVERMII 315
+ P+R L SL QK ESF+EYAQ WR AAR+ P + E E+T F+ TL+ +++RM
Sbjct: 248 MAPSRTQLQSLCQKSGESFKEYAQRWRELAARVQPPMLERELTDMFIGTLQGVFMDRMGS 307
Query: 316 AAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNASKRYGNDHHKKKETEVGMVSAGAG 375
+F ++V G R E ++ G I + V SS +SK+ +++E E +V
Sbjct: 308 CPFGSFSDVVICGERTESLIKTGKI--QDVGSS--SSKKPFAGAPRRREGETNVVQHRRD 363
Query: 376 QSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPV 435
Q+ A +P P P Q Q V QQ QQQ P
Sbjct: 364 QNRIEYRQAAAVTIPA--------------------PQPRQQQQQRVQQPQQQQQQQRPY 403
Query: 436 QQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPPPDPLLLRFRSDLKCDFHQGA 495
Q +Q R F +PM YAELLP LL G R PP L + ++++CDFH GA
Sbjct: 404 QPRQRMPDR-RFDSLPMSYAELLPELLRLGMVELRT-MAPPTVLPPGYDANVRCDFHSGA 461
Query: 496 LGHDVEGCYTLKYIVKKLIDQGKLTFENNVPYVLDNPLPNHAA--VNMIEVYE-EAPRLD 552
GH E C L++ V+ LID + F VP V++NP+P H VN IE E E ++
Sbjct: 462 PGHHTEKCRALQHKVQDLIDAKAINFAP-VPNVVNNPMPQHGGHRVNNIEGKEAEDLVVN 520
Query: 553 VHNIATPLVPLHIKLCQASLF---DHDHASCPE*FHNTLGCYVVQNEIQSLMNDNYL--- 606
V ++ T L+ + +L ++ D D C E + GC ++ IQ +M++ L
Sbjct: 521 VDDVQTSLLVVKGRLLNGGVYSGCDEDCLGCAE---SENGCDQLRAGIQGMMDEGCLQFS 577
Query: 607 -------TVSDVCVI-------------VPVFHDPPVK-SIPSKENVEPLVIRLPGPVPY 645
TVS + + P +D PV S+P + P I G
Sbjct: 578 RAVKDRGTVSTITIYFKPTEGRGQRVVSAPATNDTPVTISVPVTISA-PTTIVASGRRAV 636
Query: 646 TLEKAIPYKY-----------------NATMIENGV--EVPLASLAMVSNIAEGTTAALR 686
+ +P+KY N + G+ VP V N+ G R
Sbjct: 637 ENSRVVPWKYDNAYRSNRRAESQTRPVNQAPVTIGLPNRVPATVGPAVDNVG-GPGGFTR 695
Query: 687 SGRV--RPPLFQKKADTPTTPPIDKATLTDVSPVTKDVSRPSQSIEDSNLDEILRIIKRS 744
SGR+ PL A+ K + + PV K+ P + E +++E ++IIK+S
Sbjct: 696 SGRLFAPQPLRDNNAEALAKAK-GKQAVVEEEPVQKEA--PEGTFE-KDVEEFMKIIKKS 751
Query: 745 DYKIVDQLLQTPSKISVLSLLLSSEAHRSTLLKVLEQAYVDHEVTVDRFGGIVGNITACN 804
DYKIVDQL QTPSKIS+LSLL+ SEAHR+ LLK+L AYV E++V++ G++ N++ +
Sbjct: 752 DYKIVDQLNQTPSKISILSLLMCSEAHRNALLKMLNLAYVPQEISVNQLEGVMANVSTRH 811
Query: 805 NLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPL 864
+ F+ ++P + HN ALHI++ K +LS+VLVDTGSSLNV+P L ++ L
Sbjct: 812 GVGFTNLDLPPEGRNHNKALHITMECKGAVLSHVLVDTGSSLNVLPNQILKKIDVEGFVL 871
Query: 865 RRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAV 924
S +V+AFDGS+++V GE+ LP+ IG E F I F VMDI +YSCLLGRPWIH AGAV
Sbjct: 872 TPSDLIVRAFDGSKRSVCGEVTLPVKIGPEVFDIIFYVMDIQPAYSCLLGRPWIHAAGAV 931
Query: 925 TSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEA-GSAEGT---AFQGLTIEG--- 977
+STLH+KLK+V NG++VTV GEE LVS LSSF +E G T AF+ + +E
Sbjct: 932 SSTLHQKLKYVWNGQIVTVCGEEEILVSHLSSFKYVEVDGEIHETLCQAFETVALEKVAY 991
Query: 978 TELKKTGTAMASLKDAQKAVQEGQAAGWGKLIQLRENKHKEGLGFSP-----TSGVST*T 1032
E +K G ++ S K A++ V G+A GWGK++ L + K G+G+ P T
Sbjct: 992 AEQRKPGASITSYKQAKEVVDSGKAEGWGKMVDLPVKEDKFGVGYEPLRAEQNGQAGPST 1051
Query: 1033 FHSAGFVNAITEEAAG 1048
F SAG +N A G
Sbjct: 1052 FTSAGLMNHGDVSATG 1067
>UniRef100_Q40369 RPE15 protein [Medicago sativa]
Length = 219
Score = 256 bits (654), Expect = 6e-66
Identities = 130/192 (67%), Positives = 147/192 (75%), Gaps = 11/192 (5%)
Query: 284 AAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREGIIVFE 343
AAAR++P LDEEE+TQTFLKTLKK++ ERMI++APNNF +MVTMGTRLEEAVREGIIV E
Sbjct: 13 AAARVSPRLDEEELTQTFLKTLKKEHKERMIVSAPNNFSDMVTMGTRLEEAVREGIIVLE 72
Query: 344 KVESSVNASKRYGNDHHKKKET-EVGMVSAGAGQSMATVAPINAAQLPPSYPYAPYSQHP 402
K E S +A K+YGN HHK+KE+ +VGMVS P+NA QLP Y Y SQHP
Sbjct: 73 KGECSASAPKKYGNGHHKRKESKKVGMVSGNHD------TPVNATQLPLPYQYMQISQHP 126
Query: 403 FFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPT----FPPIPMLYAELL 458
FFPPFY QYPLP GQP VPVNA+ QQ+Q QPP QQQQ QQ P FPPIPM YAELL
Sbjct: 127 FFPPFYQQYPLPPGQPPVPVNAMAQQVQHQPPAQQQQQQQQGPRPKTYFPPIPMRYAELL 186
Query: 459 PTLLLRGHCTTR 470
PTLL +GHC TR
Sbjct: 187 PTLLAKGHCVTR 198
>UniRef100_Q60D62 Putative gag/pol polyprotein, 3'-partial [Solanum demissum]
Length = 1784
Score = 228 bits (581), Expect = 2e-57
Identities = 181/609 (29%), Positives = 282/609 (45%), Gaps = 88/609 (14%)
Query: 443 ARPTFPPIPMLYAEL---LPTLLLRGHCTTRHGKPPPDPLLLRFRSDLKCDFHQGALGHD 499
A+ F PI YA L L TL + + PPP L C + A GH+
Sbjct: 888 AKDEFTPIGESYASLFQKLRTLNVLSPIERKMSNPPPRNL----DYSQHCAYCSDAPGHN 943
Query: 500 VEGCYTLKYIVKKLIDQGKLTFEN-NVPYVLDNPLPNHAAVNMIEV---YEEA--PRLDV 553
+E C+ LK ++ LID ++ E+ N P + NPLP H NM+E+ +EE P +
Sbjct: 944 IERCWYLKKAIQDLIDTRRIIVESPNGPNINQNPLPRHTETNMLEMVNNHEEVAVPYKPI 1003
Query: 554 HNIATPLVPLHIKLCQASLFDHDHASCPE*FHNTLGCYVVQNEIQSLMNDNYLTVSDVCV 613
+ T + + + S E + + I+ + D + + + +
Sbjct: 1004 LKVETGMESSTNVIDLTKIMPSGAESTSEKLTPSSAPILT---IKGALEDVWASQREARL 1060
Query: 614 IVPVFHDPPVKSIPSKENVEPLVIRLPGPVPYTLEKAIPYKYNATMIE-NGVEVPLASLA 672
+VP D P+ + + P++IR +P T KA+P+ Y T++ G E+
Sbjct: 1061 VVPRGPDKPILIVQGAY-IPPVIIRPVSQLPMTNPKAVPWNYEPTVVTYEGKEI------ 1113
Query: 673 MVSNIAEGTTAALRSGRVRPPLFQKKADTPTTPPIDKATLTDVSPVTKDVSRPSQSIEDS 732
+ + RSGR LT++ D + + +
Sbjct: 1114 --NEEVDEIGGMTRSGRCY-------------------ALTELRKNKNDQMQVKSPVTEG 1152
Query: 733 NLDEILRIIKRSDYKIVDQLLQTPSKISVLSLLLSSEAHRSTLLKVLEQAYVDHEVTVDR 792
+E LR +K SDY IV+QL +TP++IS+LSLL+ S+AHR ++K+L +A+V +EVTV +
Sbjct: 1153 EAEEFLRKMKLSDYSIVEQLRKTPAQISLLSLLIHSDAHRKAVIKILNEAHVPNEVTVSQ 1212
Query: 793 FGGIVGNITACNNLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKS 852
I G I N + FS+DE+P GS N+ P S
Sbjct: 1213 LEKIAGRIFEVNRITFSDDELPVE--------------------------GSGANICPLS 1246
Query: 853 TLDQLSYRETPLRRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCL 912
TL +L+ +R + V+AFDG++ + +GEI+L + IGH F + FQV++INASY+ L
Sbjct: 1247 TLQKLNVNVERVRPNNVCVRAFDGTKTDAIGEIELILKIGHVDFTVNFQVLNINASYNLL 1306
Query: 913 LGRPWIHDAGAVTSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSA-EGTAFQ 971
LGRPW+H AGAV STLH+ +KF + + V VH E V + SS I+A + E +Q
Sbjct: 1307 LGRPWVHRAGAVPSTLHQMVKFEYDRQEVIVHSEGDLSVYKDSSLPFIKANNENEALVYQ 1366
Query: 972 GLTI-------EGTELKKTGTAMASLKDAQKAVQEGQAAGWGKLIQL---------RENK 1015
+ EG + K MAS+ + ++ G G G I L R++
Sbjct: 1367 AFEVVVVEHILEGNLISKPQLPMASVMMVNEMLKHGFEPGKGLGIFLQGRAYPVSPRKSL 1426
Query: 1016 HKEGLGFSP 1024
GLG+ P
Sbjct: 1427 GTFGLGYKP 1435
Score = 106 bits (265), Expect = 7e-21
Identities = 98/368 (26%), Positives = 155/368 (41%), Gaps = 60/368 (16%)
Query: 61 PTGFPYGLPPFFTPSTAAGTSGTANNGAVPGTNTISINTTLPQTTATVTEPLVHAISQGV 120
P P P F P+ A T+ T+N VP +N L T E L S
Sbjct: 598 PLNTPMMSNPLFVPT--APTNSTSNPTVVPKSNNDPSFQVLHDHGYTPEEALKIPSSYP- 654
Query: 121 NINAHHGSIP--VIKTME-ERMEELAKELR---REIKANRGNGD--SVKTHDLCLVPKVD 172
H S P + KT++ E EE+AK+++ + I+ +G G + DLC+ P V
Sbjct: 655 --QTHQYSSPFKIEKTVKNEEHEEMAKKMKSLEQSIRDMQGLGGHKGISFSDLCMFPHVH 712
Query: 173 VPKKFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLS 232
+P + L++ F +SL+ A+EW+
Sbjct: 713 LPT----------------------------GAEGKEELLMAYFGESLVGIASEWFIDQD 744
Query: 233 KDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPAL 292
+ HT+D LA F + +N + P+R L ++ +K E+FREYA WR AAR+ ++
Sbjct: 745 IANWHTWDNLARCFVQQFQYNIDIVPDRSSLANMRKKTTENFREYAVRWREQAARVKLSM 804
Query: 293 DEEEMTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNAS 352
E EM FL+ + DY ++ A F E++ +G +E ++ G IV + + +
Sbjct: 805 KESEMIDVFLQAQEPDYFHYLLSAVGKTFVEVIKVGEMVENGIKSGKIVSQAALKATTQA 864
Query: 353 KR--YGNDHHKKKETEVGMVSAGA-------GQSMAT----------VAPINAAQLPPSY 393
+ GN KK+ +V V GA G+S A+ ++PI P
Sbjct: 865 LQNGSGNIGGKKRREDVATVGNGAKDEFTPIGESYASLFQKLRTLNVLSPIERKMSNPPP 924
Query: 394 PYAPYSQH 401
YSQH
Sbjct: 925 RNLDYSQH 932
>UniRef100_Q6L419 Putative gag polyprotein [Solanum demissum]
Length = 421
Score = 141 bits (355), Expect = 3e-31
Identities = 104/364 (28%), Positives = 165/364 (44%), Gaps = 32/364 (8%)
Query: 61 PTGFPYGLPPFFTPSTAAGTSGTANNGAVPGTNTISINTTLPQTTATVTEPLVHAISQGV 120
P P P F P+ A T+ T+N VP +N L T E L S
Sbjct: 78 PLNTPMMSNPLFVPT--APTNSTSNPTVVPKSNNDPSFQVLHDHGYTPEEALKIPSSYP- 134
Query: 121 NINAHHGSIP--VIKTME----ERMEELAKELRREIKANRGNGD--SVKTHDLCLVPKVD 172
H S P + KT++ E M + K L I+ +G G + DLC+ P V
Sbjct: 135 --QTHQYSSPFKIEKTVKNEEHEEMTKKMKSLEHSIRDMQGLGGHKGISFSDLCMFPHVH 192
Query: 173 VPKKFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEWYTSLS 232
+P FK P F++Y+G P H+ +Y ++ + L++ F +SL+ A++W+
Sbjct: 193 LPTGFKTPNFEKYDGHGDPIAHLKRYYNQLRGAEGKEELLMAYFGESLVGIASKWFIDQD 252
Query: 233 KDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWRGAAARITPAL 292
+ HT+D+LA F + +N + P+R L ++ +K E+FREYA WR AAR+ P +
Sbjct: 253 IANWHTWDDLARCFVQQFQYNIDIVPDRSSLANMRKKTTENFREYAVRWREQAARVKPPM 312
Query: 293 DEEEMTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREGIIVFEKVESSVNAS 352
E EM FL+ + DY ++ A F E++ +G +E ++ G IV + + +
Sbjct: 313 KELEMIDVFLQAQEPDYFHYLLSAVGKTFTEVIKVGEMVENGIKSGKIVSQAALKATTQA 372
Query: 353 --KRYGNDHHKKKETEVGMVSAGAGQSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQ 410
GN KK+ +V V P +Y ++QH +FPP Q
Sbjct: 373 LPNGSGNIGGKKRREDVATV----------------VSAPRTYAQDNHTQH-YFPPQIPQ 415
Query: 411 YPLP 414
Y +P
Sbjct: 416 YLVP 419
>UniRef100_Q7XU13 OSJNBa0091D06.8 protein [Oryza sativa]
Length = 2657
Score = 106 bits (264), Expect = 1e-20
Identities = 84/302 (27%), Positives = 142/302 (46%), Gaps = 24/302 (7%)
Query: 685 LRSGRVRPPLFQKKADTPTTPPIDKATLTDVSPVTKDVSRPSQSIEDSNLDEILRIIKRS 744
LR G+ P + K + PI +A+ +P TK S+ I
Sbjct: 1343 LRGGKTLPDPHKAKP-SKVDKPIKEASPPGEAPETKARSKEKPDI--------------- 1386
Query: 745 DYKIVDQLLQTPSKISVLSLLLSSEAHRSTLLKVLEQAYVDHEVTVDRFGGIVGNITACN 804
DY ++ L + P+ +SV L+ R L+K L+ V +EV + + + N N
Sbjct: 1387 DYNVLAHLKRIPALLSVYDALILVPDLREALIKALQTPEV-YEVDMAKHR-LFNNPLFVN 1444
Query: 805 NLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPL 864
+ F++++ HN L+I N L +L+D GS++N++P +L + + L
Sbjct: 1445 EITFADEDNIIEGSDHNRPLYIEGNIGPAHLRRILIDPGSAVNILPVRSLTRAGFTTKDL 1504
Query: 865 RRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAV 924
+ ++ FD K+ LG I + + + +F + F V++ N SYS LLGRPWIH V
Sbjct: 1505 EPTDMVICGFDNQGKSTLGAITVKIQMSTFSFKVHFFVIEANTSYSALLGRPWIHKYRVV 1564
Query: 925 TSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTELKKTG 984
STLH+ LKF+ +GK G + +V SS++ E+ A+ + + +L +T
Sbjct: 1565 PSTLHQCLKFL-DGK-----GAQQRIVGNSSSYTIQESYHADAKYYFPVEENMQQLGRTA 1618
Query: 985 TA 986
A
Sbjct: 1619 PA 1620
Score = 51.2 bits (121), Expect = 4e-04
Identities = 77/404 (19%), Positives = 150/404 (37%), Gaps = 81/404 (20%)
Query: 163 HDLCLVPKVDVPKKFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLME 222
HDL P P KF+ +FDR + H+ + G+ ++N SL++ F SL
Sbjct: 839 HDLVAFPARWHPPKFR--QFDRTGDA---REHLAYFEAACGDTANNSSLLLRQFSGSLTG 893
Query: 223 DAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREF----LISLSQKKDESFREYA 278
A WY+ L + ++ + K H+ + ++F L + Q++DE+ +Y
Sbjct: 894 PAFHWYSRLPTGSIRSWASMKEVLKKHF-----VAMKKDFSIVELSQVRQRRDEAIDDYV 948
Query: 279 QIWRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAAPNNFFEMVTMGTRLEEAVREG 338
+R + R+ + E+ + L + T+LE
Sbjct: 949 IRFRNSFVRLAREMHLEDAIEIALSS--------------------AVAATKLEFENSPQ 988
Query: 339 IIVFEKVESSVNASKRYGNDHHKKKETEVGMVSAGAGQSMATVAPINAAQLPPSYPYAPY 398
I+ K S+ + +K + +G G A N A++ + AP
Sbjct: 989 IMELYKNASAFDPAKHFS-----------ATKPSGGGSKPKAPAEANVARV---FSTAPQ 1034
Query: 399 SQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPVQQQQHQQARPTFPPIPMLYAELL 458
Q P Q + +Q+P +Q +Q + EL+
Sbjct: 1035 GQVPMLGAKNEQ----------------ARGRQRPSIQDLLKKQY--------IFRRELV 1070
Query: 459 PTLL--LRGHCTTRHGKPPPDPLLLRFRSDLKCDFHQGALGHDVEGCYTLKYIVKKLIDQ 516
+ L+ H +P + + L C +H+ +G+ +E C K +++ +++
Sbjct: 1071 KDMFNQLKEHRALNLPEPWRPDQVTMVDNPLYCPYHR-YVGYAIEDCVAFKEWLQRAVNE 1129
Query: 517 GKLTFENNVPYVLDNPLPNHAAVNMIEVYEEAPRLDVHNIATPL 560
++ + D P++ AVNM+ V + + +I PL
Sbjct: 1130 KRINLD------ADAINPDYRAVNMVSVEPGSREQEGADIWVPL 1167
>UniRef100_Q9FE41 Oryza sativa (japonica cultivar-group) genomic DNA, chromosome 1, PAC
clone:P0433F09 [Oryza sativa]
Length = 2876
Score = 105 bits (263), Expect = 1e-20
Identities = 78/284 (27%), Positives = 133/284 (46%), Gaps = 8/284 (2%)
Query: 703 TTPPIDKATLTDVSPVTKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPSKISVL 762
T P K+ + +V K S P ++ E + DYK++ L + P+ +SV
Sbjct: 1263 TLPDPHKSKVPNVDKPAKKASPPGEAPEAPETKTGSKEKPAVDYKVLAHLKRIPALLSVY 1322
Query: 763 SLLLSSEAHRSTLLKVLEQAYVDHEVTVDRFGGIVGNITACNNLWFSEDEMPETRKYHNL 822
L+ R L+K L+ V +EV + + + N N + F++++ HN
Sbjct: 1323 DALMMVPDLREALIKALQAPEV-YEVDMAKHR-LYDNPLFVNEITFADEDNIIKGGDHNR 1380
Query: 823 ALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVKAFDGSRKNVL 882
L+I N S L +L+D GS++N++P +L + + L ++ FD K L
Sbjct: 1381 PLYIEGNIGSAHLRRILIDLGSAVNILPVRSLTRAGFTTKDLEPIDVVICGFDNQGKPTL 1440
Query: 883 GEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNGKLVT 942
G I + + + +F + F V++ N SYS LLGRPWIH V STLH+ LKF+
Sbjct: 1441 GAITIKIQMSTFSFKVRFFVIEANTSYSALLGRPWIHKYRVVPSTLHQCLKFLDG----- 1495
Query: 943 VHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTELKKTGTA 986
+G + + S S ++ E+ A+ + + +L +T A
Sbjct: 1496 -NGVQQRITSNFSPYTIQESYHADAKYYFPVEENKQQLGRTTPA 1538
>UniRef100_Q949K7 Putative gag polyprotein [Cicer arietinum]
Length = 88
Score = 99.0 bits (245), Expect = 2e-18
Identities = 41/88 (46%), Positives = 61/88 (68%)
Query: 165 LCLVPKVDVPKKFKIPEFDRYNGLTCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDA 224
+CLVP V +P+KFK+ EF++Y G TCP+NH+ Y RKM +++ ND L+IH FQDSL +
Sbjct: 1 MCLVPDVTIPRKFKVLEFEKYKGATCPKNHLTMYGRKMASHAHNDKLLIHFFQDSLSGAS 60
Query: 225 AEWYTSLSKDDVHTFDELAAAFKSHYGF 252
WY L ++ +H++ +LA AF Y +
Sbjct: 61 LSWYMHLERNRIHSWKDLADAFLRQYKY 88
>UniRef100_Q5XWL3 Gag-pol polyprotein-like [Solanum tuberosum]
Length = 1716
Score = 97.4 bits (241), Expect = 5e-18
Identities = 75/282 (26%), Positives = 128/282 (44%), Gaps = 20/282 (7%)
Query: 585 HNTLGCYVVQNEIQSLMNDNYLTVSDVCVIVPVFHDPPVKSIPSKENVEPLVIRLPGPVP 644
H+T C ++++IQ L+N N +++ V P V S P + + +
Sbjct: 473 HDTEDCINLKHKIQDLINQNVVSLQTVA--------PNVNSNPLPNHGGITINMIETDED 524
Query: 645 YTLEKAIPYKYNATMIENGVEVPLASLAM------VSNIAEGTTAAL-RSGRVRPPLFQK 697
+ + KAI + + +E +ASL++ V E A + R VRP +
Sbjct: 525 WCVMKAI-----VLIAPDKLERAVASLSIREKKEFVILTPEKVVALVPRETLVRPKFVIE 579
Query: 698 KADTPTTPPIDKATLTDVSPVTKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPS 757
T + + SI + +E R ++ DY IV L +TP+
Sbjct: 580 TTATQGMTRSGRCYTPEELAQGGQKKEKKSSINKAKAEEFWRRMQPKDYSIVKYLEKTPA 639
Query: 758 KISVLSLLLSSEAHRSTLLKVLEQAYVDHEVTVDRFGGIVGNITACNNLWFSEDEMPETR 817
+ISV +LL+SS+ HR L+K L+ YV D ++ + + + F ++E+P
Sbjct: 640 QISVWALLMSSQLHRQALMKALDDTYVPVGTNSDNLAAMINQVIRGHRISFCDEELPFEG 699
Query: 818 KYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSY 859
+ HN ALH++ + +++ VLVD GS LN+ P STL L +
Sbjct: 700 RMHNKALHVTTMCRDKIINRVLVDDGSGLNICPLSTLRHLKF 741
Score = 91.3 bits (225), Expect = 3e-16
Identities = 79/294 (26%), Positives = 124/294 (41%), Gaps = 72/294 (24%)
Query: 258 PNREFLISLSQKKDESFREYAQIWRGAAARITPALDEEEMTQTFLKTLKKDYVERMIIAA 317
P+R L + QK ES+RE+A WR AAR+ P + E+E+ + F++ K
Sbjct: 299 PDRYSLEKMKQKSTESYREFAYRWRKEAARVRPPMSEKEIVEVFVRVAK----------- 347
Query: 318 PNNFFEMVTMGTRLEEAVREGIIVFEKV--ESSVNASKRYGNDHHKKKETEVGMVSAGAG 375
F E+V +G +E+ +R G IV+ ESS+ KKK +V +S G
Sbjct: 348 ---FAEIVKIGETIEDGLRTGKIVYVAASPESSILL---------KKKRDDVSSISY-EG 394
Query: 376 QSMATVAPINAAQLPPSYPYAPYSQHPFFPPFYHQYPLPSGQPQVPVNAVVQQMQQQPPV 435
+ A + + PS+ +++P
Sbjct: 395 KKTAKKSSSCQGRSRPSH---------------------------------SSFEKKP-- 419
Query: 436 QQQQHQQARPTFPPIPMLYAELLPTLLLRGHCTTRHGKPPPDPLLLRFRSDLKCDFHQGA 495
AR F P+ +L L + G+ G P D +R D +C +H +
Sbjct: 420 -------AR-NFTPLIKSRTKLFERLTVAGYIHPV-GPKPVDTSSKFYRPDQRCAYHSNS 470
Query: 496 LGHDVEGCYTLKYIVKKLIDQGKLTFENNVPYVLDNPLPNHA--AVNMIEVYEE 547
+GHD E C LK+ ++ LI+Q ++ + P V NPLPNH +NMIE E+
Sbjct: 471 VGHDTEDCINLKHKIQDLINQNVVSLQTVAPNVNSNPLPNHGGITINMIETDED 524
>UniRef100_Q6L420 Putative polyprotein [Solanum demissum]
Length = 1483
Score = 86.7 bits (213), Expect = 8e-15
Identities = 59/163 (36%), Positives = 86/163 (52%), Gaps = 17/163 (10%)
Query: 879 KNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKLKFVKNG 938
++ +GEI+L + IG F + FQV++INASY+ LLGRPW+H AGAV STLH+ +KF +
Sbjct: 213 EDAIGEIELILKIGPVDFTVNFQVLNINASYNLLLGRPWVHRAGAVPSTLHQMVKFEYDR 272
Query: 939 KLVTVHGEEAYLVSQLSSFSCIEAGSA-EGTAFQGL-------TIEGTELKKTGTAMASL 990
+ V VH E V + SS I+A + E +Q +EG + K MAS+
Sbjct: 273 QEVIVHCERDLSVYKDSSLPFIKANNENEALVYQAFEVVVVEHNLEGNLISKPQLPMASV 332
Query: 991 KDAQKAVQEGQAAGWGKLIQL---------RENKHKEGLGFSP 1024
+ ++ G G G I L R++ GLG+ P
Sbjct: 333 MMVNEMLKHGFKLGKGLGIFLQGRAYPVSPRKSLGTFGLGYKP 375
Score = 84.7 bits (208), Expect = 3e-14
Identities = 62/174 (35%), Positives = 92/174 (52%), Gaps = 28/174 (16%)
Query: 642 PVPYTLEKAIPYKYNATMIE-NGVEVPLASLAMVSNIAEGTTAALRSGRVRPPLFQKKAD 700
P+P T KA+P+KY T++ G E+ V I EG T RS R P +K +
Sbjct: 65 PLPMTNPKAVPWKYEPTVVTYKGKEIN----EKVDEI-EGMT---RSRRCYAPAELRKNN 116
Query: 701 TPTTPPIDKATLTDVSPVTKDVSRPSQSIEDSNLDEILRIIKRSDYKIVDQLLQTPSKIS 760
+ SPVT + +E LR +K SDY IV+QL +TP++IS
Sbjct: 117 NDQ--------MQVKSPVT-----------EREAEEFLRKMKLSDYSIVEQLRKTPAQIS 157
Query: 761 VLSLLLSSEAHRSTLLKVLEQAYVDHEVTVDRFGGIVGNITACNNLWFSEDEMP 814
+LSLL+ S+ HR ++K+L +A+V +E T+ + I G I N + FS+DE+P
Sbjct: 158 LLSLLIHSDEHRKVVMKILNEAHVPNEGTMSQLEKIAGRIFEVNCITFSDDELP 211
>UniRef100_Q6ASW9 Reverse transcriptase (RNA-dependent DNA polymerase) domain
containing protein [Oryza sativa]
Length = 1372
Score = 82.8 bits (203), Expect = 1e-13
Identities = 50/154 (32%), Positives = 82/154 (52%), Gaps = 2/154 (1%)
Query: 804 NNLWFSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETP 863
N F + E E R L ++ VN K +S ++VD G+++N+MP +T +L
Sbjct: 419 NKAIFEKPEGTENRHLKPLYINGYVNGKP--MSKMMVDGGAAVNLMPYATFRKLGRNVED 476
Query: 864 LRRSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGA 923
L ++ ++K F G+ G +++ +T+G++T TF V+D SYS LLGR WIH
Sbjct: 477 LIKTNMVLKDFGGNPSETKGVLNVELTVGNKTIPTTFFVIDGKGSYSLLLGRDWIHANCC 536
Query: 924 VTSTLHKKLKFVKNGKLVTVHGEEAYLVSQLSSF 957
+ ST+H+ L + K+ V + A L+ L F
Sbjct: 537 IPSTMHQCLIQWQGDKIQIVPADRAKLIELLKEF 570
>UniRef100_Q94LN5 Putative retroelement pol polyprotein [Oryza sativa]
Length = 1580
Score = 79.0 bits (193), Expect = 2e-12
Identities = 59/206 (28%), Positives = 101/206 (48%), Gaps = 8/206 (3%)
Query: 808 FSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRS 867
F + E E R L ++ VN K +S ++VD G+++N+MP +T +L L ++
Sbjct: 350 FEKPEGTENRHLKPLYINGYVNGKP--MSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKT 407
Query: 868 TFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 927
++K F G+ +G +++ +T+G +T TF V+D SYS LLGR WIH + ST
Sbjct: 408 NMVLKDFGGNPSETMGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPST 467
Query: 928 LHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGTAFQGLTIEGTELKKTGTAM 987
+H+ L + K+ V + + S C E + T++ + K G +
Sbjct: 468 IHQCLIQWQGDKIEIVPADSQLKMENPS--YCFEGVMEGSNVYTKDTVDDLD-DKQGQGL 524
Query: 988 ASLKDAQKAVQEGQAAGWGK--LIQL 1011
S D ++ + A G G+ LI+L
Sbjct: 525 MSADDLEE-IDISPAIGQGRHLLIEL 549
>UniRef100_Q7X6I4 OSJNBa0039G19.2 protein [Oryza sativa]
Length = 1136
Score = 78.2 bits (191), Expect = 3e-12
Identities = 46/142 (32%), Positives = 79/142 (55%), Gaps = 1/142 (0%)
Query: 812 EMPE-TRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFL 870
E PE T+ H L+I+ +S ++VD G+++N+MP +T +L L +++ +
Sbjct: 364 EKPEGTKNRHLKPLYINGYVNEKPMSKMMVDGGAAVNLMPYATFRKLGRNTEDLIKTSMV 423
Query: 871 VKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHK 930
+K F G+ G ++L +T+G +T TF V+D SYS LLGR WIH + ST+H+
Sbjct: 424 LKDFGGNPSETKGVLNLELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPSTMHQ 483
Query: 931 KLKFVKNGKLVTVHGEEAYLVS 952
L + K+ V +++ V+
Sbjct: 484 CLIQWQGNKIEIVLADKSVNVA 505
>UniRef100_Q6AVG0 Putative retrotransposon gag protein [Oryza sativa]
Length = 1251
Score = 77.0 bits (188), Expect = 6e-12
Identities = 54/163 (33%), Positives = 88/163 (53%), Gaps = 8/163 (4%)
Query: 809 SEDEMPE-TRKYHNLALHIS--VNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLR 865
+E E PE T H L+I+ VN KS +S ++VD G+++N+MP +T +L L
Sbjct: 685 AEFEKPEGTGNRHLKPLYINGYVNGKS--MSKMMVDGGAAVNLMPYATFKKLGRNAEDLI 742
Query: 866 RSTFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVT 925
++ ++K F G+ G +++ +T+G +T TF V+D SYS LLGR WIH +
Sbjct: 743 KTNMVLKDFGGNPSETRGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIP 802
Query: 926 STLHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT 968
ST+H+ L + K+ V + S++ + S G EG+
Sbjct: 803 STMHQCLIQWQGDKIEIVPADSQ---SKMENPSYYFEGVVEGS 842
Score = 54.7 bits (130), Expect = 3e-05
Identities = 31/115 (26%), Positives = 61/115 (52%), Gaps = 3/115 (2%)
Query: 170 KVDVPKKFKIPEFDRYNGL--TCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEW 227
+V +P +FK+P+F +++G HI +++ + G S D+L F+ SL A W
Sbjct: 235 RVPLPNRFKVPDFSKFSGQEGVSTYEHINRFLAQCGEASAVDALRFRLFRLSLSGSAFTW 294
Query: 228 YTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWR 282
++SL ++++ +L F S++ ++ + L ++ Q+ DE EY Q +R
Sbjct: 295 FSSLPCGSINSWADLEKQFHSYF-YSWVHEMKLSDLTAIKQRHDEPVHEYIQRFR 348
>UniRef100_Q8LNE4 Putative retroelement [Oryza sativa]
Length = 1170
Score = 76.6 bits (187), Expect = 8e-12
Identities = 43/125 (34%), Positives = 72/125 (57%), Gaps = 2/125 (1%)
Query: 808 FSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRS 867
F + E E R L ++ VN K+ +S ++VD G+++N+MP +T +L L ++
Sbjct: 436 FEKPEGTENRHLKPLYINGYVNGKT--MSKMMVDGGAAVNLMPYATFRKLGRNTEALIKT 493
Query: 868 TFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 927
++K F G+ G +++ +T+G +T TF V++ N SYS LLGR WIH + ST
Sbjct: 494 NMVLKDFGGNPSETRGVLNVELTVGSKTIPTTFFVINGNGSYSLLLGRDWIHANCCIPST 553
Query: 928 LHKKL 932
+H+ L
Sbjct: 554 MHQCL 558
Score = 53.5 bits (127), Expect = 8e-05
Identities = 30/115 (26%), Positives = 61/115 (52%), Gaps = 3/115 (2%)
Query: 170 KVDVPKKFKIPEFDRYNGL--TCPQNHIIKYVRKMGNYSDNDSLMIHCFQDSLMEDAAEW 227
+V +P +FK+P+F +++G HI +++ + S D+L + F+ SL A W
Sbjct: 210 RVPLPNRFKVPDFSKFSGQEGVSTYEHISRFLAQCHEASAVDALKVRLFRLSLSGSAFTW 269
Query: 228 YTSLSKDDVHTFDELAAAFKSHYGFNTRLKPNREFLISLSQKKDESFREYAQIWR 282
++SL ++++ +L F S++ ++ + L ++ Q+ DE EY Q +R
Sbjct: 270 FSSLPYGSINSWADLEKQFHSYF-YSGVHEMKLSDLTAIKQRHDEPVHEYIQRFR 323
>UniRef100_Q7XFQ0 Putative retroelement [Oryza sativa]
Length = 1025
Score = 76.6 bits (187), Expect = 8e-12
Identities = 51/161 (31%), Positives = 84/161 (51%), Gaps = 5/161 (3%)
Query: 808 FSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRS 867
F + E E R L ++ VN K LS ++VD G+++N+MP +T +L L ++
Sbjct: 107 FEKPEGTENRHLKPLYINGYVNGKP--LSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKT 164
Query: 868 TFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 927
++K F G+ G +++ +T+G +T TF V+D SYS LLGR WIH + ST
Sbjct: 165 NMVLKDFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKGSYSLLLGRDWIHANCCIPST 224
Query: 928 LHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT 968
+H+ L + K+ V + S++ + S G EG+
Sbjct: 225 MHQCLIQWQGDKIEIVPADNQ---SKMENPSYYFEGVVEGS 262
>UniRef100_Q8S6X2 Putative pol polyprotein [Oryza sativa]
Length = 505
Score = 76.6 bits (187), Expect = 8e-12
Identities = 38/121 (31%), Positives = 67/121 (54%)
Query: 812 EMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLV 871
E P+ H L++ + +S +LVD G+++N+MP S +L + L+++ ++
Sbjct: 310 EKPDESNRHMKPLYLKGHIDGKPVSRMLVDRGAAVNLMPYSLFKRLGRGDDELKKTNMIL 369
Query: 872 KAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKK 931
F+G G + +T+G++T TF ++D+ SY+ +LGR WIH V STLH+
Sbjct: 370 NGFNGEPTEAKGIFSVELTMGNKTLPTTFFIVDVQGSYNVILGRCWIHANCCVPSTLHQC 429
Query: 932 L 932
L
Sbjct: 430 L 430
>UniRef100_Q9ZQ67 Putative retroelement pol polyprotein [Arabidopsis thaliana]
Length = 764
Score = 76.3 bits (186), Expect = 1e-11
Identities = 52/195 (26%), Positives = 89/195 (44%), Gaps = 27/195 (13%)
Query: 780 EQAYVDHEVTVDRFGGIVGNITACNN---------------LW------------FSEDE 812
+Q + D V R I+G +TAC + LW F+ ++
Sbjct: 277 DQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEMTPLTFTSED 336
Query: 813 MPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRSTFLVK 872
+ HN L I ++ ++ +L+DTGSS+NV+ K L ++ + ++ S +
Sbjct: 337 LFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLT 396
Query: 873 AFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHKKL 932
FDG+ G I LP+ +G F V+D Y+ +LG PWIHD A+ S+ H+ +
Sbjct: 397 GFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCI 456
Query: 933 KFVKNGKLVTVHGEE 947
K + + T+ G +
Sbjct: 457 KIPTSIGIETIRGNQ 471
>UniRef100_Q5W683 Hypothetical protein OSJNBa0076A09.14 [Oryza sativa]
Length = 1681
Score = 76.3 bits (186), Expect = 1e-11
Identities = 50/161 (31%), Positives = 85/161 (52%), Gaps = 5/161 (3%)
Query: 808 FSEDEMPETRKYHNLALHISVNWKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETPLRRS 867
F + E E R + L ++ VN K +S ++VD G+++N+MP +T +L L ++
Sbjct: 742 FEKPEGMENRHHKPLYVNGYVNGKP--MSKMMVDGGAAVNLMPYATFRKLGRNAEDLIKT 799
Query: 868 TFLVKAFDGSRKNVLGEIDLPMTIGHETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 927
++K F G+ G +++ +T+G +T TF V+D SYS LLGR WIH + ST
Sbjct: 800 NMVLKHFGGNPSETKGVLNVELTVGSKTIPTTFFVIDGKRSYSLLLGRDWIHTNCCIPST 859
Query: 928 LHKKLKFVKNGKLVTVHGEEAYLVSQLSSFSCIEAGSAEGT 968
+H+ L + K+ V + S++ + S G EG+
Sbjct: 860 MHQCLIQWQGHKIEVVPADSQ---SKMENPSYYFEGVVEGS 897
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.338 0.146 0.475
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,484,887,539
Number of Sequences: 2790947
Number of extensions: 139579480
Number of successful extensions: 609119
Number of sequences better than 10.0: 961
Number of HSP's better than 10.0 without gapping: 165
Number of HSP's successfully gapped in prelim test: 837
Number of HSP's that attempted gapping in prelim test: 601633
Number of HSP's gapped (non-prelim): 4404
length of query: 2330
length of database: 848,049,833
effective HSP length: 144
effective length of query: 2186
effective length of database: 446,153,465
effective search space: 975291474490
effective search space used: 975291474490
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 83 (36.6 bits)
Medicago: description of AC138465.7


