

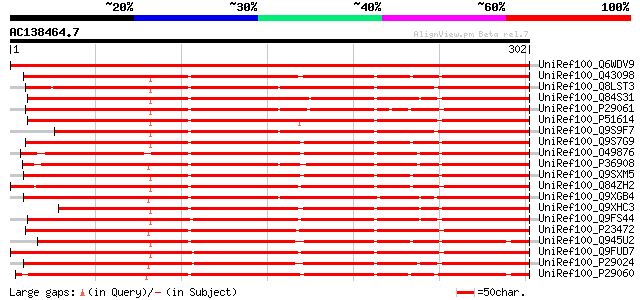
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.7 - phase: 0
(302 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6WDV9 Chitinase class III-1 [Medicago truncatula] 620 e-176
UniRef100_Q43098 Chitinase [Psophocarpus tetragonolobus] 332 7e-90
UniRef100_Q8LST3 Chitinase [Phytolacca americana] 329 5e-89
UniRef100_Q84S31 Chitinase III [Vitis vinifera] 328 1e-88
UniRef100_P29061 Basic endochitinase precursor [Nicotiana tabacum] 327 3e-88
UniRef100_P51614 Acidic endochitinase precursor [Vitis vinifera] 325 7e-88
UniRef100_Q9S9F7 Chitinase-B, PLC-B [Phytolacca americana] 323 3e-87
UniRef100_Q9S7G9 Acidic chitinase [Glycine max] 320 2e-86
UniRef100_O49876 Class III chitinase precursor [Lupinus albus] 319 5e-86
UniRef100_P36908 Acidic endochitinase precursor [Cicer arietinum] 316 4e-85
UniRef100_Q9SXM5 Acidic chitinase [Glycine max] 316 5e-85
UniRef100_Q84ZH2 Putative class III acidic chitinase [Oryza sativa] 314 2e-84
UniRef100_Q9XGB4 Chitinase [Trifolium repens] 309 5e-83
UniRef100_Q9XHC3 Class III chitinase [Sphenostylis stenocarpa] 307 3e-82
UniRef100_Q9FS44 Chitinase precursor [Vitis vinifera] 306 3e-82
UniRef100_P23472 Hevamine A precursor [Includes: Chitinase (EC 3... 306 3e-82
UniRef100_Q945U2 Chitinase 3-like protein [Trichosanthes kirilowii] 304 2e-81
UniRef100_Q9FUD7 Class III acidic chitinase [Malus domestica] 302 6e-81
UniRef100_P29024 Acidic endochitinase precursor [Phaseolus angul... 301 1e-80
UniRef100_P29060 Acidic endochitinase precursor [Nicotiana tabacum] 300 2e-80
>UniRef100_Q6WDV9 Chitinase class III-1 [Medicago truncatula]
Length = 302
Score = 620 bits (1598), Expect = e-176
Identities = 300/302 (99%), Positives = 301/302 (99%)
Query: 1 MSNKYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLS 60
MSNKYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDG+LRDTCNTGLFNIVNIAFLS
Sbjct: 1 MSNKYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGALRDTCNTGLFNIVNIAFLS 60
Query: 61 TFGSGRQPQLNLAGHCNPPNCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQ 120
TFGSGRQPQLNLAGHCNPPNCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQ
Sbjct: 61 TFGSGRQPQLNLAGHCNPPNCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQ 120
Query: 121 LADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLT 180
LADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLT
Sbjct: 121 LADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLT 180
Query: 181 AAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKK 240
AAPLCIFQDNILQ AINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKK
Sbjct: 181 AAPLCIFQDNILQKAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKK 240
Query: 241 VFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKA 300
VFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKA
Sbjct: 241 VFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKA 300
Query: 301 SV 302
SV
Sbjct: 301 SV 302
>UniRef100_Q43098 Chitinase [Psophocarpus tetragonolobus]
Length = 298
Score = 332 bits (851), Expect = 7e-90
Identities = 169/296 (57%), Positives = 210/296 (70%), Gaps = 8/296 (2%)
Query: 9 SLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQP 68
SL+L I + F +NA + +YWGQ+ G+GSL DTCNTG + VNIAFLSTFGSG+ P
Sbjct: 8 SLVLFPILVLSLFNHSNAAGIAVYWGQNGGEGSLADTCNTGNYEFVNIAFLSTFGSGQTP 67
Query: 69 QLNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIW 126
QLNLAGHC+P + C I CQ+RGIKV+LS+GG TYS +S +DA QLA+Y+W
Sbjct: 68 QLNLAGHCDPSSNGCTGFSSEIQTCQNRGIKVLLSLGG-SAGTYSLNSADDATQLANYLW 126
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
+NFLGG S SRP GDA+LDGVDFDIE G HY+ LA LN SS+KK YL+AAP CI
Sbjct: 127 DNFLGGQSGSRPLGDAVLDGVDFDIESGGSNHYDDLARALNS--LSSQKKVYLSAAPQCI 184
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
D L AI TGLFDYVWVQFYN+P +C + + T+ NSW+QWI ++ VF+GLP
Sbjct: 185 IPDQHLDAAIQTGLFDYVWVQFYNNP-SCQYSNGGTTNLINSWNQWI-TVPASLVFMGLP 242
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
AS + AAPSGGFV L +Q+LP++K S KYGGVMLW+R DV +GYS+ I S+
Sbjct: 243 ASDA-AAPSGGFVSTDVLTSQVLPVIKQSSKYGGVMLWDRFNDVQTGYSAAIIGSI 297
>UniRef100_Q8LST3 Chitinase [Phytolacca americana]
Length = 301
Score = 329 bits (844), Expect = 5e-89
Identities = 170/296 (57%), Positives = 219/296 (73%), Gaps = 9/296 (3%)
Query: 10 LLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQ 69
L+L S+ L L ++S+ AG + IYWGQ+ G+G+LRDTCN+GL++ VNI FLSTFG+G+ PQ
Sbjct: 12 LVLTSLLLALPWKSS-AGGIAIYWGQNGGEGTLRDTCNSGLYSYVNIGFLSTFGNGQTPQ 70
Query: 70 LNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWN 127
LNLAGHC+P + CK L +SI CQS+GIKVMLSIGG +YS +S ++ +A+Y+W+
Sbjct: 71 LNLAGHCDPSSGGCKQLSNSIKQCQSQGIKVMLSIGG-GGGSYSIASADEGRNVANYLWD 129
Query: 128 NFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIF 187
NFLGG S +RP GDAILDG+DFDIE G+ +Y TLA L+ H + S +K YLTAAP C F
Sbjct: 130 NFLGGQSSNRPLGDAILDGIDFDIEQGTD-NYVTLAKTLSQHGQQSGRKVYLTAAPQCPF 188
Query: 188 QDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPA 247
D+ L + TGLFD+VWVQFYN+P CNF + NP FK++W+QW + + +K FVGLPA
Sbjct: 189 PDHWLNKGLKTGLFDFVWVQFYNNP-QCNFDAGNPQGFKDAWNQWTSQIPAQKFFVGLPA 247
Query: 248 SSSNAAPSGGFVEAQDLINQLLPIVKPS-PKYGGVMLWNRRFDVTSGYSSKIKASV 302
S AA GFV +Q LINQ+LP VK S KYGGVMLW+R D SGYS++IK SV
Sbjct: 248 --SRAAAGNGFVPSQTLINQVLPFVKGSGQKYGGVMLWDRFNDKNSGYSTRIKGSV 301
>UniRef100_Q84S31 Chitinase III [Vitis vinifera]
Length = 297
Score = 328 bits (841), Expect = 1e-88
Identities = 164/294 (55%), Positives = 215/294 (72%), Gaps = 8/294 (2%)
Query: 11 LLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQL 70
LL+S+S+ Q++ AG + IYWGQ+ +G+L TCNTG ++ VNIAFL+ FG+G+ P++
Sbjct: 10 LLISLSVLALLQTSYAGGIAIYWGQNGNEGTLTQTCNTGKYSYVNIAFLNKFGNGQTPEI 69
Query: 71 NLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNN 128
NLAGHCNP + C ++ I CQ+RGIKVMLSIGG +YS SS DA +A+Y+WNN
Sbjct: 70 NLAGHCNPASNGCTSVSTGIRNCQNRGIKVMLSIGG-GVGSYSLSSSNDAQNVANYLWNN 128
Query: 129 FLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQ 188
FLGG S SRP GDA+LDG+DFDIE GS LH++ LA L+ K R K YLTAAP C F
Sbjct: 129 FLGGQSSSRPLGDAVLDGIDFDIELGSTLHWDDLARALSGFSKRGR-KVYLTAAPQCPFP 187
Query: 189 DNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPAS 248
D L TA+NTGLFDYVWVQFYN+P C + S N + NSW++W +S+++ ++F+GLPAS
Sbjct: 188 DKFLGTALNTGLFDYVWVQFYNNP-QCQYSSGNTNNLLNSWNRWTSSINS-QIFMGLPAS 245
Query: 249 SSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
S AA GF+ A L +Q+LP++K SPKYGGVMLW++ +D SGYSS IK+SV
Sbjct: 246 S--AAAGSGFIPANVLTSQILPVIKRSPKYGGVMLWSKYYDDQSGYSSSIKSSV 297
>UniRef100_P29061 Basic endochitinase precursor [Nicotiana tabacum]
Length = 294
Score = 327 bits (837), Expect = 3e-88
Identities = 168/295 (56%), Positives = 213/295 (71%), Gaps = 10/295 (3%)
Query: 10 LLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQ 69
L +L I L L AGD+V+YWGQD G+G L DTCN+GL+NIVNIAFLS+FG+ + P+
Sbjct: 8 LFILPIFLLLLTSKVKAGDIVVYWGQDVGEGKLIDTCNSGLYNIVNIAFLSSFGNFQTPK 67
Query: 70 LNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWN 127
LNLAGHC P + C+ L SI CQS GIK+MLSIGG TY+ SS +DA Q+ADY+WN
Sbjct: 68 LNLAGHCEPSSGGCQQLTKSIRHCQSIGIKIMLSIGG-GTPTYTLSSVDDARQVADYLWN 126
Query: 128 NFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIF 187
NFLGG S RP GDA+LDG+DFDIE G HY LA +L++H + KK YLTAAP C F
Sbjct: 127 NFLGGQSSFRPLGDAVLDGIDFDIELGQP-HYIALARRLSEHGQQG-KKLYLTAAPQCPF 184
Query: 188 QDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPA 247
D +L A+ TGLFDYVWVQFYN+P C F+SN+ +FK W+QW S+ KK+++GLPA
Sbjct: 185 PDKLLNGALQTGLFDYVWVQFYNNP-ECEFMSNS-ENFKRRWNQW-TSIPAKKLYIGLPA 241
Query: 248 SSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
+ + A G++ Q L++Q+LP +K S KYGGVMLWNR+FDV GYSS I+ +V
Sbjct: 242 AKT--AAGNGYIPKQVLMSQVLPFLKGSSKYGGVMLWNRKFDVQCGYSSAIRGAV 294
>UniRef100_P51614 Acidic endochitinase precursor [Vitis vinifera]
Length = 301
Score = 325 bits (834), Expect = 7e-88
Identities = 161/296 (54%), Positives = 214/296 (71%), Gaps = 8/296 (2%)
Query: 11 LLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQL 70
LL+S+S+ Q++ AG + IYWGQ+ +G+L TCNTG ++ VNIAFL+ FG+G+ P++
Sbjct: 10 LLISLSVLALLQTSYAGGIAIYWGQNGNEGTLTQTCNTGKYSYVNIAFLNKFGNGQTPEI 69
Query: 71 NLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNN 128
NLAGHCNP + C ++ I CQ+RGIKVMLSIGG +YS SS DA +A+Y+WNN
Sbjct: 70 NLAGHCNPASNGCTSVSTGIRNCQNRGIKVMLSIGG-GAGSYSLSSSNDAQNVANYLWNN 128
Query: 129 FLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLN--DHYKSSRKKFYLTAAPLCI 186
FLGG S SRP GDA+LDG+DFDIE GS LH++ LA L+ + + +K YLTAAP C
Sbjct: 129 FLGGQSSSRPLGDAVLDGIDFDIELGSTLHWDDLARALSRIEFQQERGRKVYLTAAPQCP 188
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
F D + TA+NTGLFDYVWVQFYN+P C + S N + NSW++W +S+++ F+GLP
Sbjct: 189 FPDKVPGTALNTGLFDYVWVQFYNNP-PCQYSSGNTNNLLNSWNRWTSSINSTGSFMGLP 247
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
ASS AA GF+ A L +Q+LP++K SPKYGGVMLW++ +D SGYSS IK+SV
Sbjct: 248 ASS--AAAGRGFIPANVLTSQILPVIKRSPKYGGVMLWSKYYDDQSGYSSSIKSSV 301
>UniRef100_Q9S9F7 Chitinase-B, PLC-B [Phytolacca americana]
Length = 274
Score = 323 bits (829), Expect = 3e-87
Identities = 163/279 (58%), Positives = 207/279 (73%), Gaps = 8/279 (2%)
Query: 27 GDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQLNLAGHCNPPN--CKNL 84
G + IYWGQ+ G+G+LRDTCN+GL++ VNI FLSTFG+G+ PQLNLAGHC+P + CK L
Sbjct: 1 GGIAIYWGQNGGEGTLRDTCNSGLYSYVNIGFLSTFGNGQTPQLNLAGHCDPSSGGCKQL 60
Query: 85 RDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNNFLGGNSPSRPFGDAIL 144
+SI CQS+GIKVMLSIGG +YS +S ++ +A+Y+W+NFLGG S +RP GDAIL
Sbjct: 61 SNSIKQCQSQGIKVMLSIGG-GGGSYSIASADEGRNVANYLWDNFLGGQSSNRPLGDAIL 119
Query: 145 DGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQDNILQTAINTGLFDYV 204
DG+DFDIE G+ +Y TLA L+ H + S +K YLTAAP C F D+ L + TGLFD+V
Sbjct: 120 DGIDFDIEQGTD-NYVTLAKTLSQHGQQSGRKVYLTAAPQCPFPDHWLNKGLKTGLFDFV 178
Query: 205 WVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPASSSNAAPSGGFVEAQDL 264
WVQFYN+P CNF + NP FK++W+QW + + +K FVGLPA S AA GFV +Q L
Sbjct: 179 WVQFYNNP-QCNFDAGNPQGFKDAWNQWTSQIPAQKFFVGLPA--SRAAAGNGFVPSQTL 235
Query: 265 INQLLPIVKPS-PKYGGVMLWNRRFDVTSGYSSKIKASV 302
INQ+LP VK S KYGGVMLW+R D SGYS++IK SV
Sbjct: 236 INQVLPFVKGSGQKYGGVMLWDRFNDKNSGYSTRIKGSV 274
>UniRef100_Q9S7G9 Acidic chitinase [Glycine max]
Length = 299
Score = 320 bits (821), Expect = 2e-86
Identities = 165/295 (55%), Positives = 205/295 (68%), Gaps = 8/295 (2%)
Query: 10 LLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQ 69
LL +SL+LF ++A + +YWGQ+ G+G+L + CNTG + VNIAFLSTFG+G+ PQ
Sbjct: 10 LLFPLLSLSLFINHSHAAGIAVYWGQNGGEGTLAEACNTGNYQYVNIAFLSTFGNGQTPQ 69
Query: 70 LNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWN 127
LNLAGHC+P N C L IN CQ GIKV+LS+GG +YS SS +DA QLA+Y+W
Sbjct: 70 LNLAGHCDPNNNGCTGLSSDINTCQDLGIKVLLSLGG-GAGSYSLSSADDATQLANYLWE 128
Query: 128 NFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIF 187
NFLGG + S P GD ILDG+DFDIE G HY+ LA LN SS+ K YL+AAP CI
Sbjct: 129 NFLGGQTGSGPLGDVILDGIDFDIESGGSDHYDDLARALNSF--SSQSKVYLSAAPQCII 186
Query: 188 QDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPA 247
D L AI TGLFDYVWVQFYN+P +C + S N NSW+QWI ++ VF+GLPA
Sbjct: 187 PDAHLDAAIQTGLFDYVWVQFYNNP-SCQYSSGNTNDLINSWNQWI-TVPASLVFMGLPA 244
Query: 248 SSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
S + AAPSGGFV A L +Q+LP++K S YGGVMLW+R DV +GYS+ I SV
Sbjct: 245 SEA-AAPSGGFVPADVLTSQILPVIKQSSNYGGVMLWDRFNDVQNGYSNAIIGSV 298
>UniRef100_O49876 Class III chitinase precursor [Lupinus albus]
Length = 293
Score = 319 bits (818), Expect = 5e-86
Identities = 168/296 (56%), Positives = 206/296 (68%), Gaps = 14/296 (4%)
Query: 7 YPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGR 66
+P LLL+S S F+ +NA +VIYWGQ+ +GSL D CNT + VNIAFLSTFG+G+
Sbjct: 12 FPLLLLISSS----FKLSNAAGIVIYWGQNGNEGSLADACNTNNYQYVNIAFLSTFGNGQ 67
Query: 67 QPQLNLAGHCNPPNCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIW 126
P+LNLAGH L I CQ +GIKV+LS+GG +YS +S +DA LA+Y+W
Sbjct: 68 TPELNLAGHSRD----GLNADIKGCQGKGIKVLLSLGG-GAGSYSLNSADDATNLANYLW 122
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
NNFLGG S SRPFGDA+LDG+DFDIE G HY+ LA LN SS+KK YL AAP C
Sbjct: 123 NNFLGGTSDSRPFGDAVLDGIDFDIEAGGAQHYDELARALNGF--SSQKKVYLGAAPQCP 180
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
D L AINTGLFDYVWVQFYN+P C + N + NSW+QW +S + K+VF+GLP
Sbjct: 181 IPDAHLDAAINTGLFDYVWVQFYNNP-QCQYACGNTNNLINSWNQWTSSQA-KQVFLGLP 238
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
AS + AAPSGGF+ LI+Q+LP +K SPKYGGVMLWN D+ +GYS IKASV
Sbjct: 239 ASEA-AAPSGGFIPTDVLISQVLPTIKTSPKYGGVMLWNGFNDIQTGYSDAIKASV 293
>UniRef100_P36908 Acidic endochitinase precursor [Cicer arietinum]
Length = 293
Score = 316 bits (810), Expect = 4e-85
Identities = 161/297 (54%), Positives = 211/297 (70%), Gaps = 14/297 (4%)
Query: 8 PSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQ 67
PSLLL+S L +S+NA + +YWGQ+ +GSL+D CNT + VNIAFLSTFG+G+
Sbjct: 9 PSLLLIS----LLIKSSNAAGIAVYWGQNGNEGSLQDACNTNNYQFVNIAFLSTFGNGQN 64
Query: 68 PQLNLAGHCNPP--NCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYI 125
PQ+NLAGHC+P C I CQ++GIKV+LS+GG +YS +S E+A LA+Y+
Sbjct: 65 PQINLAGHCDPSTNGCTKFSPEIQACQAKGIKVLLSLGG-GAGSYSLNSAEEATTLANYL 123
Query: 126 WNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLC 185
WNNFLGG S SRP GDA+LDG+DFDIE G + HY+ LA LN S++K YL+AAP C
Sbjct: 124 WNNFLGGTSTSRPLGDAVLDGIDFDIESGGQ-HYDELAKALNGF---SQQKVYLSAAPQC 179
Query: 186 IFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGL 245
+ D L +AI TGLFDYVWVQFYN+P C + + N + N+W+QW +S + K+VF+G+
Sbjct: 180 PYPDAHLDSAIQTGLFDYVWVQFYNNP-QCQYSNGNINNLVNAWNQWTSSQA-KQVFLGV 237
Query: 246 PASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
PAS + AAPSGG + A L +Q+LP +K SPKYGGVM+W+R D SGYS+ IK SV
Sbjct: 238 PASDA-AAPSGGLIPADVLTSQVLPAIKTSPKYGGVMIWDRFNDAQSGYSNAIKGSV 293
>UniRef100_Q9SXM5 Acidic chitinase [Glycine max]
Length = 298
Score = 316 bits (809), Expect = 5e-85
Identities = 162/296 (54%), Positives = 206/296 (68%), Gaps = 8/296 (2%)
Query: 9 SLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQP 68
SL+L + F+ ++A + IYWGQ+ G+G+L + CNT + VNIAFLSTFG+G+ P
Sbjct: 8 SLILFPLLFLSLFKHSHAAGIAIYWGQNGGEGTLAEACNTRNYQYVNIAFLSTFGNGQTP 67
Query: 69 QLNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIW 126
QLNLAGHC+P N C L I CQ GIKV+LS+GG +YS SS +DA QLA+Y+W
Sbjct: 68 QLNLAGHCDPNNNGCTGLSSDIKTCQDLGIKVLLSLGG-GAGSYSLSSADDATQLANYLW 126
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
NFLGG + S P G+ ILDG+DFDIE G HY+ LA LN SS++K YL+AAP CI
Sbjct: 127 QNFLGGQTGSGPLGNVILDGIDFDIESGGSDHYDDLARALNSF--SSQRKVYLSAAPQCI 184
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
D L AI TGLFDYVWVQFYN+P +C + S N + NSW+QWI ++ ++F+GLP
Sbjct: 185 IPDAHLDRAIQTGLFDYVWVQFYNNP-SCQYSSGNTNNLINSWNQWI-TVPASQIFMGLP 242
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
AS + AAPSGGFV A L +Q+LP++K S KYGGVMLWNR DV +GYS+ I SV
Sbjct: 243 ASEA-AAPSGGFVPADVLTSQVLPVIKQSSKYGGVMLWNRFNDVQNGYSNAIIGSV 297
>UniRef100_Q84ZH2 Putative class III acidic chitinase [Oryza sativa]
Length = 297
Score = 314 bits (805), Expect = 2e-84
Identities = 157/304 (51%), Positives = 211/304 (68%), Gaps = 9/304 (2%)
Query: 1 MSNKYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLS 60
M+NK S LLL++ ++ F S+ AG + IYWGQ+NG+G+L DTC TG + VNIAFL+
Sbjct: 1 MANKSSLLQLLLIA-AVASQFVSSQAGSIAIYWGQNNGEGTLADTCATGNYKFVNIAFLA 59
Query: 61 TFGSGRQPQLNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDA 118
FG+G+ P NLAGHC+P N C + I CQSRG+K+MLSIGG +Y SS EDA
Sbjct: 60 AFGNGQPPVFNLAGHCDPTNGGCASQSSDIKSCQSRGVKIMLSIGG-GAGSYYLSSSEDA 118
Query: 119 NQLADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFY 178
+A Y+WNNFLGG S SRP GDA+LDG+DFDIEGG+ H++ LA L Y +S ++ Y
Sbjct: 119 KNVATYLWNNFLGGQSSSRPLGDAVLDGIDFDIEGGTNQHWDDLARYLKG-YSNSGRRVY 177
Query: 179 LTAAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMST 238
LTAAP C F D + A+NTGLFDYVWVQFYN+P C + S + ++ ++W QW+ S+
Sbjct: 178 LTAAPQCPFPDACIGDALNTGLFDYVWVQFYNNP-PCQYSSGSTSNLADAWKQWL-SVPA 235
Query: 239 KKVFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKI 298
K++F+GLPAS A GF+ A DL +Q+LP++K S KYGG+MLW++ +D YSS +
Sbjct: 236 KQIFLGLPASPQ--AAGSGFIPADDLKSQVLPVIKSSGKYGGIMLWSKYYDDQDDYSSSV 293
Query: 299 KASV 302
K+ V
Sbjct: 294 KSDV 297
>UniRef100_Q9XGB4 Chitinase [Trifolium repens]
Length = 298
Score = 309 bits (792), Expect = 5e-83
Identities = 155/296 (52%), Positives = 207/296 (69%), Gaps = 7/296 (2%)
Query: 9 SLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQP 68
S+LL + L F+S++A + +YWGQ+ G+GSL D CNT + VNIAFLSTFG+G+ P
Sbjct: 8 SILLFPLFLISLFKSSSAAGIAVYWGQNGGEGSLEDACNTNNYQFVNIAFLSTFGNGQTP 67
Query: 69 QLNLAGHCNPP--NCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIW 126
LNLAGHCNP C I CQ++G+K++LS+GG +YS SS +DA Q+A+Y+W
Sbjct: 68 PLNLAGHCNPAANGCAIFSSQIQACQAKGVKILLSLGG-GAGSYSLSSSDDATQVANYLW 126
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
+NFLGG S SRP GDA+LDG+DFDIE G + H++ LA LN S+K+ YL+AAP C
Sbjct: 127 DNFLGGTSSSRPLGDAVLDGIDFDIEAGGE-HFDELAKALNGFSSQSQKQIYLSAAPQCP 185
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
+ D L +AI TGLFDYV VQFYN+P + + N + N+W+QW +S +T +VF+G+P
Sbjct: 186 YPDAHLDSAIPTGLFDYVGVQFYNNPQR-QYSNGNTANLVNAWNQWTSSQAT-QVFLGVP 243
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
A + AAPSGGF+ + LINQ+LP +K S KYGGVM+W+R D SGYS IK SV
Sbjct: 244 A-NEGAAPSGGFIPSDVLINQVLPAIKSSAKYGGVMIWDRFNDGQSGYSDAIKGSV 298
>UniRef100_Q9XHC3 Class III chitinase [Sphenostylis stenocarpa]
Length = 294
Score = 307 bits (786), Expect = 3e-82
Identities = 156/276 (56%), Positives = 196/276 (70%), Gaps = 8/276 (2%)
Query: 29 LVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQLNLAGHCNPPN--CKNLRD 86
L +YWG++ G+G+L DTCNT F VNIAFLSTFG+G+ PQLNLAGHC+P N C L
Sbjct: 24 LAVYWGENGGEGTLADTCNTQNFQFVNIAFLSTFGNGQTPQLNLAGHCDPLNNGCTGLSS 83
Query: 87 SINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNNFLGGNSPSRPFGDAILDG 146
I CQ+ G+KV+LS+GG +YS +S +A QLA Y+WNNFLG S SRP GDAILDG
Sbjct: 84 DITTCQNGGVKVLLSLGG-SAGSYSLNSASEATQLATYLWNNFLGVQSDSRPLGDAILDG 142
Query: 147 VDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQDNILQTAINTGLFDYVWV 206
+DFDIE G H++ L LND SS++K YL+AAP CI D L +AI TGLFDYVWV
Sbjct: 143 IDFDIESGGGEHWDELVKALND--LSSQRKVYLSAAPQCIIPDQHLNSAIQTGLFDYVWV 200
Query: 207 QFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPASSSNAAPSGGFVEAQDLIN 266
QFYN+P +C + S N NSW+QWI +T +VF+GLPAS + AAPSGGF+ LI+
Sbjct: 201 QFYNNP-SCQYSSGNTNDLINSWNQWITVPAT-QVFMGLPASEA-AAPSGGFIPLDVLIS 257
Query: 267 QLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
Q+LP +K S KYGG+MLW+R D+ + YS+ I SV
Sbjct: 258 QVLPQIKQSSKYGGIMLWDRFNDIQNSYSNAIVGSV 293
>UniRef100_Q9FS44 Chitinase precursor [Vitis vinifera]
Length = 297
Score = 306 bits (785), Expect = 3e-82
Identities = 152/294 (51%), Positives = 211/294 (71%), Gaps = 8/294 (2%)
Query: 11 LLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQL 70
LL+S+S+ F Q+T AG + IYWGQ+ +G+L TCNTG ++ VNIAFL+ FG+G+ P++
Sbjct: 10 LLISLSVLAFLQTTYAGGIAIYWGQNGNEGTLTQTCNTGKYSYVNIAFLNKFGNGQTPEI 69
Query: 71 NLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNN 128
NLAGHC+ + C ++ I+ CQS+G+KVMLSIGG +YS SS +DA +A+Y+WNN
Sbjct: 70 NLAGHCDSASNGCTSVSTDISNCQSQGVKVMLSIGGA-IGSYSLSSSDDAQNVANYLWNN 128
Query: 129 FLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQ 188
FLGG S SRP GDA+LDG+DF I GS +++ LA L+ + +K YLTAAP C F
Sbjct: 129 FLGGRSSSRPLGDAVLDGIDFVILLGSTQYWDDLARALSG-FSQRGRKVYLTAAPQCPFP 187
Query: 189 DNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPAS 248
D + TA+NTG FD VW QFYN+P C + S N T+ NSW++W +S+++ ++F+GLPA+
Sbjct: 188 DKFMGTALNTGRFDNVWAQFYNNP-PCQYTSGNTTNLLNSWNRWTSSINS-RIFLGLPAA 245
Query: 249 SSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
S AA GF+ L +Q+LP++K SPKYGGVMLW++ +D SGYSS IK+SV
Sbjct: 246 S--AAAGSGFIPPNVLTSQILPVIKTSPKYGGVMLWSKYYDDQSGYSSTIKSSV 297
>UniRef100_P23472 Hevamine A precursor [Includes: Chitinase (EC 3.2.1.14); Lysozyme
(EC 3.2.1.17)] [Hevea brasiliensis]
Length = 311
Score = 306 bits (785), Expect = 3e-82
Identities = 155/295 (52%), Positives = 204/295 (68%), Gaps = 7/295 (2%)
Query: 10 LLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQ 69
LLLL+ISL + + G + IYWGQ+ +G+L TC+T ++ VNIAFL+ FG+G+ PQ
Sbjct: 10 LLLLAISLIMSSSHVDGGGIAIYWGQNGNEGTLTQTCSTRKYSYVNIAFLNKFGNGQTPQ 69
Query: 70 LNLAGHCNPP--NCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWN 127
+NLAGHCNP C + + I CQ +GIKVMLS+GG +Y+ +S DA +ADY+WN
Sbjct: 70 INLAGHCNPAAGGCTIVSNGIRSCQIQGIKVMLSLGG-GIGSYTLASQADAKNVADYLWN 128
Query: 128 NFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIF 187
NFLGG S SRP GDA+LDG+DFDIE GS L+++ LA L+ Y KK YLTAAP C F
Sbjct: 129 NFLGGKSSSRPLGDAVLDGIDFDIEHGSTLYWDDLARYLS-AYSKQGKKVYLTAAPQCPF 187
Query: 188 QDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPA 247
D L TA+NTGLFDYVWVQFYN+P C + S N + NSW++W S++ K+F+GLPA
Sbjct: 188 PDRYLGTALNTGLFDYVWVQFYNNP-PCQYSSGNINNIINSWNRWTTSINAGKIFLGLPA 246
Query: 248 SSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
+ A G+V LI+++LP +K SPKYGGVMLW++ +D +GYSS I SV
Sbjct: 247 APE--AAGSGYVPPDVLISRILPEIKKSPKYGGVMLWSKFYDDKNGYSSSILDSV 299
>UniRef100_Q945U2 Chitinase 3-like protein [Trichosanthes kirilowii]
Length = 292
Score = 304 bits (779), Expect = 2e-81
Identities = 161/288 (55%), Positives = 201/288 (68%), Gaps = 13/288 (4%)
Query: 17 LTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQPQLNLAGHC 76
L FQS++A + IYWGQ+ +GSL TC+TG + VNIAFLS+FGSGR P LNLAGHC
Sbjct: 16 LAPIFQSSHAAGIAIYWGQNGNEGSLSFTCSTGNYQFVNIAFLSSFGSGRTPVLNLAGHC 75
Query: 77 NPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIWNNFLGGNS 134
NP N C L I CQSRGIKV+LSIGG +YS SS +DA Q+A++IWNNFLGG S
Sbjct: 76 NPSNNGCAFLSSQIKACQSRGIKVLLSIGG-GAGSYSLSSADDARQVANFIWNNFLGGRS 134
Query: 135 PSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCIFQDNILQT 194
SRP GDA+LDGVDFDIE GS ++TLA +L + L AAP C D L
Sbjct: 135 SSRPLGDAVLDGVDFDIESGSGQFWDTLARQL-----KGLGRVLLAAAPQCPIPDAHLDA 189
Query: 195 AINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLPASSSNAAP 254
AI TGLFD+VWVQFYN+P C F + + NSW++W+ S K+F+GLPA+++ AAP
Sbjct: 190 AIKTGLFDFVWVQFYNNP-PCMFANGKANNLLNSWNRWL-SFPVGKLFMGLPAAAA-AAP 246
Query: 255 SGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
SGGF+ A LI+Q+LP +K SPKYGGVMLW++ FD +GYS+ IK S+
Sbjct: 247 SGGFIPANVLISQVLPKIKTSPKYGGVMLWSKFFD--NGYSNAIKGSL 292
>UniRef100_Q9FUD7 Class III acidic chitinase [Malus domestica]
Length = 299
Score = 302 bits (774), Expect = 6e-81
Identities = 150/304 (49%), Positives = 204/304 (66%), Gaps = 7/304 (2%)
Query: 1 MSNKYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLS 60
M++K + L LLS+ + NAG + IYWGQ+ +G+L +TC +G + VN+AFL+
Sbjct: 1 MASKSTATFLALLSLVTLVLALGANAGGIAIYWGQNGNEGTLAETCASGNYQFVNVAFLT 60
Query: 61 TFGSGRQPQLNLAGHCNPPN--CKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDA 118
TFG+G+ P +NLAGHC+P C L I CQ++GIKV+LSIGG +YS +S +DA
Sbjct: 61 TFGNGQTPAINLAGHCDPTTEECTKLSPEIKSCQAKGIKVILSIGGAS-GSYSLTSADDA 119
Query: 119 NQLADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFY 178
Q+A Y+WNNFLGG S SRP G A+LDG+DFDIEGG+ H++ LA L+ Y KK Y
Sbjct: 120 RQVATYLWNNFLGGQSSSRPLGAAVLDGIDFDIEGGTDQHWDDLARYLSG-YSKRGKKVY 178
Query: 179 LTAAPLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMST 238
LTAAP C F D + A+ TGLFD VWVQFYN+P C + S + T+ +++W QW +++
Sbjct: 179 LTAAPQCPFPDAYVGNALKTGLFDNVWVQFYNNP-PCQYASGDVTNLEDAWKQWTSAIPA 237
Query: 239 KKVFVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKI 298
K+F+GLPA+ A GF+ A DL +Q+LP +K S KYGGVMLW++ +D GYSS I
Sbjct: 238 DKIFLGLPAAPQ--AAGSGFIPATDLSSQVLPAIKSSAKYGGVMLWSKYYDDPDGYSSSI 295
Query: 299 KASV 302
K V
Sbjct: 296 KNDV 299
>UniRef100_P29024 Acidic endochitinase precursor [Phaseolus angularis]
Length = 298
Score = 301 bits (771), Expect = 1e-80
Identities = 160/296 (54%), Positives = 198/296 (66%), Gaps = 11/296 (3%)
Query: 9 SLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFGSGRQP 68
S LLL + FF+ ++AG + +YWGQ+ +GSL D CNTG + VNIAFL TFG G+ P
Sbjct: 12 SALLLPLLFISFFKPSHAGGISVYWGQNGNEGSLADACNTGNYKYVNIAFLFTFGGGQTP 71
Query: 69 QLNLAGHCNPP--NCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQLADYIW 126
QLNLAGHCNP NC D I CQS+ IKV+LS+GG +YS +S +DA Q+A+YIW
Sbjct: 72 QLNLAGHCNPSINNCNVFSDQIKECQSKDIKVLLSLGGAS-GSYSLTSADDATQVANYIW 130
Query: 127 NNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTAAPLCI 186
NNFLGG S SRP GDAILDGVDFDIE G+ H++ LA L K + LTAAP C
Sbjct: 131 NNFLGGQSSSRPLGDAILDGVDFDIESGTGEHWDDLARAL----KGFNSQLLLTAAPQCP 186
Query: 187 FQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKVFVGLP 246
D L TAI TGLFD VWVQFYN+P C + S N +SW+QW +S + K++F+G+P
Sbjct: 187 IPDAHLDTAIKTGLFDIVWVQFYNNP-PCQYSSGNTNDLISSWNQWTSSQA-KQLFLGVP 244
Query: 247 ASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKASV 302
AS+ AA GF+ A L +Q+LP +K S KYGGVMLW+R D SGYS I SV
Sbjct: 245 AST--AAAGSGFIPADVLTSQVLPTIKGSSKYGGVMLWDRFNDGQSGYSGAIIGSV 298
>UniRef100_P29060 Acidic endochitinase precursor [Nicotiana tabacum]
Length = 291
Score = 300 bits (769), Expect = 2e-80
Identities = 155/301 (51%), Positives = 208/301 (68%), Gaps = 14/301 (4%)
Query: 4 KYSYPSLLLLSISLTLFFQSTNAGDLVIYWGQDNGDGSLRDTCNTGLFNIVNIAFLSTFG 63
KYS+ LL ++ L L AGD+VIYWGQ+ +GSL DTC T + IVNIAFL FG
Sbjct: 3 KYSF---LLTALVLFLRALKLEAGDIVIYWGQNGNEGSLADTCATNNYAIVNIAFLVVFG 59
Query: 64 SGRQPQLNLAGHCNP--PNCKNLRDSINICQSRGIKVMLSIGGEDRNTYSFSSPEDANQL 121
+G+ P LNLAGHC+P C L + I CQ++GIKVMLS+GG +Y SS +DA +
Sbjct: 60 NGQNPVLNLAGHCDPNAGACTGLSNDIRACQNQGIKVMLSLGG-GAGSYFLSSADDARNV 118
Query: 122 ADYIWNNFLGGNSPSRPFGDAILDGVDFDIEGGSKLHYETLALKLNDHYKSSRKKFYLTA 181
A+Y+WNN+LGG S +RP GDA+LDG+DFDIEGG+ H++ LA L+ S ++K YLTA
Sbjct: 119 ANYLWNNYLGGQSNTRPLGDAVLDGIDFDIEGGTTQHWDELAKTLSQF--SQQRKVYLTA 176
Query: 182 APLCIFQDNILQTAINTGLFDYVWVQFYNSPGACNFVSNNPTSFKNSWSQWINSMSTKKV 241
AP C F D L A++TGLFDYVWVQFYN+P C + + + KN W+QW N++ K+
Sbjct: 177 APQCPFPDTWLNGALSTGLFDYVWVQFYNNP-PCQYSGGSADNLKNYWNQW-NAIQAGKI 234
Query: 242 FVGLPASSSNAAPSGGFVEAQDLINQLLPIVKPSPKYGGVMLWNRRFDVTSGYSSKIKAS 301
F+GLPA + A GF+ + L++Q+LP++ SPKYGGVMLW++ +D +GYSS IKA+
Sbjct: 235 FLGLPA--AQGAAGSGFIPSDVLVSQVLPLINGSPKYGGVMLWSKFYD--NGYSSAIKAN 290
Query: 302 V 302
V
Sbjct: 291 V 291
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 554,417,520
Number of Sequences: 2790947
Number of extensions: 24405009
Number of successful extensions: 53630
Number of sequences better than 10.0: 202
Number of HSP's better than 10.0 without gapping: 141
Number of HSP's successfully gapped in prelim test: 61
Number of HSP's that attempted gapping in prelim test: 52753
Number of HSP's gapped (non-prelim): 227
length of query: 302
length of database: 848,049,833
effective HSP length: 127
effective length of query: 175
effective length of database: 493,599,564
effective search space: 86379923700
effective search space used: 86379923700
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 74 (33.1 bits)
Medicago: description of AC138464.7


