

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.6 + phase: 0
(250 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
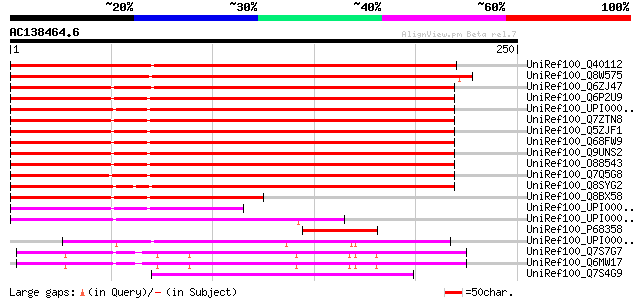
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q40112 Hypothetical protein LC15 [Lycopersicon chilense] 329 4e-89
UniRef100_Q8W575 COP9 signalosome complex subunit 3 [Arabidopsis... 292 5e-78
UniRef100_Q6ZJ47 Putative COP9 complex subunit 3, FUS11 [Oryza s... 288 1e-76
UniRef100_Q6P2U9 COP9 signalosome complex subunit 3 [Brachydanio... 193 3e-48
UniRef100_UPI0000360B09 UPI0000360B09 UniRef100 entry 191 1e-47
UniRef100_Q7ZTN8 COP9 signalosome complex subunit 3 [Xenopus lae... 189 4e-47
UniRef100_Q5ZJF1 Hypothetical protein [Gallus gallus] 188 1e-46
UniRef100_Q68FW9 COP9 constitutive photomorphogenic homolog subu... 187 3e-46
UniRef100_Q9UNS2 COP9 signalosome complex subunit 3 [Homo sapiens] 187 3e-46
UniRef100_O88543 COP9 signalosome complex subunit 3 [Mus musculus] 185 1e-45
UniRef100_Q7Q5G8 ENSANGP00000004286 [Anopheles gambiae str. PEST] 180 3e-44
UniRef100_Q8SYG2 COP9 signalosome complex subunit 3 [Drosophila ... 165 9e-40
UniRef100_Q8BX58 Mus musculus 0 day neonate cerebellum cDNA, RIK... 97 3e-19
UniRef100_UPI000027CFB0 UPI000027CFB0 UniRef100 entry 87 5e-16
UniRef100_UPI00003C1341 UPI00003C1341 UniRef100 entry 74 5e-12
UniRef100_P68358 COP9 signalosome complex subunit 3 [Brassica ol... 73 6e-12
UniRef100_UPI00002198D7 UPI00002198D7 UniRef100 entry 63 6e-09
UniRef100_Q7S7G7 Predicted protein [Neurospora crassa] 63 8e-09
UniRef100_Q6MW17 Related to JAB1-containing signalosome subunit ... 63 8e-09
UniRef100_Q7S4G9 Hypothetical protein [Neurospora crassa] 60 7e-08
>UniRef100_Q40112 Hypothetical protein LC15 [Lycopersicon chilense]
Length = 250
Score = 329 bits (844), Expect = 4e-89
Identities = 164/220 (74%), Positives = 190/220 (85%), Gaps = 1/220 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
M+CIGQK+F+KAL+LLHNVVTAPM+ +NAI +EAYKKYILVSLI GQFSTS PKY S +
Sbjct: 1 MVCIGQKQFRKALELLHNVVTAPMSTLNAIAVEAYKKYILVSLIHLGQFSTSFPKYTSSV 60
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQRNLK F Q P +EL+ +Y G+++E+E FV N +FE+DNNLGL QVVSSMYKRNI
Sbjct: 61 AQRNLKNFSQ-PYLELSNSYGTGRISELETFVQTNKEKFESDNNLGLVMQVVSSMYKRNI 119
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLTQTYLTLSL+DIANTV L K+AEMHVLQMI+DGEIYATINQKDGMVRFLEDPEQY
Sbjct: 120 QRLTQTYLTLSLQDIANTVQLRGPKQAEMHVLQMIEDGEIYATINQKDGMVRFLEDPEQY 179
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKV 220
KTC MIEHIDSSI+R+MA+SKKLT+ DE +SCD +YL KV
Sbjct: 180 KTCGMIEHIDSSIKRLMAVSKKLTSMDELMSCDPMYLGKV 219
>UniRef100_Q8W575 COP9 signalosome complex subunit 3 [Arabidopsis thaliana]
Length = 429
Score = 292 bits (748), Expect = 5e-78
Identities = 156/230 (67%), Positives = 178/230 (76%), Gaps = 3/230 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MICIG KRFQKAL+LL+NVVTAPM +NAI +EAYKKYILVSLI +GQF+ +LPK AS
Sbjct: 189 MICIGLKRFQKALELLYNVVTAPMHQVNAIALEAYKKYILVSLIHNGQFTNTLPKCASTA 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQR+ K + P IEL YN+GK+ E+EA V A FE D NLGL KQ VSS+YKRNI
Sbjct: 249 AQRSFKNYT-GPYIELGNCYNDGKIGELEALVVARNAEFEEDKNLGLVKQAVSSLYKRNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
RLTQ YLTLSL+DIAN V L +AKEAEMHVLQMIQDG+I+A INQKDGMVRFLEDPEQY
Sbjct: 308 LRLTQKYLTLSLQDIANMVQLGNAKEAEMHVLQMIQDGQIHALINQKDGMVRFLEDPEQY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKV--SVQSFSFG 228
K+ EMIE +DS IQR + LSK L A DE +SCD LYL KV Q + FG
Sbjct: 368 KSSEMIEIMDSVIQRTIGLSKNLLAMDESLSCDPLYLGKVGRERQRYDFG 417
>UniRef100_Q6ZJ47 Putative COP9 complex subunit 3, FUS11 [Oryza sativa]
Length = 426
Score = 288 bits (736), Expect = 1e-76
Identities = 148/219 (67%), Positives = 177/219 (80%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI IG K+F ALD LHN VT PM+ +NAI +EAYKKYILVSLI++GQ S PKY S
Sbjct: 188 MIYIGLKKFTIALDFLHNAVTVPMSSLNAIAVEAYKKYILVSLIQNGQVP-SFPKYTSST 246
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
AQRNLK Q ++L+ Y+ G +E+E ++ NA +F++DNNLGL KQV+SSMYKRNI
Sbjct: 247 AQRNLKNHAQ-VYVDLSTCYSKGNYSELEEYIQLNAEKFQSDNNLGLVKQVLSSMYKRNI 305
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLTQTYLTLSLEDIA++V L + KEAEMHVL+MI+DGEI+ATINQKDGMV F EDPEQY
Sbjct: 306 QRLTQTYLTLSLEDIASSVQLKTPKEAEMHVLRMIEDGEIHATINQKDGMVSFHEDPEQY 365
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
K+CEM+EHIDSSIQR+MALSKKL++ DE ISCD YL K
Sbjct: 366 KSCEMVEHIDSSIQRLMALSKKLSSIDENISCDPAYLMK 404
>UniRef100_Q6P2U9 COP9 signalosome complex subunit 3 [Brachydanio rerio]
Length = 423
Score = 193 bits (491), Expect = 3e-48
Identities = 104/219 (47%), Positives = 145/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFFEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y AE+ A VN ++ F DNN GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQIYATNNPAELRALVNKHSETFTRDNNTGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEIYA+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ ID + + + L +KL + D++I+ + ++ K
Sbjct: 368 NNPAMLHKIDQEMLKCIELDEKLKSMDQEITVNPQFVQK 406
>UniRef100_UPI0000360B09 UPI0000360B09 UniRef100 entry
Length = 397
Score = 191 bits (486), Expect = 1e-47
Identities = 103/219 (47%), Positives = 144/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 164 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQP-LPKYTSQI 222
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y AE+ VN ++ F DNN GL KQ +SS+YK+NI
Sbjct: 223 VGRFIKPL-SNAYHELAQVYTTNNPAELRNLVNKHSETFTRDNNTGLVKQCLSSLYKKNI 281
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEIYA+INQKDGMV F ++PE+Y
Sbjct: 282 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIYASINQKDGMVCFHDNPEKY 341
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ ID + + + L +KL + D++I+ + ++ K
Sbjct: 342 NNPAMLHKIDQEMLKCIELDEKLKSMDQEITVNPQFVQK 380
>UniRef100_Q7ZTN8 COP9 signalosome complex subunit 3 [Xenopus laevis]
Length = 423
Score = 189 bits (481), Expect = 4e-47
Identities = 102/219 (46%), Positives = 145/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++A+ +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERAMYFYEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-QLPKYTSQV 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ AE+ V+ + F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPAELRNLVSKHNETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ A+EAE +VL MI+DGEI+A+INQKDGMV F + PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGAQEAEKYVLYMIEDGEIFASINQKDGMVCFHDSPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + R + L +L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLRCIDLDDRLKAMDQEITVNPQFVQK 406
>UniRef100_Q5ZJF1 Hypothetical protein [Gallus gallus]
Length = 423
Score = 188 bits (477), Expect = 1e-46
Identities = 100/219 (45%), Positives = 147/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ K +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNKPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVCFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>UniRef100_Q68FW9 COP9 constitutive photomorphogenic homolog subunit 3 [Rattus
norvegicus]
Length = 423
Score = 187 bits (474), Expect = 3e-46
Identities = 99/219 (45%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>UniRef100_Q9UNS2 COP9 signalosome complex subunit 3 [Homo sapiens]
Length = 423
Score = 187 bits (474), Expect = 3e-46
Identities = 99/219 (45%), Positives = 146/219 (66%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ VN ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVNKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>UniRef100_O88543 COP9 signalosome complex subunit 3 [Mus musculus]
Length = 423
Score = 185 bits (469), Expect = 1e-45
Identities = 98/219 (44%), Positives = 146/219 (65%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ V+ ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVSKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL+D+A+ V L+ +EAE +VL MI+DGEI+A+INQKDGMV F ++PE+Y
Sbjct: 308 QRLTKTFLTLSLQDMASRVQLSGPQEAEKYVLHMIEDGEIFASINQKDGMVSFHDNPEKY 367
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
M+ +ID + + + L ++L A D++I+ + ++ K
Sbjct: 368 NNPAMLHNIDQEMLKCIELDERLKAMDQEITVNPQFVQK 406
>UniRef100_Q7Q5G8 ENSANGP00000004286 [Anopheles gambiae str. PEST]
Length = 432
Score = 180 bits (457), Expect = 3e-44
Identities = 96/219 (43%), Positives = 140/219 (63%), Gaps = 2/219 (0%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI K +++AL V+ P M+ I++E+YKKYILVSLI HG+ +PKY+S +
Sbjct: 193 MIYSAVKNYERALYFFEVAVSTPALAMSHIMLESYKKYILVSLILHGKV-LPIPKYSSQV 251
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K + +L+ YN EV VN F+ D N+GL KQVV+S+YK+NI
Sbjct: 252 ITRFMKPL-SHAYHDLSSAYNTSSADEVRNVVNKYRDTFQRDTNMGLVKQVVASLYKKNI 310
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL D+A+ V L+ EAE ++L MI+ GEI+ATINQKDGMV F +DP++Y
Sbjct: 311 QRLTKTFLTLSLADVASRVQLSGPAEAEKYILNMIKSGEIFATINQKDGMVVFKDDPQEY 370
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
E+ E + I +M L+K++ DE+I + +++ K
Sbjct: 371 NDQEVFEMVQHQISLVMGLNKQILKMDEEIMLNPVFVKK 409
>UniRef100_Q8SYG2 COP9 signalosome complex subunit 3 [Drosophila melanogaster]
Length = 445
Score = 165 bits (418), Expect = 9e-40
Identities = 91/219 (41%), Positives = 138/219 (62%), Gaps = 3/219 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI K +++AL +T P M+ I++EAYKK+++VSLI G+ + +PK +
Sbjct: 207 MIYTAVKNYERALYFFEVCITTPAMAMSHIMLEAYKKFLMVSLIVEGKIAY-IPKNTQVI 265
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N +L Y N E+ + + F DNN+GLAKQV +S+YKRNI
Sbjct: 266 G-RFMKPMA-NHYHDLVNVYANSSSEELRIIILKYSEAFTRDNNMGLAKQVATSLYKRNI 323
Query: 121 QRLTQTYLTLSLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQY 180
QRLT+T+LTLSL D+A+ V L SA EAE ++L MI+ GEIYA+INQKDGMV F +DPE+Y
Sbjct: 324 QRLTKTFLTLSLSDVASRVQLASAVEAERYILNMIKSGEIYASINQKDGMVLFKDDPEKY 383
Query: 181 KTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSK 219
+ EM ++ ++I ++ +++ +E+I + +Y+ K
Sbjct: 384 NSPEMFLNVQNNITHVLDQVRQINKMEEEIILNPMYVKK 422
>UniRef100_Q8BX58 Mus musculus 0 day neonate cerebellum cDNA, RIKEN full-length
enriched library, clone:C230080H11 product:COP9
(constitutive photomorphogenic) homolog, subunit 3
(Arabidopsis thaliana), full insert sequence [Mus
musculus]
Length = 313
Score = 97.4 bits (241), Expect = 3e-19
Identities = 55/125 (44%), Positives = 77/125 (61%), Gaps = 2/125 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++E+YKKYILVSLI G+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLESYKKYILVSLILLGKVQ-QLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R +K N ELAQ Y+ +E+ V+ ++ F DNN+GL KQ +SS+YK+NI
Sbjct: 249 VGRFIKPL-SNAYHELAQVYSTNNPSELRNLVSKHSETFTRDNNMGLVKQCLSSLYKKNI 307
Query: 121 QRLTQ 125
QRLT+
Sbjct: 308 QRLTK 312
>UniRef100_UPI000027CFB0 UPI000027CFB0 UniRef100 entry
Length = 302
Score = 86.7 bits (213), Expect = 5e-16
Identities = 50/115 (43%), Positives = 67/115 (57%), Gaps = 2/115 (1%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
MI G K F++AL +T P ++ I++EAYKKYILVSLI HG+ LPKY S +
Sbjct: 190 MIYTGLKNFERALYFYEQAITTPAMAVSHIMLEAYKKYILVSLILHGKVQ-PLPKYTSQI 248
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSM 115
R +K N ELAQ Y +E+ + VN ++ F DNN GL KQ +SS+
Sbjct: 249 VGRFIKPL-SNAYHELAQIYTTNNPSELRSLVNKHSETFTRDNNTGLVKQCLSSL 302
>UniRef100_UPI00003C1341 UPI00003C1341 UniRef100 entry
Length = 487
Score = 73.6 bits (179), Expect = 5e-12
Identities = 49/168 (29%), Positives = 83/168 (49%), Gaps = 4/168 (2%)
Query: 1 MICIGQKRFQKALDLLHNVVTAPMTMMNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPL 60
+I I R A+D L +++P ++AI ++AYKK +LV L+ G+ S +P+Y L
Sbjct: 226 LIYIKLDRLHDAIDALETCISSPAVAVSAIHMDAYKKLVLVQLLADGKTSR-VPRYTPQL 284
Query: 61 AQRNLKLFCQNPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNI 120
R + Q + +A G EV FE D N+GL ++ ++ +R I
Sbjct: 285 ITRMFNMSAQPYTLFVAAYEKWGDTQEVYRLAEEYHNAFEKDRNVGLIRRCLAVYRQRKI 344
Query: 121 QRLTQTYLTLSLEDIANTVHL---NSAKEAEMHVLQMIQDGEIYATIN 165
Q+L Q Y LSL IA + L ++ V ++++ G + AT++
Sbjct: 345 QKLAQVYSALSLGAIAQKIGLEGDDAVPSVYADVQEVVRKGWVRATLS 392
>UniRef100_P68358 COP9 signalosome complex subunit 3 [Brassica oleracea]
Length = 55
Score = 73.2 bits (178), Expect = 6e-12
Identities = 34/37 (91%), Positives = 36/37 (96%)
Query: 145 KEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYK 181
KEAEMHVLQMIQDG+I+A INQKDGMVRFLEDPEQYK
Sbjct: 19 KEAEMHVLQMIQDGQIHALINQKDGMVRFLEDPEQYK 55
>UniRef100_UPI00002198D7 UPI00002198D7 UniRef100 entry
Length = 497
Score = 63.2 bits (152), Expect = 6e-09
Identities = 51/210 (24%), Positives = 94/210 (44%), Gaps = 20/210 (9%)
Query: 27 MNAIVIEAYKKYILVSLIRHGQFST--SLPKYASPLAQRNLKLFCQNPCIELAQTYNNGK 84
++ +++EA+KK++LVSL+ G+ +T Y S + R + + P +A+ + +
Sbjct: 254 VSKVMLEAFKKWVLVSLLAEGRITTPGGFAPYISQASARTFGIMAR-PYAAVAKRFESAD 312
Query: 85 VAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNIQRLTQTYLTLSLEDI-------AN 137
+ V ++ +F D N GL +V+ S K I RL + Y +SL +I
Sbjct: 313 ADALRLAVEVSSQQFAEDGNEGLVAEVLESHQKWQIVRLREVYSKVSLAEINAKTKSAVT 372
Query: 138 TVHLNSAKEAEMHVLQMIQDGEIYATINQK-----DG-----MVRFLEDPEQYKTCEMIE 187
+ S ++ + MI G + A I DG + FL + E+ +
Sbjct: 373 GAPVGSEEDMTQLIETMIDSGMLRAAIEPPRATGGDGATTPACLAFLAEEADITEQEVAQ 432
Query: 188 HIDSSIQRIMALSKKLTATDEQISCDQLYL 217
I S++RI L AT E+++ + Y+
Sbjct: 433 RIAQSVERIKQLGAVYNATKERLATSREYI 462
>UniRef100_Q7S7G7 Predicted protein [Neurospora crassa]
Length = 426
Score = 62.8 bits (151), Expect = 8e-09
Identities = 66/239 (27%), Positives = 107/239 (44%), Gaps = 21/239 (8%)
Query: 4 IGQKRFQKALDLLHNVVTAPMTM--MNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPLA 61
I ++ +Q+A D VT P + I+ EAY K+ILV L+ G+ T LP+ S A
Sbjct: 155 IERRMWQQAFDAFERCVTYPTRDGGCSKIMTEAYNKWILVGLLLTGKPPT-LPETTSQAA 213
Query: 62 QRNLKLFCQN--PCIELAQTYNNGKVAE-VEAFVNANAGRFEADNNLGLAKQVVSSMYKR 118
+ K+F P AQ + + + V F N+ + N+ LAK V++ +
Sbjct: 214 K---KIFATQGKPYKLFAQAFKSETAGDLVREFEVINSELLPNEGNVELAKLVLAHYQRW 270
Query: 119 NIQRLTQTYLTLSLEDIANTVH-------LNSAKEAEMHVLQMIQDGEIYATINQ-KDG- 169
I L Y +SLE I L + + + V MI DG + I + KDG
Sbjct: 271 QIINLRNIYTNISLEKIQERTQSAETGAPLPTVEAVDQLVQSMIADGSLQGAIERPKDGS 330
Query: 170 --MVRFLEDPEQ-YKTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKVSVQSF 225
+ FL P Q E ++ +Q I AL + AT+++++ ++ Y+S + F
Sbjct: 331 PAYLTFLSSPAQGMSEVEFSAQVNKVMQGIKALEPIIEATNKRLASNREYISHTVKRQF 389
>UniRef100_Q6MW17 Related to JAB1-containing signalosome subunit 3 [Neurospora
crassa]
Length = 497
Score = 62.8 bits (151), Expect = 8e-09
Identities = 66/239 (27%), Positives = 107/239 (44%), Gaps = 21/239 (8%)
Query: 4 IGQKRFQKALDLLHNVVTAPMTM--MNAIVIEAYKKYILVSLIRHGQFSTSLPKYASPLA 61
I ++ +Q+A D VT P + I+ EAY K+ILV L+ G+ T LP+ S A
Sbjct: 226 IERRMWQQAFDAFERCVTYPTRDGGCSKIMTEAYNKWILVGLLLTGKPPT-LPETTSQAA 284
Query: 62 QRNLKLFCQN--PCIELAQTYNNGKVAE-VEAFVNANAGRFEADNNLGLAKQVVSSMYKR 118
+ K+F P AQ + + + V F N+ + N+ LAK V++ +
Sbjct: 285 K---KIFATQGKPYKLFAQAFKSETAGDLVREFEVINSELLPNEGNVELAKLVLAHYQRW 341
Query: 119 NIQRLTQTYLTLSLEDIANTVH-------LNSAKEAEMHVLQMIQDGEIYATINQ-KDG- 169
I L Y +SLE I L + + + V MI DG + I + KDG
Sbjct: 342 QIINLRNIYTNISLEKIQERTQSAETGAPLPTVEAVDQLVQSMIADGSLQGAIERPKDGS 401
Query: 170 --MVRFLEDPEQ-YKTCEMIEHIDSSIQRIMALSKKLTATDEQISCDQLYLSKVSVQSF 225
+ FL P Q E ++ +Q I AL + AT+++++ ++ Y+S + F
Sbjct: 402 PAYLTFLSSPAQGMSEVEFSAQVNKVMQGIKALEPIIEATNKRLASNREYISHTVKRQF 460
>UniRef100_Q7S4G9 Hypothetical protein [Neurospora crassa]
Length = 552
Score = 59.7 bits (143), Expect = 7e-08
Identities = 36/129 (27%), Positives = 64/129 (48%)
Query: 71 NPCIELAQTYNNGKVAEVEAFVNANAGRFEADNNLGLAKQVVSSMYKRNIQRLTQTYLTL 130
+P L + G + + E + +A F D L ++ ++ K I+ ++ +Y +
Sbjct: 371 HPYFLLVRAVRVGNLEDFETTIAQHADTFRRDGTYSLILRLRQNVIKTGIRMMSLSYSRI 430
Query: 131 SLEDIANTVHLNSAKEAEMHVLQMIQDGEIYATINQKDGMVRFLEDPEQYKTCEMIEHID 190
SL DI +HL S + AE V + I+DG I AT++++ G ++ E + Y T E E
Sbjct: 431 SLRDICIRLHLGSEESAEYIVAKAIRDGVIEATLDRERGFMKSKEVGDVYATSEPAEAFH 490
Query: 191 SSIQRIMAL 199
I+ +AL
Sbjct: 491 DRIRACLAL 499
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.133 0.370
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 355,340,348
Number of Sequences: 2790947
Number of extensions: 12837112
Number of successful extensions: 34669
Number of sequences better than 10.0: 132
Number of HSP's better than 10.0 without gapping: 62
Number of HSP's successfully gapped in prelim test: 71
Number of HSP's that attempted gapping in prelim test: 34572
Number of HSP's gapped (non-prelim): 142
length of query: 250
length of database: 848,049,833
effective HSP length: 124
effective length of query: 126
effective length of database: 501,972,405
effective search space: 63248523030
effective search space used: 63248523030
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138464.6


