

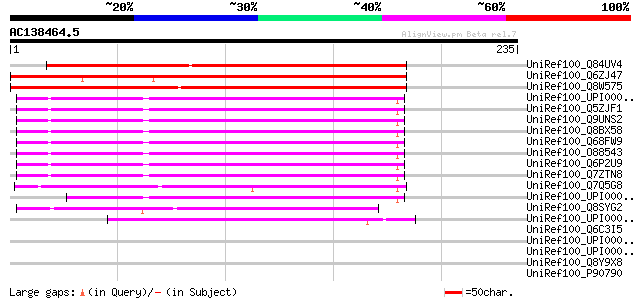
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.5 + phase: 0
(235 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84UV4 CSN3 [Nicotiana benthamiana] 209 5e-53
UniRef100_Q6ZJ47 Putative COP9 complex subunit 3, FUS11 [Oryza s... 205 7e-52
UniRef100_Q8W575 COP9 signalosome complex subunit 3 [Arabidopsis... 182 5e-45
UniRef100_UPI000027CFB0 UPI000027CFB0 UniRef100 entry 93 5e-18
UniRef100_Q5ZJF1 Hypothetical protein [Gallus gallus] 93 5e-18
UniRef100_Q9UNS2 COP9 signalosome complex subunit 3 [Homo sapiens] 93 5e-18
UniRef100_Q8BX58 Mus musculus 0 day neonate cerebellum cDNA, RIK... 93 6e-18
UniRef100_Q68FW9 COP9 constitutive photomorphogenic homolog subu... 93 6e-18
UniRef100_O88543 COP9 signalosome complex subunit 3 [Mus musculus] 93 6e-18
UniRef100_Q6P2U9 COP9 signalosome complex subunit 3 [Brachydanio... 92 1e-17
UniRef100_Q7ZTN8 COP9 signalosome complex subunit 3 [Xenopus lae... 91 2e-17
UniRef100_Q7Q5G8 ENSANGP00000004286 [Anopheles gambiae str. PEST] 90 4e-17
UniRef100_UPI0000360B09 UPI0000360B09 UniRef100 entry 89 9e-17
UniRef100_Q8SYG2 COP9 signalosome complex subunit 3 [Drosophila ... 64 2e-09
UniRef100_UPI00003C1341 UPI00003C1341 UniRef100 entry 47 4e-04
UniRef100_Q6C3I5 Similar to AAH45415 Danio rerio Similar to COP9... 42 0.013
UniRef100_UPI0000235718 UPI0000235718 UniRef100 entry 40 0.065
UniRef100_UPI00002198D7 UPI00002198D7 UniRef100 entry 39 0.11
UniRef100_Q8Y9X8 Low temperature requirement protein A [Listeria... 37 0.42
UniRef100_P90790 Hypothetical protein D2030.3 [Caenorhabditis el... 35 1.6
>UniRef100_Q84UV4 CSN3 [Nicotiana benthamiana]
Length = 175
Score = 209 bits (532), Expect = 5e-53
Identities = 99/167 (59%), Positives = 129/167 (76%), Gaps = 1/167 (0%)
Query: 18 SADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLDAFSAASVSNQQQAE 77
S+DI +LH LK S+E L SD RL L +DP+ HSLGFLY+L+A + + +Q E
Sbjct: 1 SSDITQLHNFLKQSEELLHSDFGRLLSSLAELDPNTHSLGFLYILEACMSFPAAKEQVNE 60
Query: 78 EAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGVAPMFTALRKLQVSA 137
V ++ RFIN+C+ EQIRLAP+KF+SVCKR KD + LLEAPIRGVA M TA+RKLQ S+
Sbjct: 61 LLVSVV-RFINSCAAEQIRLAPDKFISVCKRFKDQIILLEAPIRGVASMLTAVRKLQSSS 119
Query: 138 EHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLYCYYG 184
E LT LH +FL+LC+LAKCYKTG+++L+DD++E+D PRD +LYCYYG
Sbjct: 120 EQLTTLHPDFLLLCVLAKCYKTGITVLEDDIYEIDQPRDFFLYCYYG 166
>UniRef100_Q6ZJ47 Putative COP9 complex subunit 3, FUS11 [Oryza sativa]
Length = 426
Score = 205 bits (522), Expect = 7e-52
Identities = 103/186 (55%), Positives = 138/186 (73%), Gaps = 2/186 (1%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSD-ETLRSDSSRLYPILQLIDPSIHSLGFL 59
M+ +E L+A IQGLS S ++A LH +L+ +D E LR+ S+ L P L + PS HSLGFL
Sbjct: 1 METVETLVAHIQGLSGSGEELAHLHNLLRQADGEPLRAHSAALLPFLAQLHPSAHSLGFL 60
Query: 60 YLLDAF-SAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEA 118
YLL+AF S+AS Q + + A F+ +CS EQIRLAP+KF+SVC+ K+ V L A
Sbjct: 61 YLLEAFASSASNLRAQGGGDFLVTTADFLVSCSAEQIRLAPDKFLSVCRVFKNEVMQLNA 120
Query: 119 PIRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY 178
PIRG+AP+ A+RK+Q S+E LTP+HA++L+LCLLAK YK GLS+L+DD+ EVD P+DL+
Sbjct: 121 PIRGIAPLRAAIRKIQTSSEELTPIHADYLLLCLLAKQYKAGLSVLEDDILEVDQPKDLF 180
Query: 179 LYCYYG 184
LYCYYG
Sbjct: 181 LYCYYG 186
>UniRef100_Q8W575 COP9 signalosome complex subunit 3 [Arabidopsis thaliana]
Length = 429
Score = 182 bits (463), Expect = 5e-45
Identities = 92/184 (50%), Positives = 126/184 (68%), Gaps = 1/184 (0%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY 60
++ +E +I IQGLS S D++ LH +L+ + ++LR++ + L +D S HSLG+LY
Sbjct: 5 VNSVEAVITSIQGLSGSPEDLSALHDLLRGAQDSLRAEPGVNFSTLDQLDASKHSLGYLY 64
Query: 61 LLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPI 120
L+ + VS ++ A E +PIIARFIN+C QIRLA KFVS+CK LKD V L P+
Sbjct: 65 FLEVLTCGPVSKEKAAYE-IPIIARFINSCDAGQIRLASYKFVSLCKILKDHVIALGDPL 123
Query: 121 RGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLY 180
RGV P+ A++KLQVS++ LT LH + L LCL AK YK+G SIL DD+ E+D PRD +LY
Sbjct: 124 RGVGPLLNAVQKLQVSSKRLTALHPDVLQLCLQAKSYKSGFSILSDDIVEIDQPRDFFLY 183
Query: 181 CYYG 184
YYG
Sbjct: 184 SYYG 187
>UniRef100_UPI000027CFB0 UPI000027CFB0 UniRef100 entry
Length = 302
Score = 93.2 bits (230), Expect = 5e-18
Identities = 58/184 (31%), Positives = 93/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RGV
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGV 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L L+ D+ ++ Y
Sbjct: 122 VILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q5ZJF1 Hypothetical protein [Gallus gallus]
Length = 423
Score = 93.2 bits (230), Expect = 5e-18
Identities = 57/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 SILRQAIDKMQMNTNQLTSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q9UNS2 COP9 signalosome complex subunit 3 [Homo sapiens]
Length = 423
Score = 93.2 bits (230), Expect = 5e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q8BX58 Mus musculus 0 day neonate cerebellum cDNA, RIKEN full-length
enriched library, clone:C230080H11 product:COP9
(constitutive photomorphogenic) homolog, subunit 3
(Arabidopsis thaliana), full insert sequence [Mus
musculus]
Length = 313
Score = 92.8 bits (229), Expect = 6e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q68FW9 COP9 constitutive photomorphogenic homolog subunit 3 [Rattus
norvegicus]
Length = 423
Score = 92.8 bits (229), Expect = 6e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_O88543 COP9 signalosome complex subunit 3 [Mus musculus]
Length = 423
Score = 92.8 bits (229), Expect = 6e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q6P2U9 COP9 signalosome complex subunit 3 [Brachydanio rerio]
Length = 423
Score = 91.7 bits (226), Expect = 1e-17
Identities = 56/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDIQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K + L+ D+ ++ Y
Sbjct: 122 SILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPAVPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q7ZTN8 COP9 signalosome complex subunit 3 [Xenopus laevis]
Length = 423
Score = 90.9 bits (224), Expect = 2e-17
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LSS + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSSQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++A LT +H + L LLAKC+K L+ LD D+ ++ Y
Sbjct: 122 CVIRQAIDKMQMNANQLTSIHGDLCQLSLLAKCFKPALAYLDVDMMDICKENGAYDAKPF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>UniRef100_Q7Q5G8 ENSANGP00000004286 [Anopheles gambiae str. PEST]
Length = 432
Score = 90.1 bits (222), Expect = 4e-17
Identities = 59/190 (31%), Positives = 96/190 (50%), Gaps = 10/190 (5%)
Query: 3 PLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLL 62
PLE ++ ++ SS+ + L L S E + + + L +++ +D HSLG+L++L
Sbjct: 4 PLEYFVSFVRS-QSSAGNFRELVVYLIDSVELVTKNGNILDNVMETLDIQQHSLGYLFVL 62
Query: 63 DAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKD-LVTLLEAPIR 121
A S SN E + + FI +C EQ+R AP+ + +C L LV + I+
Sbjct: 63 SAKFNDS-SNVDDTENVLRSVREFITSCDAEQVRYAPQVYYELCHHLTTALVKNKQHIIQ 121
Query: 122 GVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY--- 178
G+ + AL K+++ LTP+HA+ LCL AK + + +LD D+ + D Y
Sbjct: 122 GIHVLVLALEKIRLFNAQLTPIHADLCQLCLCAKVFNPAIRLLDCDIAAIATTDDNYADT 181
Query: 179 ----LYCYYG 184
LY YYG
Sbjct: 182 KYFLLYYYYG 191
>UniRef100_UPI0000360B09 UPI0000360B09 UniRef100 entry
Length = 397
Score = 89.0 bits (219), Expect = 9e-17
Identities = 53/161 (32%), Positives = 82/161 (50%), Gaps = 6/161 (3%)
Query: 27 ILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLDAFSAASVSNQQQAEEAVPIIARF 86
++ S E L + S L +L +D HSLG L +L F S+ N E + F
Sbjct: 1 LINKSGELLAKNLSHLDTVLGALDIQEHSLGVLAVL--FVKFSMPNIPDFETLFSQVQLF 58
Query: 87 INACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGVAPMFTALRKLQVSAEHLTPLHAE 146
I+ C+ E IR A + F +C +L + + + P+RGV + A+ K+Q++ LT +HA+
Sbjct: 59 ISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGVVILKQAIDKMQMNTNQLTSVHAD 118
Query: 147 FLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----LYCYY 183
LCLLAKC+K L L+ D+ ++ Y CYY
Sbjct: 119 LCQLCLLAKCFKPALPFLELDMMDICKENGAYDAKHFLCYY 159
>UniRef100_Q8SYG2 COP9 signalosome complex subunit 3 [Drosophila melanogaster]
Length = 445
Score = 64.3 bits (155), Expect = 2e-09
Identities = 49/169 (28%), Positives = 82/169 (47%), Gaps = 3/169 (1%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY-LL 62
LE + Q++ LS+S + L L S L + S L +L+ +D HSLG LY LL
Sbjct: 5 LENYVNQVRTLSASGS-YRELAEELPESLSLLARNWSILDNVLETLDMQQHSLGVLYVLL 63
Query: 63 DAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRG 122
+AS +N + + + ++ F+ + EQ+R A F C + V I G
Sbjct: 64 AKLHSASTANPEPV-QLIQLMRDFVQRNNNEQLRYAVCAFYETCHLFTEFVVQKNLSILG 122
Query: 123 VAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEV 171
+ + A+ +++ LTP+HA+ +L L AK + L LD D+ ++
Sbjct: 123 IRIISRAIDQIRQLETQLTPIHADLCLLSLKAKNFSVVLPYLDADITDI 171
>UniRef100_UPI00003C1341 UPI00003C1341 UniRef100 entry
Length = 487
Score = 47.0 bits (110), Expect = 4e-04
Identities = 34/151 (22%), Positives = 64/151 (41%), Gaps = 9/151 (5%)
Query: 46 LQLIDPSIHSLGFLYLLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSV 105
L +DP++HS+G LY+L A ++ + + + A +P I F+ ++ Q++LA +K +
Sbjct: 79 LSALDPAVHSVGMLYILAARASQAATTVEGAVVLLPHITAFVQRFNLAQVQLAADKVTQL 138
Query: 106 CKRLKDLVTLLEAPIRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSIL- 164
L P + + + S+E +T LH L Y ++L
Sbjct: 139 AASFTALGDRGTNPESALQLLQALASRFLTSSESITTLHPMLAYQYLKTARYAEAATMLI 198
Query: 165 -------DDDVFEVDHPRDLYLYCYYGCVAF 188
D + + H D+ Y YY + +
Sbjct: 199 DLPLIDADTSITPLTH-SDVLQYFYYAALIY 228
>UniRef100_Q6C3I5 Similar to AAH45415 Danio rerio Similar to COP9 constitutive
photomorph [Yarrowia lipolytica]
Length = 403
Score = 42.0 bits (97), Expect = 0.013
Identities = 30/132 (22%), Positives = 57/132 (42%), Gaps = 7/132 (5%)
Query: 70 VSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGVAPMFTA 129
+SN+ + + F+ S++++ P S+ + V + P+ G++
Sbjct: 60 LSNELDTSIFLYYVDTFLQQFSIQRLSAFPHILSSLLDQFSHHVIRAKRPLLGLS----L 115
Query: 130 LRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDH---PRDLYLYCYYGCV 186
L+ +TP+HA+ + L A GL +LD + +D DL L+ +YG +
Sbjct: 116 LQPRLPDKNTITPMHAQLMRLSFEANKCSVGLEVLDREFLSLDKCATRNDLALFHFYGAL 175
Query: 187 AFFFSFELEKRD 198
+ E EK D
Sbjct: 176 VYIAYKEWEKAD 187
>UniRef100_UPI0000235718 UPI0000235718 UniRef100 entry
Length = 487
Score = 39.7 bits (91), Expect = 0.065
Identities = 35/128 (27%), Positives = 56/128 (43%), Gaps = 9/128 (7%)
Query: 46 LQLIDPSIHSLGFLYL-------LDAFSAASVSNQQQAEEAV-PIIARFINACSVEQIRL 97
L+ I P++HSL +LYL L +A V N Q + +F+ + QIR
Sbjct: 53 LEAISPAVHSLSYLYLLRIRIQQLQEKTAVGVPNDLQPGGTLWNQTVKFLRSFDPIQIRY 112
Query: 98 APEKFVSVCKRLKDLVTLLEAPIRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCY 157
++ + + + + PI V + AL +L +A T LH + L LL+ Y
Sbjct: 113 VGHEWRELVDSVANAALSVSKPILAVKMIRDALERLN-TAGVFTSLHLMLVKLALLSSSY 171
Query: 158 KTGLSILD 165
L +LD
Sbjct: 172 TYVLPVLD 179
>UniRef100_UPI00002198D7 UPI00002198D7 UniRef100 entry
Length = 497
Score = 38.9 bits (89), Expect = 0.11
Identities = 36/134 (26%), Positives = 58/134 (42%), Gaps = 12/134 (8%)
Query: 19 ADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLDAFSAASVSNQQQAEE 78
A I+ ++++ K + ++ Y +L L++P+I+SL +LYLL A N + +E
Sbjct: 30 AHISNINSLFKERSAVIGDNA---YKLLDLLNPAINSLSYLYLLAALLPQGTKNAEYDKE 86
Query: 79 AVPIIARFINACSVEQIR---LAPEKFVSVCKRLKDLVTLLEAPIRGVAPMFTALRKLQV 135
+ F QIR +A S R + + PI + AL +L
Sbjct: 87 FAERVILFFRRFDPRQIRYMGVAFSDLFSATGRRNVMPPTIAVPI-----LADALLRLDP 141
Query: 136 SAEHLTPLHAEFLM 149
S LT H FLM
Sbjct: 142 SGSVLTSNHI-FLM 154
>UniRef100_Q8Y9X8 Low temperature requirement protein A [Listeria monocytogenes]
Length = 371
Score = 37.0 bits (84), Expect = 0.42
Identities = 34/132 (25%), Positives = 55/132 (40%), Gaps = 25/132 (18%)
Query: 112 LVTLLEAPIRGVAPMFTA--LRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVF 169
LV L + + P+F A LRK+ V HL F+++ G SI+
Sbjct: 159 LVMYLGIAVDIILPLFLAKTLRKVPVDFPHLAERFGLFVIITF-------GESIVAITTI 211
Query: 170 EVDHPRDLYLYCYYGC---------VAFFFSFELEKRDRCRICSYISVLHFSYITYCNFF 220
V H DLY CY ++F+SFE +I + V H ++ Y +FF
Sbjct: 212 LVGHTLDLYTICYTLLGFLIICTLWASYFYSFE-------KIVDHHKVTHGQFLIYGHFF 264
Query: 221 NILDLFFTSFSI 232
I+ + + ++
Sbjct: 265 IIVSVMLLAVNL 276
>UniRef100_P90790 Hypothetical protein D2030.3 [Caenorhabditis elegans]
Length = 508
Score = 35.0 bits (79), Expect = 1.6
Identities = 53/162 (32%), Positives = 68/162 (41%), Gaps = 22/162 (13%)
Query: 27 ILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLDAFSAASVSNQQQAEEAVPIIARF 86
I+K S +TLR+ SRL L I G LY + V N Q E PI+ +
Sbjct: 50 IIKCSSQTLRAVQSRLANRLAHISDHFQLCGSLYTRELL----VRNPQAGELHSPIVFKI 105
Query: 87 INAC-------SVEQIR-LAPEKF------VSVCKRLKDLVTLLEA-PIRGVAPMFTALR 131
I C S IR LA E F ++ L L LL A I+ V+ LR
Sbjct: 106 ILNCMDAAAQTSDPNIRGLAAELFGLRISNQNLTMLLNTLEVLLSATKIKNVSNDLLQLR 165
Query: 132 KLQVSA--EHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEV 171
L VS+ + LT L E L L L K GL + D+ +V
Sbjct: 166 TLGVSSHRDSLTSLLFEVLALS-LGYASKPGLFVERKDIIQV 206
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.329 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 362,020,499
Number of Sequences: 2790947
Number of extensions: 13962546
Number of successful extensions: 40562
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 40539
Number of HSP's gapped (non-prelim): 24
length of query: 235
length of database: 848,049,833
effective HSP length: 123
effective length of query: 112
effective length of database: 504,763,352
effective search space: 56533495424
effective search space used: 56533495424
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138464.5


