

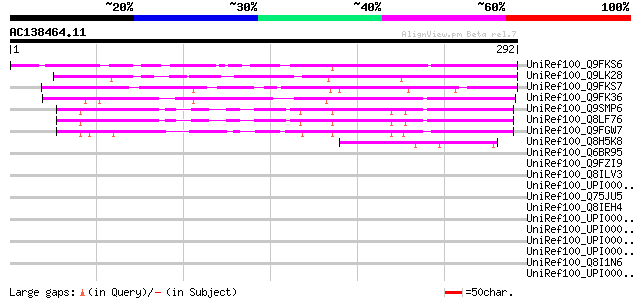
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.11 - phase: 0
(292 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9FKS6 Emb|CAB62340.1 [Arabidopsis thaliana] 179 1e-43
UniRef100_Q9LK28 Emb|CAB62340.1 [Arabidopsis thaliana] 175 1e-42
UniRef100_Q9FKS7 Emb|CAB62340.1 [Arabidopsis thaliana] 140 4e-32
UniRef100_Q9FK36 Emb|CAB62340.1 [Arabidopsis thaliana] 126 8e-28
UniRef100_Q9SMP6 Hypothetical protein T8P19.20 [Arabidopsis thal... 120 4e-26
UniRef100_Q8LF76 Hypothetical protein [Arabidopsis thaliana] 119 7e-26
UniRef100_Q9FGW7 Emb|CAB62340.1 [Arabidopsis thaliana] 117 3e-25
UniRef100_Q8H5K8 Hypothetical protein OJ1060_D03.127 [Oryza sativa] 50 9e-05
UniRef100_Q6BR95 Similar to CA5154|CaRGA2 Candida albicans CaRGA... 44 0.004
UniRef100_Q9FZI9 F17L21.26 [Arabidopsis thaliana] 42 0.025
UniRef100_Q8ILV3 Hypothetical protein [Plasmodium falciparum] 39 0.12
UniRef100_UPI00003164E9 UPI00003164E9 UniRef100 entry 37 0.47
UniRef100_Q75JU5 Similar to cell wall biosynthesis kinase; Cbk1p... 37 0.47
UniRef100_Q8IEH4 Hypothetical protein MAL13P1.72 [Plasmodium fal... 37 0.47
UniRef100_UPI000042D640 UPI000042D640 UniRef100 entry 37 0.61
UniRef100_UPI000034D452 UPI000034D452 UniRef100 entry 37 0.61
UniRef100_UPI000030D582 UPI000030D582 UniRef100 entry 37 0.61
UniRef100_UPI000033F052 UPI000033F052 UniRef100 entry 37 0.80
UniRef100_Q8I1N6 Hypothetical protein PFD0985w [Plasmodium falci... 37 0.80
UniRef100_UPI00002BCF4E UPI00002BCF4E UniRef100 entry 36 1.0
>UniRef100_Q9FKS6 Emb|CAB62340.1 [Arabidopsis thaliana]
Length = 272
Score = 179 bits (453), Expect = 1e-43
Identities = 116/301 (38%), Positives = 168/301 (55%), Gaps = 38/301 (12%)
Query: 1 MESEH-VWRVGACNRNPSLNNDNNIFNGIMLRFRPIAPKPVT-GEQTPANQSTTASLPGK 58
M+ +H +WR C D + +ML++RPIAPKP T G+ + S+T +
Sbjct: 1 MDQDHELWRTLRCAGKAQ---DKTSVDTLMLKYRPIAPKPTTTGQPLVGDTSST-----R 52
Query: 59 RTKRKYVRIRRNNGYIKKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAAT 118
RTKRKYVR+ +NN K +C+ + S+S D +VTLQL+PE++
Sbjct: 53 RTKRKYVRVSKNN----KATCRSKTNGFR-----SSSTDPENGREDIVTLQLLPERSTPL 103
Query: 119 SNVDLNLNLNLTVENIQILDNLNRSQNNPDLETMNAGKFLDLLPAVEKAVVESWITVESV 178
S +D N NL+ TVE I N + + N N G A+++ VE+W+TVESV
Sbjct: 104 S-LDHN-NLDPTVETI----NGDETCNTDTWLKFNGGD-----DALQQVPVETWVTVESV 152
Query: 179 TGMCMV-------VEKVKNIETDTCPCFITDGNGKVLRVNDAYKKMVVNEESEEVEVVIW 231
+ E ++ DTCP FI+DG+ +V+ VN+AY+++V + EV++W
Sbjct: 153 NSGLVSHAVGLTDEELTYALDKDTCPGFISDGSNRVVMVNEAYRRIVTGDGGFGREVIVW 212
Query: 232 LKVKQSVAWCYSYPAFTCGVRLQYTWRNEKCLKMVPCDVWRLECGGFAWRLDVKAALSLG 291
L V Q+ +C Y FTC VR++YTWR K K +PCDVW++E GGFAWRLD AAL+L
Sbjct: 213 LVVDQTATFC-DYRTFTCKVRMEYTWRETKYTKTLPCDVWKMEFGGFAWRLDTTAALTLW 271
Query: 292 L 292
L
Sbjct: 272 L 272
>UniRef100_Q9LK28 Emb|CAB62340.1 [Arabidopsis thaliana]
Length = 282
Score = 175 bits (443), Expect = 1e-42
Identities = 110/284 (38%), Positives = 159/284 (55%), Gaps = 41/284 (14%)
Query: 26 NGIMLRFRPIAPKPVTGEQTPANQSTTASLPG--KRTKRKYVRIRRNNGYIKKNSCKKSS 83
+ +MLR+RPIAPKP TG+ + S G KRTKRKYVR+ +NN K +C+ S
Sbjct: 23 DALMLRYRPIAPKPTTGQPCGVADNNNNSSYGMSKRTKRKYVRVSKNN----KGTCRGKS 78
Query: 84 PEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQILDNLNRS 143
S+ +D+ ++ VVTLQL+PEK+ + ++ + ++
Sbjct: 79 --------RSDLSDDREQT-DVVTLQLLPEKSDISGEYS-------PLDQDSLDPSVKSI 122
Query: 144 QNNPDLETMNAGKFLDLLPAVEKAVVESWITVESVTGMC-----------MVVEKVKNIE 192
ET G F + A +E+W+TVESVT +C VE V N+
Sbjct: 123 IGEETQETNTWGMFNGSVTAE----METWVTVESVTSVCEGSLSSHAVGITDVEIVDNLG 178
Query: 193 TDTCPCFITDGNGKVLRVNDAYKKMVVNEESE---EVEVVIWLKVKQSVAWCY-SYPAFT 248
DTCP F++DG+ +V+ VN+AY++ V ++S EVV+WL +++ A + +Y AFT
Sbjct: 179 KDTCPAFVSDGSNRVVWVNEAYRRNVSGDDSTASVSPEVVVWLVAEEATAAMHCNYQAFT 238
Query: 249 CGVRLQYTWRNEKCLKMVPCDVWRLECGGFAWRLDVKAALSLGL 292
C VR+QYTW+ K K VPCDVW++E GGFAWRLD AAL+L L
Sbjct: 239 CRVRMQYTWKETKYTKTVPCDVWKMEFGGFAWRLDTTAALTLWL 282
>UniRef100_Q9FKS7 Emb|CAB62340.1 [Arabidopsis thaliana]
Length = 303
Score = 140 bits (353), Expect = 4e-32
Identities = 101/307 (32%), Positives = 158/307 (50%), Gaps = 54/307 (17%)
Query: 19 NNDNNIFNGIMLRFRPIAPKPVTGEQTPANQSTTASLPGKRTKRKYVRIRRNNGYIKKNS 78
+ D + + IM RFRPIAPKP GE + S +R+KRKYVR+R KKNS
Sbjct: 18 DQDKTVISKIMQRFRPIAPKPAVGESSDDTNSDRFLGRNRRSKRKYVRVRD-----KKNS 72
Query: 79 CKKSSPEEEIVVQHSNSNDEMKRNLA----------VVTLQLMPEKAAATSNVDLNLNLN 128
++ ++ ++ +K +L +VTLQL+PEK + D+ N +
Sbjct: 73 SGSNNKKDITGKKNGCDRGNIKTDLDKKIDGDDRTDIVTLQLLPEK-----DRDIGNNGD 127
Query: 129 LTVENIQILDNLNRSQNNPDLETMNAGKFLDLLPAVEKAVVESWITVESVTGMCM----- 183
E L +++ P N+ + L + ++ VVESW+TVE V+ C
Sbjct: 128 KAGEFCSDLSDMD-----PKKSLYNS--IIGLSSSFDRKVVESWLTVECVSDTCTDLGWY 180
Query: 184 -VVEKV-----------KNIETDTCPCFITDGNGKVLRVNDAYKKMVVNEESEEVEV--V 229
++E++ + +E DTCP ++DG+ +V VN AY++M+ + + + V V
Sbjct: 181 HILEQLGRMDQAEERVMRMLEVDTCPWLVSDGSNRVCWVNRAYRRMMGAPDVDVIRVWLV 240
Query: 230 IWLKVKQSVAWCYS-YPAFTCGVRLQY---TWRNEKCLKMVPCDVWRLECGGFAWRLDVK 285
+ + + + +A Y A TC VR++Y TWR VPCDVWR+ GGFAWRLDV+
Sbjct: 241 VAMDLMEEIACMVELYGAVTCRVRVRYEPSTWRK----MTVPCDVWRIRSGGFAWRLDVE 296
Query: 286 AALSLGL 292
+AL LG+
Sbjct: 297 SALRLGM 303
>UniRef100_Q9FK36 Emb|CAB62340.1 [Arabidopsis thaliana]
Length = 291
Score = 126 bits (316), Expect = 8e-28
Identities = 100/294 (34%), Positives = 143/294 (48%), Gaps = 44/294 (14%)
Query: 20 NDNNIFNGIMLRFRPIAPKPVTG----EQTPANQS--TTASLPGKRTKRKYVRIRRNNGY 73
ND + + IMLR+RPIAP+P +G T N S T S +R KRKY + ++
Sbjct: 19 NDMSKVDRIMLRYRPIAPRPDSGGSPASPTEKNGSVITNVSSRSRRGKRKYSKENNSSST 78
Query: 74 IKKNSCKKSSPEEEIVVQHSNSNDEMKRNLA----VVTLQLMPEKAAAT-SNVDLNLNLN 128
NS S + NDE K +VTL L+PE S +
Sbjct: 79 GSVNSNGNSKRQR---------NDETKNGSGGGREIVTLPLLPETPEKKDSPLKAKAAPE 129
Query: 129 LTVENIQILDNLNRSQNNPDLETMNAGKFLDLLPAVEKAVVESWITVESVTGM------- 181
L + + N S N + + + VV S +TVE VT
Sbjct: 130 LGAAALWLSFNDGASYNRRYQTEL-----------MTETVVSSLLTVECVTERLMEGEYE 178
Query: 182 --CMVVEKVKNIETDTCPCFITDGNGKVLRVNDAYKKMVVNEESEEV-EVVIWLKVKQSV 238
C E+ N+E DTCP FI+DG G+V+ N +Y+++VV ++ E+ ++ +WL +K+
Sbjct: 179 LGCTDEERKMNLERDTCPGFISDGLGRVIWTNGSYRELVVGKDHEQCSKMSVWLVMKEKP 238
Query: 239 AWCYSYPAFTCGVRLQYTWRNEKCLKMVP-CDVWRLECGGFAWRLDVKAALSLG 291
+Y FTC +RLQYT R+++ + CDVWR+ GGFAWRLDV AAL LG
Sbjct: 239 --LVTYRTFTCRMRLQYTCRDKEVSSITSFCDVWRMSDGGFAWRLDVDAALCLG 290
>UniRef100_Q9SMP6 Hypothetical protein T8P19.20 [Arabidopsis thaliana]
Length = 294
Score = 120 bits (301), Expect = 4e-26
Identities = 103/299 (34%), Positives = 145/299 (48%), Gaps = 67/299 (22%)
Query: 28 IMLRFRPIAPKP--------VTGEQTPANQSTTASLPGKRTKRKYVRIRRNNGYIKKNSC 79
IMLRFRPIAPKP V+ + S + R KRK +++ N G K+ +
Sbjct: 27 IMLRFRPIAPKPASPGGVNPVSSGDSGGGSSDVSFRSASRRKRKCHQLKENGGNAKRCTR 86
Query: 80 KKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQILDN 139
+K+S + V H ++N VTL L+PEK ++L VE
Sbjct: 87 RKTSDKP---VVHGSAN--------AVTLSLLPEKP---------IDLKAAVEK------ 120
Query: 140 LNRSQNNPDLETMNAGKFLDLLPAVEK-------AVVESWITVESVTGMCMV-------- 184
+ Q L + G L PA + V+ S +TVE VT +
Sbjct: 121 -QKRQGPLWLSFKDGGGMLT--PAYQTPEIVQRTVVISSCMTVERVTDAWIDGYGLGRSD 177
Query: 185 VEKVKNIETDTCPCFITDGNGKVLRVNDAYKKMV-----VNEESEEV------EVVIWLK 233
E+ N+ DTCP FI+DG+G+V NDAY+KM V E + E+ V++ L
Sbjct: 178 EERKMNLVRDTCPGFISDGSGRVTWTNDAYRKMARDIIPVEEGAPEITSGDSFHVIVRLV 237
Query: 234 VKQSVAWCYSYPAFTCGVRLQYTWRN-EKCLKMVPCDVWRLEC-GGFAWRLDVKAALSL 290
+++ + P FTC ++LQYT +N E+ VPCDVWR++ GGFAWRLDVKAAL L
Sbjct: 238 MRERP--MLTSPGFTCRMKLQYTCQNRERGSVTVPCDVWRMDVGGGFAWRLDVKAALCL 294
>UniRef100_Q8LF76 Hypothetical protein [Arabidopsis thaliana]
Length = 294
Score = 119 bits (299), Expect = 7e-26
Identities = 103/299 (34%), Positives = 145/299 (48%), Gaps = 67/299 (22%)
Query: 28 IMLRFRPIAPKP--------VTGEQTPANQSTTASLPGKRTKRKYVRIRRNNGYIKKNSC 79
IMLRFRPIAPKP V+ + S + R KRK +++ N G K+ +
Sbjct: 27 IMLRFRPIAPKPASPVGVNPVSSGDSGGGGSDVSFRSASRRKRKCHQLKENGGNAKRCTR 86
Query: 80 KKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQILDN 139
+K+S + V H ++N VTL L+PEK ++L VE
Sbjct: 87 RKTSDKP---VVHGSAN--------AVTLSLLPEKP---------IDLKAAVEK------ 120
Query: 140 LNRSQNNPDLETMNAGKFLDLLPAVEK-------AVVESWITVESVTGMCMV-------- 184
+ Q L + G L PA + V+ S +TVE VT +
Sbjct: 121 -QKRQGPLWLSFKDGGGMLT--PAYQTPEIVQRTVVISSCMTVERVTDAWIDGYGLGRSD 177
Query: 185 VEKVKNIETDTCPCFITDGNGKVLRVNDAYKKMV-----VNEESEEV------EVVIWLK 233
E+ N+ DTCP FI+DG+G+V NDAY+KM V E + E+ V++ L
Sbjct: 178 EERKMNLVRDTCPGFISDGSGRVTWTNDAYRKMARDIIPVEEGAPEITSGDSFHVIVRLV 237
Query: 234 VKQSVAWCYSYPAFTCGVRLQYTWRN-EKCLKMVPCDVWRLEC-GGFAWRLDVKAALSL 290
+++ + P FTC ++LQYT +N E+ VPCDVWR++ GGFAWRLDVKAAL L
Sbjct: 238 MRERP--MLTSPGFTCRMKLQYTCQNRERGSVTVPCDVWRMDVGGGFAWRLDVKAALCL 294
>UniRef100_Q9FGW7 Emb|CAB62340.1 [Arabidopsis thaliana]
Length = 302
Score = 117 bits (294), Expect = 3e-25
Identities = 108/306 (35%), Positives = 141/306 (45%), Gaps = 73/306 (23%)
Query: 28 IMLRFRPIAPKP--------VTGEQ--TPANQSTTASLPGK--RTKRKYVRIRRNNGYIK 75
IMLRFRPIAPKP +TG+ T ++ L GK R KRKY + +
Sbjct: 27 IMLRFRPIAPKPASDGGSVSLTGKSGSTTTTSGGSSDLSGKSGRGKRKYQKDCSGGNSRR 86
Query: 76 KNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQ 135
N K+ + VTL L+PE DLN VE
Sbjct: 87 CNKKKRDLSGDTATT-------------TAVTLSLLPETPEKRVFPDLNA---FPVEK-- 128
Query: 136 ILDNLNRSQNNPDLETMNAGKFLDLLPAVEKA-------VVESWITVESVTGMCMV---- 184
+ +N P + N G ++L + A VV S +TVE VT +
Sbjct: 129 ------QKRNGPLWLSFNGGG--EILTPYKTAEISRRTVVVSSCVTVERVTDAWIDGYGL 180
Query: 185 ----VEKVKNIETDTCPCFITDGNGKVLRVNDAYKKMV---VNEESEE------------ 225
E+ N+ DTCP FI+DG G+V N+AYKKM +N EE
Sbjct: 181 GETNQERKMNLVEDTCPGFISDGVGRVTWTNEAYKKMAREDINIPMEEGVPEDISYDNFH 240
Query: 226 VEVVIWLKVKQSVAWCYSYPAFTCGVRLQYTWRN-EKCLKMVPCDVWRLECGGFAWRLDV 284
V V + +K + + +YPAFTC VRLQYT ++ E+ VPCDVWR++ GGFAWRLDV
Sbjct: 241 VNVRLVMKERPML----TYPAFTCRVRLQYTCQDRERGSVTVPCDVWRMDGGGFAWRLDV 296
Query: 285 KAALSL 290
KAAL L
Sbjct: 297 KAALCL 302
>UniRef100_Q8H5K8 Hypothetical protein OJ1060_D03.127 [Oryza sativa]
Length = 343
Score = 49.7 bits (117), Expect = 9e-05
Identities = 33/110 (30%), Positives = 51/110 (46%), Gaps = 19/110 (17%)
Query: 191 IETDTCPCFITDGNGKVLRVNDAYKKMVVNEESEEVEVVIWL---KVKQSVAWCYSYPA- 246
+E+D+ P ++D + +V VNDAYK+MV E ++ V ++ VA S PA
Sbjct: 216 LESDSLPAVVSDSSNRVRLVNDAYKRMVGQPECPWLDAVATAASRRISGEVALVVSEPAA 275
Query: 247 -----------FTCGVRLQYTWRNEKCLKMVPCDVWRLECGG----FAWR 281
F+C ++ + + PCDV RL+C FAWR
Sbjct: 276 AAAALPETCKGFSCSAKIAWERDGKWSSVHAPCDVTRLQCESRDYVFAWR 325
>UniRef100_Q6BR95 Similar to CA5154|CaRGA2 Candida albicans CaRGA2 [Debaryomyces
hansenii]
Length = 1245
Score = 44.3 bits (103), Expect = 0.004
Identities = 30/111 (27%), Positives = 55/111 (49%)
Query: 56 PGKRTKRKYVRIRRNNGYIKKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKA 115
P RT + + NNG+I++++ S V+ + SND +K +L + +L+ +
Sbjct: 679 PLPRTSLSNHQFQTNNGFIREHNRSTSDTPFSSVLDNRYSNDIVKNDLDMRSLKSEVYQL 738
Query: 116 AATSNVDLNLNLNLTVENIQILDNLNRSQNNPDLETMNAGKFLDLLPAVEK 166
+ V N N L EN ++ + L QNN +E+ N + + + A+EK
Sbjct: 739 DSQKQVLFNENKALKNENEKLSEQLRNLQNNISVESSNHDRLTEEIIALEK 789
>UniRef100_Q9FZI9 F17L21.26 [Arabidopsis thaliana]
Length = 1034
Score = 41.6 bits (96), Expect = 0.025
Identities = 31/110 (28%), Positives = 51/110 (46%), Gaps = 14/110 (12%)
Query: 186 EKVKNIETDTCPCFITDGNGKVLRVNDAYKKMVVNEESEEVEVVIWLK---VKQSVAWCY 242
E +++E+D P ITD N +V VN AYK+M+ E ++ ++ K + + +C
Sbjct: 917 EVEEDVESDDLPSVITDSNSRVRLVNSAYKEMMGQPECSWLDSMVRGKRICGEVMINFCE 976
Query: 243 S-------YPAFTCGVRLQYTWRNEKCLKMVPCDVWRLECGG----FAWR 281
S F+C VR+ + ++ CDV +L C F WR
Sbjct: 977 SKIPVMTENNGFSCWVRIDWGRDGKEEYMHAFCDVTKLACDSKDYVFTWR 1026
>UniRef100_Q8ILV3 Hypothetical protein [Plasmodium falciparum]
Length = 407
Score = 39.3 bits (90), Expect = 0.12
Identities = 35/149 (23%), Positives = 63/149 (41%), Gaps = 18/149 (12%)
Query: 13 NRNPSLNNDNNIFNGIMLRFRPIAPKPVTGEQTPANQSTTASLPGKRTKR--KYVRIRRN 70
N N + NNDNN N +++ N S + KR Y
Sbjct: 91 NNNDNDNNDNNNNNN----------DNNNDDESQHNNSNNNDVDNNTFKRYNNYNEKYEE 140
Query: 71 NGYIKK---NSCKKSSPEEEIVVQHSNSNDEMK-RNLAVVTLQLMPEKAAATSNVDLN-- 124
N ++K ++C SPE V ++N+N+ N +T Q+ E ++N+ ++
Sbjct: 141 NDNMEKEVYHTCSIQSPERSAVHNNNNNNNNNNSNNYYYMTYQMNDEHIKNSNNIKIHNN 200
Query: 125 LNLNLTVENIQILDNLNRSQNNPDLETMN 153
+N+N++ N+ D+ N +N D M+
Sbjct: 201 VNINMSSNNVDNNDDANSIDSNDDSNNMD 229
>UniRef100_UPI00003164E9 UPI00003164E9 UniRef100 entry
Length = 138
Score = 37.4 bits (85), Expect = 0.47
Identities = 28/99 (28%), Positives = 49/99 (49%), Gaps = 6/99 (6%)
Query: 76 KNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQ 135
K+ KK E + + N+ D++ R+LA + Q + +D N EN+
Sbjct: 15 KSEEKKEKKELSVEEKLKNTEDKLLRSLAELENQRNRFEKDIKEAIDFG-GFNFARENLS 73
Query: 136 ILDNLNRS----QNNPDL-ETMNAGKFLDLLPAVEKAVV 169
ILDNL+R+ +N+P L + + KFL + +EK ++
Sbjct: 74 ILDNLHRAHISIKNDPSLKDNKDLDKFLKNIEIIEKDLI 112
>UniRef100_Q75JU5 Similar to cell wall biosynthesis kinase; Cbk1p [Dictyostelium
discoideum]
Length = 2254
Score = 37.4 bits (85), Expect = 0.47
Identities = 42/166 (25%), Positives = 62/166 (37%), Gaps = 27/166 (16%)
Query: 13 NRNPSLNNDNNIFNGIMLRFRPI---APKPVTGEQTPANQSTTASLPGKRT--------- 60
N N + NN+ N +G P AP V+ Q A S SLP
Sbjct: 768 NNNNNNNNNGNTASGSSCNTTPNLLPAPTNVSPIQNRARSSPMTSLPLPNNLLIPIPVFS 827
Query: 61 -KRKYVRIRRNNGYIKKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATS 119
+Y I NN I+ N+ ++ ++N+N+ N +
Sbjct: 828 LNDEYNNINNNNSNIESNNNNNNNNNNNNNNNNNNNNNNNNNN----------NNNNNNN 877
Query: 120 NVDLNLNLNLTVE----NIQILDNLNRSQNNPDLETMNAGKFLDLL 161
N + N N N T E NI+ DN N + NN + N+ LDL+
Sbjct: 878 NNNNNNNNNNTTELVNNNIESNDNDNNNSNNNNNNNNNSNNSLDLI 923
>UniRef100_Q8IEH4 Hypothetical protein MAL13P1.72 [Plasmodium falciparum]
Length = 1205
Score = 37.4 bits (85), Expect = 0.47
Identities = 30/143 (20%), Positives = 69/143 (47%), Gaps = 24/143 (16%)
Query: 69 RNNGYIKKNSCKKSSPEEEIVVQHSNSN--DEMKRNLA--------------VVTLQLMP 112
R Y+K NS + +E+ ++ ++N+N ++ K N++ + L+ +
Sbjct: 48 RRTNYLKANSLNVKNIKEKNIILNNNTNKLNDQKNNVSTKNIKPSGRDKNNTIYNLKYIS 107
Query: 113 EKAAATSNVDLNLNLNLTVENI-QILDNLNRSQNNPDLETMNAGKFLDLLPAVEKAVVES 171
+K+ +N++++ N N+ + + LD N+S+N E + K L+ L ++
Sbjct: 108 QKSDRNNNINVSNNPNVYINKLHDYLDISNKSEN----EKVKINKLLNKLKTENLSI--- 160
Query: 172 WITVESVTGMCMVVEKVKNIETD 194
I + S+ +C ++ K +I+ D
Sbjct: 161 HIILNSLKDLCSLLLKKNSIDLD 183
>UniRef100_UPI000042D640 UPI000042D640 UniRef100 entry
Length = 1111
Score = 37.0 bits (84), Expect = 0.61
Identities = 27/136 (19%), Positives = 68/136 (49%), Gaps = 11/136 (8%)
Query: 88 IVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQILDNLNRSQNNP 147
+V+Q + SN ++ LA++ L+++P S+ ++NL NL + + +++N N
Sbjct: 722 LVMQETYSNVDLVLYLAIL-LEILPYLLLVKSSTNINLLKNLRLNLLHLINNFNN----- 775
Query: 148 DLETMNAGKFLDLLPAVEKAVVESWITVESVTGMCMVVEKVKNIETDTCPCFITDGNGKV 207
+L+ +++ KF +E++ + + + + +++ +K I D C ++
Sbjct: 776 ELKKLDSPKF-----PIERSTIFLVLVKKLINLNDLLITFIKLINRDNCKTTMSSDIENF 830
Query: 208 LRVNDAYKKMVVNEES 223
L++N K+ ++ S
Sbjct: 831 LKINSQTVKLDDDDNS 846
>UniRef100_UPI000034D452 UPI000034D452 UniRef100 entry
Length = 191
Score = 37.0 bits (84), Expect = 0.61
Identities = 29/100 (29%), Positives = 53/100 (53%), Gaps = 10/100 (10%)
Query: 75 KKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENI 134
KK++ K+ S EE++ ++ +++ R+LA + Q + ++ N EN+
Sbjct: 18 KKDNAKEISIEEKL----KDTEEKLLRSLAELENQRKRFEKEIKEAIEFG-GFNFAKENL 72
Query: 135 QILDNLNRS----QNNPDLETMN-AGKFLDLLPAVEKAVV 169
ILDNL R+ +N+P+L+ N KFL + +EK V+
Sbjct: 73 VILDNLQRAYISIKNDPNLKNNNDLDKFLTNIEIIEKDVI 112
>UniRef100_UPI000030D582 UPI000030D582 UniRef100 entry
Length = 182
Score = 37.0 bits (84), Expect = 0.61
Identities = 28/96 (29%), Positives = 49/96 (50%), Gaps = 8/96 (8%)
Query: 81 KSSPEEEIVVQHS--NSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENIQILD 138
K S E+E+ ++ + D++ R+LA + Q + ++ N EN+ ILD
Sbjct: 8 KKSDEKELTLEEKLKETEDKLLRSLAEIENQRSRFEKDIKEAINFG-GFNFAKENLAILD 66
Query: 139 NLNRS----QNNPDL-ETMNAGKFLDLLPAVEKAVV 169
NL R+ +N+PDL + + KFL + +EK +V
Sbjct: 67 NLQRAHTSIKNDPDLKDNKDIDKFLKNIEIIEKDLV 102
>UniRef100_UPI000033F052 UPI000033F052 UniRef100 entry
Length = 175
Score = 36.6 bits (83), Expect = 0.80
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 9/78 (11%)
Query: 74 IKKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVEN 133
I+K+ S EE+ S N+ +K+N ++ TL EK + SNV +NL+ +N
Sbjct: 7 IEKSDIVNLSKNEELFSNESFENNNLKQNKSLSTL----EKTSENSNVKFEVNLDKNKDN 62
Query: 134 -----IQILDNLNRSQNN 146
+ I DNL+ + NN
Sbjct: 63 SNQSILDIGDNLHSTINN 80
>UniRef100_Q8I1N6 Hypothetical protein PFD0985w [Plasmodium falciparum]
Length = 2148
Score = 36.6 bits (83), Expect = 0.80
Identities = 32/139 (23%), Positives = 59/139 (42%), Gaps = 28/139 (20%)
Query: 28 IMLRFRPIAPKPVTGEQTPANQSTTASLPGKRTKRKYVRI--------------RRNNGY 73
++++F + K ++ P N++ + +L K KY+ + +RNN
Sbjct: 317 LIIKFDKVNRKAYIVKRVPVNKNISFNL-AKFIAEKYINMLRKNLRKREIKMERKRNNNI 375
Query: 74 IKKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVEN 133
+ + + SS +E I N+ND + N + + +N D N N+N +N
Sbjct: 376 LNSRTKRGSSKKEHIESNMENNNDNINNN---------DDDNNSNNNDDNNDNINNNNDN 426
Query: 134 I----QILDNLNRSQNNPD 148
I ++N N S NN D
Sbjct: 427 INNNNDNINNNNNSNNNDD 445
>UniRef100_UPI00002BCF4E UPI00002BCF4E UniRef100 entry
Length = 188
Score = 36.2 bits (82), Expect = 1.0
Identities = 28/97 (28%), Positives = 45/97 (45%), Gaps = 6/97 (6%)
Query: 75 KKNSCKKSSPEEEIVVQHSNSNDEMKRNLAVVTLQLMPEKAAATSNVDLNLNLNLTVENI 134
+K K EE I + N+ D++ R+LA + Q + +D N EN+
Sbjct: 10 EKKESKDKKKEESIEDRLKNTEDKLLRSLAELENQRSRFEKEIKEAIDFG-GFNFARENL 68
Query: 135 QILDNLNRS----QNNPDL-ETMNAGKFLDLLPAVEK 166
LDNL R+ +N+P L + + KFL + +EK
Sbjct: 69 STLDNLQRAYNSIKNDPALKDNKDLDKFLKNIDIIEK 105
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.132 0.398
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 495,324,531
Number of Sequences: 2790947
Number of extensions: 20107005
Number of successful extensions: 64001
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 63898
Number of HSP's gapped (non-prelim): 123
length of query: 292
length of database: 848,049,833
effective HSP length: 126
effective length of query: 166
effective length of database: 496,390,511
effective search space: 82400824826
effective search space used: 82400824826
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC138464.11


