

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138452.7 + phase: 0 /pseudo
(677 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
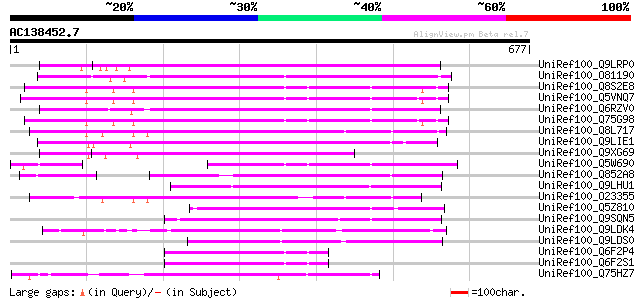
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LRP0 Emb|CAB39405.1 [Arabidopsis thaliana] 311 5e-83
UniRef100_O81190 Putative transposase [Arabidopsis thaliana] 293 1e-77
UniRef100_Q8S2E8 P0022F10.3 protein [Oryza sativa] 291 3e-77
UniRef100_Q5VNQ7 HAT dimerisation domain-containing protein-like... 290 1e-76
UniRef100_Q6RZV0 Transposase-like protein [Musa acuminata] 283 9e-75
UniRef100_Q75G98 Hypothetical protein OSJNBa0042F15.17 [Oryza sa... 282 3e-74
UniRef100_Q8L717 Hypothetical protein At4g15020 [Arabidopsis tha... 279 2e-73
UniRef100_Q9LIE1 Transposase-like protein [Arabidopsis thaliana] 254 6e-66
UniRef100_Q9XG69 Hypothetical protein [Nicotiana tabacum] 242 2e-62
UniRef100_Q5W690 Hypothetical protein OSJNBa0076A09.5 [Oryza sat... 221 7e-56
UniRef100_Q852A8 Hypothetical protein OSJNBb0081B07.17 [Oryza sa... 213 1e-53
UniRef100_Q9LHU1 Similar to Arabidopsis thaliana chromosome 1 [O... 206 1e-51
UniRef100_O23355 Hypothetical protein AT4g15020 [Arabidopsis tha... 202 3e-50
UniRef100_Q5Z810 HAT dimerisation domain-containing protein-like... 194 9e-48
UniRef100_Q9SQN5 Hypothetical protein F19K16.28 [Arabidopsis tha... 191 6e-47
UniRef100_Q9LDK4 Transposase-like protein [Arabidopsis thaliana] 190 1e-46
UniRef100_Q9LDS0 Transposase-like protein [Arabidopsis thaliana] 184 7e-45
UniRef100_Q6F2P4 Hypothetical protein OSJNBa0075J06.6 [Oryza sat... 167 7e-40
UniRef100_Q6F2S1 Hypothetical protein OSJNBa0060G17.6 [Oryza sat... 164 8e-39
UniRef100_Q75HZ7 Hypothetical protein OSJNBb0035N21.20 [Oryza sa... 164 1e-38
>UniRef100_Q9LRP0 Emb|CAB39405.1 [Arabidopsis thaliana]
Length = 883
Score = 311 bits (796), Expect = 5e-83
Identities = 185/559 (33%), Positives = 296/559 (52%), Gaps = 36/559 (6%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDV-----K 93
DP WE+ I+ + ++V+C +C +G R K HLA +VA C + P +V +
Sbjct: 133 DPGWEHGIAQDERKKKVKCNYCNKIVSGGINRFKQHLARIPGEVAPCKTAPEEVYVKIKE 192
Query: 94 KLMQDKVNKLQKKLLEKANLIVV-----DADGDKDGA------ADADLEIISGGR---GK 139
+ + K Q + ++ + D D ++D D ++ GR K
Sbjct: 193 NMKWHRAGKRQNRPDDEMGALTFRTVSQDPDQEEDREDHDFYPTSQDRLMLGNGRFSKDK 252
Query: 140 RKSAGEIAHKNMLKE-------------DSLR*NKAQSMVCGRRSTGKMLVRLLLSFFYN 186
RKS +++ + S + K S R + K + + F ++
Sbjct: 253 RKSFDSTNMRSVSEAKTKRARMIPFQSPSSSKQRKLYSSCSNRVVSRKDVTSSISKFLHH 312
Query: 187 NAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEK 246
+ A S + KM EL+ +G GF S + L I L +R+ W
Sbjct: 313 VGVPTEAANSLYFQKMIELIGMYGEGFVVPSSQLFSGRLLQEEMSTIKSYLREYRSSWVV 372
Query: 247 CGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGE 306
GC IM D WT+ + +++FLV P+G +F SID +DI + A +FK +D +V++IGE
Sbjct: 373 TGCSIMADTWTNTEGKKMISFLVSCPRGVYFHSSIDATDIVEDALSLFKCLDKLVDDIGE 432
Query: 307 ENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGK 366
ENVVQV+T N A +++AG+LL EKR +LYWTPCA HC +L+LEDF K+ ++ + K +
Sbjct: 433 ENVVQVITQNTAIFRSAGKLLEEKRKNLYWTPCAIHCTELVLEDF-SKLEFVSECLEKAQ 491
Query: 367 TITTYIYGRTSLISLL-HKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEW 425
IT +IY +T L++L+ ++FT +D +RPA+ A+ + TL L +H+ L LF+S+ W
Sbjct: 492 RITRFIYNQTWLLNLMKNEFTQGLDLLRPAVMRHASGFTTLQSLMDHKASLRGLFQSDGW 551
Query: 426 KTSK-LAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMV-DWEEKAAMGYIYEE 483
S+ AKS++G+ V+ +VLS VFWK V L+ P+++V+ M+ D ++ +M Y Y
Sbjct: 552 ILSQTAAKSEEGREVEKMVLSAVFWKKVQYVLKSVDPVMQVIHMINDGGDRLSMPYAYGY 611
Query: 484 MDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFTVDDE 543
M AK I++ S AR Y P W++I+ RW+ H PL+ A F NP +Y P+F E
Sbjct: 612 MCCAKMAIKSIHSDDARKYGPFWRVIEYRWNPLFHHPLYVAAYFFNPAYKYRPDFMAQSE 671
Query: 544 IVNGMYACLRRMVADAEKR 562
+V G+ C+ R+ D +R
Sbjct: 672 VVRGVNECIVRLEPDNTRR 690
Score = 55.1 bits (131), Expect = 7e-06
Identities = 23/69 (33%), Positives = 39/69 (56%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKKLMQD 98
DP WE+ ++ + ++V+C +C +G YR+K HLA S +V C P +V M++
Sbjct: 11 DPGWEHGVAQDQRKKKVKCNYCGKIVSGGIYRLKQHLARVSGEVTYCDKSPEEVCMRMKE 70
Query: 99 KVNKLQKKL 107
+ + KKL
Sbjct: 71 NLVRSTKKL 79
>UniRef100_O81190 Putative transposase [Arabidopsis thaliana]
Length = 729
Score = 293 bits (750), Expect = 1e-77
Identities = 180/550 (32%), Positives = 285/550 (51%), Gaps = 19/550 (3%)
Query: 37 KTDPAWEYAIST--EPGSRRVQCKFCQLAFTGSAY-RVKHHLAGTSRDVAVCPSVPLDVK 93
K+D AW Y I + E G ++C FC G R+KHHLAG + C + D++
Sbjct: 22 KSDIAWSYVIQSKDEKGKTVLECAFCHKKKRGGGINRMKHHLAGVKGNTDACLKISADIR 81
Query: 94 KLMQDKVNKLQKKLLEKANLIVVDADGDKDGAADAD---LEIISGGRGKRKSAGEIAH-- 148
+ + + + + K +K N+++ D + D D D + + KRK G H
Sbjct: 82 FKIVNALKEAENK--KKQNIVLDDTNLDGPEIEDVDGDDIRVDVRPSQKRKKQGIDLHDY 139
Query: 149 -KNMLKEDSLR*NKAQSMVCGRRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVA 207
K + + + KA R M V L +FY+ I N S + M +VA
Sbjct: 140 FKRGVHDQTQPSIKACMQSKERIHAVDMSVAL---WFYDACIPMNAVNSPLFQPMMSMVA 196
Query: 208 KHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNF 267
G G+ SY +R L ++ ++ ++ W GC +M DGW D R+R ++NF
Sbjct: 197 SMGHGYVGPSYHALRVGLLRDAKLQVSLIIDKFKSSWASTGCTLMADGWKDTRQRPLINF 256
Query: 268 LVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELL 327
LV+ PKG FLKS+D SDI +A+ + + ++V IG EN+V VTD+A NYKAAG+LL
Sbjct: 257 LVYCPKGITFLKSVDASDIYASAENLCNLFAELVGIIGSENIVHFVTDSAPNYKAAGKLL 316
Query: 328 MEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTN 387
+EK P + W+PC+AHCI+L+LED K +H + + +T ++Y ++ + K +
Sbjct: 317 VEKFPTIAWSPCSAHCINLILEDVAKLPHVH-HIVRRMSKVTIFVYNHKPALNWVRKRSG 375
Query: 388 DVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKV 447
+ IRP T FAT+++ L L +H+ L L S + + +L K+ K+ ++V+L +
Sbjct: 376 WREIIRPGETRFATTFIALQSLYQHKEDLQALVTSADPELKQLFKTSKAKVAKSVILDER 435
Query: 448 FWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWK 507
W D L ++ P++++L + D +EK ++ Y+YE M RA+ I+ F + Y P
Sbjct: 436 MWNDCLIIVKVMTPIIRLLRICDADEKPSLPYVYEGMYRARLGIKNIFQEKETLYKPYTN 495
Query: 508 IIDDRWDRQLHRPLHAAGLFLNPMLRY-APNFTVDDEIVNGMYACLRRMVADAEKRKKKL 566
IID RWDR L LHAA +LNP Y P F E+++G+ + D+ K KL
Sbjct: 496 IIDRRWDRMLRHDLHAAAYYLNPAFMYDQPTFCEKPEVMSGLMNLFEKQKNDS---KTKL 552
Query: 567 ISNLRISTQR 576
LR+ +R
Sbjct: 553 FQELRVYRER 562
>UniRef100_Q8S2E8 P0022F10.3 protein [Oryza sativa]
Length = 965
Score = 291 bits (746), Expect = 3e-77
Identities = 187/588 (31%), Positives = 289/588 (48%), Gaps = 39/588 (6%)
Query: 20 ASSSAAARVRPKNAKGNKTDPAWEYAISTEPGSRR-VQCKFCQLAFTGSAYRVKHHLAGT 78
+++S + PK A N DPAW + + + ++CK+C T R+K+HLAG
Sbjct: 171 SAASVSVEAGPKQAGINSDDPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKYHLAGI 230
Query: 79 SR-DVAVCPSVPLDVKKLMQD--------KVNKLQKKLLEKANLIVVDADGDK--DGAAD 127
+ C VP V + M D K K ++K + +A + + ++D + +G+
Sbjct: 231 KGFNTTKCQKVPSPVHQEMFDLLTKKTSEKEQKNKEKEVARAQVDIENSDCESGSEGSDH 290
Query: 128 ADLEII------SGGRGKRKSAGEIAHKNMLKEDSLR*------------NKAQSMVCG- 168
+ ++ +G R AG K S+ NK Q+ +
Sbjct: 291 GNNVLVVKPKETTGSSSSRSVAGGHTIDKYYKPPSIEESASMTQRVIKLSNKVQTALTTQ 350
Query: 169 -RRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLN 227
R + +FY +I N + M E + + G K S E+ +L
Sbjct: 351 KREERRNRTCEYICQWFYEASIPHNTVTLPSFAHMLEAIGQFGRSLKGPSPYEMSGSFLQ 410
Query: 228 YFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDIT 287
++ + H+ WE GC IMTD WTDR+ R ++N +VHS G FL S++ S
Sbjct: 411 KRKEKVMDGFKEHKESWELTGCSIMTDAWTDRKGRGVMNLVVHSAHGVLFLDSVECSGDR 470
Query: 288 KTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLM 347
K IFK++D +EEIGE++VVQVVTDNA+ A LL KRP ++W CAAHC+DLM
Sbjct: 471 KDGKYIFKLVDRYIEEIGEQHVVQVVTDNASVNTIAASLLTAKRPSIFWNGCAAHCLDLM 530
Query: 348 LEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLG 407
LED K P+ + I+ + +T ++Y T ++ L+ KF N D +R +T FAT+YL L
Sbjct: 531 LEDIGKLGPVE-ETIANARQVTVFLYAHTRVLDLMRKFLNR-DLVRSGVTRFATAYLNLK 588
Query: 408 CLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLC 467
L +++ +L+ LFKS+E + K GK V+ S+ FWK+V + PL VL
Sbjct: 589 SLLDNKKELVRLFKSDEMEQLGYLKQTKGKKASKVIRSETFWKNVDIAVNYFEPLANVLR 648
Query: 468 MVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLF 527
+D + +MG+ + M AK++I F + +W IID RWD +L PLH AG +
Sbjct: 649 RMD-SDVPSMGFFHGLMLEAKKEISQRFDNDKSRFIEVWDIIDKRWDNKLKTPLHLAGYY 707
Query: 528 LNPMLRYAPN---FTVDDEIVNGMYACLRRMVADAEKRKKKLISNLRI 572
LNP Y PN D G+ +C+ ++V D E + K+I L +
Sbjct: 708 LNPYYYYYPNKQEIESDGSFRAGVISCIDKLV-DDEDIQDKIIEELNL 754
>UniRef100_Q5VNQ7 HAT dimerisation domain-containing protein-like [Oryza sativa]
Length = 622
Score = 290 bits (741), Expect = 1e-76
Identities = 188/593 (31%), Positives = 292/593 (48%), Gaps = 40/593 (6%)
Query: 15 SSAPVASSSAAARVRPKNAKGNKTDPAWEYAISTEPGSRR-VQCKFCQLAFTGSAYRVKH 73
S P +++S + PK A N DPAW + + + ++CK+C T R+K+
Sbjct: 5 SGNPSSAASVSVEAGPKQAGINSDDPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKY 64
Query: 74 HLAGTSR-DVAVCPSVPLDVKKLMQD--------KVNKLQKKLLEKANLIVVDADGDK-- 122
HLAG + C VP V++ M D K K ++K + +A + + ++D +
Sbjct: 65 HLAGIKGFNTTKCQKVPSPVQQEMFDLLTKKTSEKEQKNKEKEVARAEVDIENSDCESGS 124
Query: 123 DGAADADLEII------SGGRGKRKSAGEIAHKNMLKEDSLR*------------NKAQS 164
+G+ + ++ +G R AG K S+ NK Q+
Sbjct: 125 EGSDHGNNVLVVKPKETTGSSSSRSVAGGHTIDKYYKPPSIEESASMTQRVIKLSNKVQT 184
Query: 165 MVCG--RRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIR 222
+ R + +FY +I N + M E + + G K S E+
Sbjct: 185 TLTTQKREERRNRTCEYICQWFYEASIPHNTVTLPSFAHMLEAIGQFGRSLKGPSPYEMS 244
Query: 223 NKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSID 282
+L ++ + H+ WE GC IMTD WTDR+ R ++N +VHS G FL S++
Sbjct: 245 GSFLQKRKEKVMDGFKEHKESWELTGCSIMTDAWTDRKGRGVMNLVVHSAHGVLFLDSVE 304
Query: 283 PSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAH 342
S K IF+++D +EEIGE++VVQVVTDNA+ A LL KRP ++W CAAH
Sbjct: 305 CSGDRKDGKYIFELVDRYIEEIGEQHVVQVVTDNASVNTTAASLLTAKRPSIFWNGCAAH 364
Query: 343 CIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATS 402
C+DLMLED K P+ + I+ + +T ++Y T ++ L+ KF N D +R +T FAT+
Sbjct: 365 CLDLMLEDIGKLGPVE-ETIANARQVTVFLYAHTRVLDLMRKFLNR-DLVRSGVTRFATA 422
Query: 403 YLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPL 462
YL L L +++ +L+ LFKSNE + K GK V++S+ FWK+V + PL
Sbjct: 423 YLNLKSLLDNKKELVRLFKSNEMEQLGYLKQAKGKKASKVIISETFWKNVDIAVNYFEPL 482
Query: 463 VKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLH 522
VL +D + +MG+ + M AK++I F + +W IID RWD +L LH
Sbjct: 483 ANVLRRMD-SDVPSMGFFHGLMLEAKKEISQRFDNDKSRFIEVWDIIDKRWDNKLKTLLH 541
Query: 523 AAGLFLNPMLRYAPN---FTVDDEIVNGMYACLRRMVADAEKRKKKLISNLRI 572
AG +LNP Y PN D G+ +C+ ++V D E + K+I L +
Sbjct: 542 LAGYYLNPYY-YYPNKQEIESDGSFRAGVISCIDKLV-DDEDIQDKIIEELNL 592
>UniRef100_Q6RZV0 Transposase-like protein [Musa acuminata]
Length = 670
Score = 283 bits (725), Expect = 9e-75
Identities = 168/529 (31%), Positives = 276/529 (51%), Gaps = 14/529 (2%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGT-SRDVAVCPSVPLDVKKLMQ 97
D WE+ + + ++V+C +C F+G YR+K HLA ++D+ C VP DV+ L+
Sbjct: 6 DACWEHCVLVDATRQKVRCNYCHREFSGGVYRMKFHLAQIKNKDIVPCSEVPDDVRNLIH 65
Query: 98 DKVNKLQKKLLEKANLIVVDADGDKDGAADADLEIISGGRGKRKSAGEIAHKNMLKEDSL 157
+ +K+ K I A+G + ++ A + G G +L S
Sbjct: 66 SILTTPRKQKAPKKLKIDHTANGPQHSSSSAS-GYNAKNAGSSGQHGSTCPSLLLPLPSP 124
Query: 158 ----R*NKAQSMVCGRRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGF 213
N AQ L FF++N+I F+ ++S Y M + +A G G+
Sbjct: 125 GAQPTANDAQKQKYDNADNKIAL------FFFHNSIPFSASKSIYYQAMIDAIADCGAGY 178
Query: 214 KPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPK 273
KP +Y+ +R+ L EI ++ + W+ GC I++D W+D R +++L V SPK
Sbjct: 179 KPPTYEGLRSTLLEKVKEEINENHRKLKDEWKDTGCTILSDNWSDGRSKSLLVLSVASPK 238
Query: 274 GTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPH 333
GT FLK +D S A +F+++D V+ E+G ENVVQV+TD+A +Y A LL++K P
Sbjct: 239 GTQFLKLVDISSRADDAYYLFELLDSVIMEVGAENVVQVITDSATSYTYAAGLLLKKYPS 298
Query: 334 LYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIR 393
L+W PCA++ I+ MLED K+ + + + +TI +I ++SL+ K T + +R
Sbjct: 299 LFWFPCASYSIEKMLEDI-SKLEWVSTTLEETRTIARFICSDGWILSLMKKLTGGRELVR 357
Query: 394 PAMTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVL 453
P + F T +LTL + ++ L F +W +S ++ D +++++ + FWK
Sbjct: 358 PKVARFMTHFLTLRSIVNQEDDLKHFFSHADWLSSVHSRRPDALAIKSLLYLERFWKSAH 417
Query: 454 NCLRGAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRW 513
+ + PL+K+L +VD + AMGYIYE ++RAK I+ + G Y + +II+ RW
Sbjct: 418 EIIGMSEPLLKLLRLVD-GDMPAMGYIYEGIERAKMAIKAFYKGCEEKYMSVLEIIERRW 476
Query: 514 DRQLHRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEKR 562
H LHAA FLNP + Y P+F D + NG +A + +M + R
Sbjct: 477 SMHCHSHLHAAAAFLNPSIFYDPSFKFDVNMRNGFHAAMWKMFPEENDR 525
>UniRef100_Q75G98 Hypothetical protein OSJNBa0042F15.17 [Oryza sativa]
Length = 1005
Score = 282 bits (721), Expect = 3e-74
Identities = 183/588 (31%), Positives = 289/588 (49%), Gaps = 40/588 (6%)
Query: 20 ASSSAAARVRPKNAKGNKTDPAWEYAISTEPGSRR-VQCKFCQLAFTGSAYRVKHHLAGT 78
+++S + PK A N DPAW + + + ++CK+C T R+K+HLAG
Sbjct: 216 SAASVSVEAGPKQAGINSDDPAWAHCFCPDITKKHHLRCKYCDKVCTAGITRIKYHLAGI 275
Query: 79 SR-DVAVCPSVPLDVKKLMQD--------KVNKLQKKLLEKANLIVVDADGDK--DGAAD 127
+ C VP V++ M D K K ++K + +A + + ++D + +G+
Sbjct: 276 KGFNTTKCQKVPSPVQQEMFDLLTKKTSEKEQKNKEKEVARAEVDIENSDCESGSEGSDH 335
Query: 128 ADLEII------SGGRGKRKSAGEIAHKNMLKEDSLR*------------NKAQSMVCG- 168
+ ++ +G R AG K S+ NK Q+ +
Sbjct: 336 GNNVLVVKPKETTGSSSSRSVAGGHTIDKYYKPPSIEESASMTQRVIKLSNKVQTALTTQ 395
Query: 169 -RRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLN 227
R + +FY +I N + M E + + G K S E+ +L
Sbjct: 396 KREERRNRTCEYICQWFYEASIPHNTVTLPSFAHMLEAIGQFGRSLKGPSPYEMSGSFLQ 455
Query: 228 YFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDIT 287
++ + H+ WE GC IMTD WTDR+ R ++N +VHS G FL S++ S
Sbjct: 456 KRKEKVMDGFKEHKESWELTGCSIMTDAWTDRKGRGVMNLVVHSAHGVLFLDSVECSGDR 515
Query: 288 KTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLM 347
K IF+++D +EEIGE++VVQVVTDNA+ A LL KRP ++W AAHC+DLM
Sbjct: 516 KDGKYIFELVDRYIEEIGEQHVVQVVTDNASVNTTAASLLTAKRPSIFWNGYAAHCLDLM 575
Query: 348 LEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLG 407
LED K P+ + I+ + +T ++Y T ++ L+ KF N D +R +T FAT+YL L
Sbjct: 576 LEDIGKLGPVE-ETIANARQVTVFLYAHTRVLDLMRKFLNR-DLVRSGVTRFATAYLNLK 633
Query: 408 CLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLC 467
L +++ +L+ LFKS+E + K GK V+ S+ FWK+V + PL VL
Sbjct: 634 SLLDNKKELVRLFKSDEMEQLGYLKQAKGKKASKVIRSETFWKNVDIAVNYFEPLANVLR 693
Query: 468 MVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLF 527
+D + +MG+ + M AK++I F + +W IID RWD ++ PLH AG +
Sbjct: 694 RMD-SDVPSMGFFHGLMLEAKKEISQRFDNDKSRFIEVWDIIDKRWDNKIKAPLHLAGYY 752
Query: 528 LNPMLRYAPN---FTVDDEIVNGMYACLRRMVADAEKRKKKLISNLRI 572
LNP Y PN D G+ +C+ +++ D E + K+I L +
Sbjct: 753 LNPYY-YFPNKQEIESDGSFRAGVISCIDKLI-DDEDIQDKIIEELNL 798
>UniRef100_Q8L717 Hypothetical protein At4g15020 [Arabidopsis thaliana]
Length = 768
Score = 279 bits (714), Expect = 2e-73
Identities = 183/586 (31%), Positives = 298/586 (50%), Gaps = 45/586 (7%)
Query: 26 ARVRPKNAKGNKTDPAWEYAISTEPGSR-RVQCKFCQLAFTGSAY-RVKHHLAGTSRDVA 83
A + P K D AW++ + G R +++C +C+ F G RVK HLAG
Sbjct: 3 AELEPVALTPQKQDNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVKEHLAGKKGQGT 62
Query: 84 VCPSVPLDVKKLMQD----------KVNKLQKKLLEKANLIVVDAD----------GDKD 123
+C VP DV+ +Q K +K + L A+L ++ D G K
Sbjct: 63 ICDQVPEDVRLFLQQCIDGTVRRQRKRHKSSSEPLSVASLPPIEGDMMVVQPDVNDGFKS 122
Query: 124 -GAADADLEIISGGRGKRKSAGEIAHKNMLKEDSLR*N------KAQSMVCGRRSTGKML 176
G++D ++ S G+ K + KN + S N +++ S+ K +
Sbjct: 123 PGSSDVVVQNESLLSGRTKQRTYRSKKNAFENGSASNNVDLIGRDMDNLIPVAISSVKNI 182
Query: 177 VR------------LLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNK 224
V + F + F+ S + M + +A G G ++D++R
Sbjct: 183 VHPSFRDRENTIHMAIGRFLFGIGADFDAVNSVNFQPMIDAIASGGFGVSAPTHDDLRGW 242
Query: 225 YLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPS 284
L E+ K+++ +A+W++ GC I+ + + +LNFLV+ P+ FLKS+D S
Sbjct: 243 ILKNCVEEMAKEIDECKAMWKRTGCSILVEELNSDKGFKVLNFLVYCPEKVVFLKSVDAS 302
Query: 285 DITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCI 344
++ +ADK+F+++ ++VEE+G NVVQV+T Y AG+ LM P LYW PCAAHCI
Sbjct: 303 EVLSSADKLFELLSELVEEVGSTNVVQVITKCDDYYVDAGKRLMLVYPSLYWVPCAAHCI 362
Query: 345 DLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYL 404
D MLE+F K+ ++ I + + IT ++Y + +++L+ KFT+ D + PA + AT++
Sbjct: 363 DQMLEEF-GKLGWISETIEQAQAITRFVYNHSGVLNLMWKFTSGNDILLPAFSSSATNFA 421
Query: 405 TLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVK 464
TLG + E ++ L + S EW ++ G +V N + + FWK V PL++
Sbjct: 422 TLGRIAELKSNLQAMVTSAEWNECSYSEEPSG-LVMNALTDEAFWKAVALVNHLTSPLLR 480
Query: 465 VLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAA 524
L +V E++ AMGY+Y + RAK+ I+T+ + Y WKIID W++Q H PL AA
Sbjct: 481 ALRIVCSEKRPAMGYVYAALYRAKDAIKTHLVNR-EDYIIYWKIIDRWWEQQQHIPLLAA 539
Query: 525 GLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRKKKLISNL 570
G FLNP L Y N + E++ + C+ R+V D +K + K+I L
Sbjct: 540 GFFLNPKLFYNTNEEMRSELILSVLDCIERLVPD-DKIQDKIIKEL 584
>UniRef100_Q9LIE1 Transposase-like protein [Arabidopsis thaliana]
Length = 761
Score = 254 bits (649), Expect = 6e-66
Identities = 162/559 (28%), Positives = 275/559 (48%), Gaps = 42/559 (7%)
Query: 37 KTDPAWEYAISTEPGSR-RVQCKFCQLAFTGSAY-RVKHHLAGTSRDVAVCPSVPLDVKK 94
K D AW++ + G R +++C +C+ F G RVK HLAG +C VP +V+
Sbjct: 14 KQDSAWKHCEVYKYGDRVQMRCLYCRKMFKGGGITRVKEHLAGKKGQGTICDQVPDEVRL 73
Query: 95 LMQDKVN---KLQKKL---------------LEKANLIVVDADGDKDGAADADLEIISGG 136
+Q ++ + Q+K E + +D + + + ++
Sbjct: 74 FLQQCIDGTVRRQRKRRKSSPEPLPIAYFPPCEVETQVAASSDVNNGFKSPSSDVVVGQS 133
Query: 137 RGKRKSAGEIAHKNMLKEDS----------------LR*NKAQSMVCGRRSTGKMLVRLL 180
G+ K + KN E + + + +++V + V +
Sbjct: 134 TGRTKQRTYRSRKNNAFERNDLANVEVDRDMDNLIPVAISSVKNIVHPTSKEREKTVHMA 193
Query: 181 LS-FFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEA 239
+ F ++ F+ A S + + G G +++++R L E+ K+++
Sbjct: 194 MGRFLFDIGADFDAANSVNVQPFIDAIVSGGFGVSIPTHEDLRGWILKSCVEEVKKEIDE 253
Query: 240 HRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDD 299
+ +W++ GC ++ IL FLV+ P+ FLKS+D S+I + DK+++++ +
Sbjct: 254 CKTLWKRTGCSVLVQELNSNEGPLILKFLVYCPEKVVFLKSVDASEILDSEDKLYELLKE 313
Query: 300 VVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHT 359
VVEEIG+ NVVQV+T +Y AAG+ LM+ P LYW PCAAHCID MLE+F K+
Sbjct: 314 VVEEIGDTNVVQVITKCEDHYAAAGKKLMDVYPSLYWVPCAAHCIDKMLEEF-GKMDWIR 372
Query: 360 DAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDL 419
+ I + +T+T IY + +++L+ KFT D ++P T AT++ T+G + + + L +
Sbjct: 373 EIIEQARTVTRIIYNHSGVLNLMRKFTFGNDIVQPVCTSSATNFTTMGRIADLKPYLQAM 432
Query: 420 FKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGY 479
S+EW +K G + + + FWK + P+++VL +V E K AMGY
Sbjct: 433 VTSSEWNDCSYSKEAGGLAMTETINDEDFWKALTLANHITAPILRVLRIVCSERKPAMGY 492
Query: 480 IYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFT 539
+Y M RAKE I+TN + Y WKIID W L +PL+AAG +LNP Y+ +
Sbjct: 493 VYAAMYRAKEAIKTNLA-HREEYIVYWKIIDRWW---LQQPLYAAGFYLNPKFFYSIDEE 548
Query: 540 VDDEIVNGMYACLRRMVAD 558
+ EI + C+ ++V D
Sbjct: 549 MRSEIHLAVVDCIEKLVPD 567
>UniRef100_Q9XG69 Hypothetical protein [Nicotiana tabacum]
Length = 593
Score = 242 bits (618), Expect = 2e-62
Identities = 140/448 (31%), Positives = 240/448 (53%), Gaps = 37/448 (8%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKKLMQD 98
DP WE+ + + ++V+C +C+ +G R K HLA +VA C S P +V +++
Sbjct: 135 DPGWEHGVPQDERKKKVKCNYCEKIISGGINRFKQHLARIPGEVAPCKSAPEEVYLRIKE 194
Query: 99 KVN------KLQKKLLEKANLIVVDADGDKD--------------------GAADADLEI 132
+ + ++ ++ + +++D +++ G D +
Sbjct: 195 NMKWHRTGRRHRRPHTKELSSFYMNSDNEEEDEDQEEEALHHHMSNEKLLIGDKRLDRDC 254
Query: 133 ISGGRGKRKSAGEIAHKNMLKEDSLR*NKAQSM---------VCGRRSTGKMLVRLLLSF 183
+G G + K ++L + +S+ VC + + K ++ + F
Sbjct: 255 RRSFKGMSPGIGSESLLKRPKYETLGTKEPKSLFQASGKHVKVCSNKKSRKEVISSICKF 314
Query: 184 FYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAV 243
FY+ IS + A S + KM ELV ++G G S + + L I L ++A
Sbjct: 315 FYHAGISPHAASSPYFQKMLELVGQYGEGLVGPSSRVLSGRLLQDEIVSIRNYLSEYKAS 374
Query: 244 WEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEE 303
W G I+ D W D + RT++N LV P G +F+ S+D + + + A IFK++D VVE+
Sbjct: 375 WAVTGYSILADSWQDTQGRTLINVLVSCPHGMYFVCSVDATGVVEDATYIFKLLDRVVED 434
Query: 304 IGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAIS 363
+GEENVVQV+T N NY+AAG++L EKR +L+WTPCAA+CID +LEDF KI + +
Sbjct: 435 MGEENVVQVITQNTPNYQAAGKMLEEKRRNLFWTPCAAYCIDRILEDF-VKIKWVRECME 493
Query: 364 KGKTITTYIYGRTSLISLLHK-FTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKS 422
K + IT +IY L+SL+ K FT + ++P+ T +++++ T+ L +H+N L +F+S
Sbjct: 494 KAQKITKFIYNSFWLLSLMKKEFTAGQELLKPSFTRYSSTFATVQSLLDHRNGLKRMFQS 553
Query: 423 NEWKTSKLAKSKDGKIVQNVVLSKVFWK 450
N+W +S+ +K +DGK V+ +VL+ FW+
Sbjct: 554 NKWLSSRYSKLEDGKEVEKIVLNATFWR 581
Score = 54.7 bits (130), Expect = 9e-06
Identities = 24/70 (34%), Positives = 39/70 (55%), Gaps = 2/70 (2%)
Query: 39 DPAWEYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKKLMQD 98
DP WE+ ++ + ++V+C +C +G YR+K HLA S +V C P DV M++
Sbjct: 11 DPGWEHGVAQDERKKKVRCNYCGKVVSGGIYRLKQHLARVSGEVTYCDKAPEDVCLKMRE 70
Query: 99 KVN--KLQKK 106
+ +L KK
Sbjct: 71 NLEGCRLSKK 80
>UniRef100_Q5W690 Hypothetical protein OSJNBa0076A09.5 [Oryza sativa]
Length = 1030
Score = 221 bits (562), Expect = 7e-56
Identities = 121/327 (37%), Positives = 185/327 (56%), Gaps = 4/327 (1%)
Query: 259 RRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAA 318
RR R+++N + H +G F+ +ID S I+ ++ ++EIG + VVQVVTDNA+
Sbjct: 521 RRGRSLMNLVAHCARGMCFIDAIDASLEVHDGKYIYSLVSSCIDEIGPKKVVQVVTDNAS 580
Query: 319 NYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSL 378
N +A ++L K PH++WT CAAH IDLMLED K +H + I GK+IT +Y + L
Sbjct: 581 NNMSASKMLQVKHPHIFWTSCAAHYIDLMLEDIGKITMVH-NIIRDGKSITNLLYAQVRL 639
Query: 379 ISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKI 438
++++ +FT D +R T FATSYL L L + +N+L LF S +W S AK G+
Sbjct: 640 LAIMRQFTKG-DLVRAGTTRFATSYLNLKSLYDKRNELKQLFASQDWAKSSWAKKIKGQN 698
Query: 439 VQNVVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQ 498
N+V++ FW +L + PL VL VD + AMGYIY ++ +AK+ + +G
Sbjct: 699 AHNLVMNNKFWSQMLEVINYFEPLAYVLRRVD-GDVPAMGYIYGDLIKAKKDVAACLNGN 757
Query: 499 ARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFTV-DDEIVNGMYACLRRMVA 557
+ Y+ IWKIID RWD +L LH AG FLNP Y + +D ++ + C M
Sbjct: 758 EKKYSHIWKIIDARWDSKLKTTLHKAGYFLNPCFFYENKREIKEDFLMQAVVECATCMYR 817
Query: 558 DAEKRKKKLISNLRISTQRLENLVMTL 584
D + ++ L + T+ +++ T+
Sbjct: 818 DDITVQDICVAQLSLYTEAMDSFGTTM 844
Score = 54.3 bits (129), Expect = 1e-05
Identities = 35/101 (34%), Positives = 50/101 (48%), Gaps = 9/101 (8%)
Query: 2 ASTSNVNETSSVASS------APVASSSAAARVRPKNAKGNKTDPAWEYAI--STEPGSR 53
A+ + + TS +ASS + SSAA+ R + K DPAWE+A+ +
Sbjct: 329 AAAAELKSTSHLASSIRWPNISCCTMSSAASTARSSQSTSRK-DPAWEFALPLTGNETKN 387
Query: 54 RVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKK 94
+V C +C+ F G R+K HLAG C VP DVK+
Sbjct: 388 QVGCLYCKNYFKGGITRLKRHLAGVRGQSVYCTQVPDDVKE 428
>UniRef100_Q852A8 Hypothetical protein OSJNBb0081B07.17 [Oryza sativa]
Length = 779
Score = 213 bits (543), Expect = 1e-53
Identities = 124/382 (32%), Positives = 194/382 (50%), Gaps = 19/382 (4%)
Query: 183 FFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRA 242
F Y+ +S S + M E VA G + SY + R L E+ LE ++
Sbjct: 234 FLYDAGVSLEAVNSVYFQPMLEAVASAGGKPEAFSYHDFRGSILKKSLDEVTAQLEFYKG 293
Query: 243 VWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVE 302
W + GC ++ D WT R RT++NF V+ P+ D +++++ +VVE
Sbjct: 294 SWTRTGCTLLADEWTTDRGRTLINFSVYCPE-----------------DPLYELLKNVVE 336
Query: 303 EIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAI 362
E+GE+NVVQV+T+N+ + AG+ L E P L+W+ C+ CID MLEDF K+ + I
Sbjct: 337 EVGEKNVVQVITNNSEIHAVAGKRLCETFPTLFWSQCSFQCIDGMLEDF-SKVGAINEII 395
Query: 363 SKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKS 422
K IT +IY +L+ + + D + PA T A +++TL + ++ L + S
Sbjct: 396 CNAKVITGFIYNSAFAFNLMKRHLHGKDLLVPAETRAAMNFVTLKNMYNLKDSLEAMISS 455
Query: 423 NEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGYIYE 482
+EW L K G V N++ + FW +R PLV +L +V ++ +MGY+Y
Sbjct: 456 DEWIHYLLPKKPGGVEVTNLIGNLQFWSSCAAVVRITEPLVHLLKLVGSNKRPSMGYVYA 515
Query: 483 EMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFTVDD 542
+ +AK I+ + Y W IID RW++ RPLH AG FLNP+
Sbjct: 516 GLYQAKAAIKKELV-RKNDYMAYWDIIDWRWNKDAPRPLHLAGFFLNPLFFDGVRGGTSS 574
Query: 543 EIVNGMYACLRRMVADAEKRKK 564
EI +GM C+ R+V+D + + K
Sbjct: 575 EIFSGMLDCVERLVSDVKIQDK 596
Score = 48.5 bits (114), Expect = 6e-04
Identities = 31/103 (30%), Positives = 49/103 (47%), Gaps = 4/103 (3%)
Query: 13 VASSAPVASSSAAARVRPKNAKGNKTDPAWEYAISTEPGSR-RVQCKFCQLAFTGSA-YR 70
+AS A ++ V P A+ K DPAW++ R R++C +C F G +R
Sbjct: 1 MASEAEAEVAAGPEVVLPIGAQ--KHDPAWKHCQMVRSAGRVRLKCVYCHKHFLGGGIHR 58
Query: 71 VKHHLAGTSRDVAVCPSVPLDVKKLMQDKVNKLQKKLLEKANL 113
K HLA + CP VP +V++ M ++ + K K +L
Sbjct: 59 FKEHLANRPGNACCCPKVPREVQETMLHSLDAVAAKKKRKQSL 101
>UniRef100_Q9LHU1 Similar to Arabidopsis thaliana chromosome 1 [Oryza sativa]
Length = 521
Score = 206 bits (525), Expect = 1e-51
Identities = 110/354 (31%), Positives = 191/354 (53%), Gaps = 3/354 (0%)
Query: 210 GIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLV 269
G GF+ S + ++ K+L+ E+ + + W GC I+ D WTD + + ++NF V
Sbjct: 7 GQGFRGPSAEVLKTKWLHKLKSEVLQKTKEIEKDWATTGCTILADSWTDNKSKALINFSV 66
Query: 270 HSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLME 329
SP GTFFLK++D S K+ +++++ DDV+ E+G +NVVQ++TD NY + +L+M+
Sbjct: 67 SSPLGTFFLKTVDASPHIKS-HQLYELFDDVIREVGPDNVVQIITDRNINYGSVDKLIMQ 125
Query: 330 KRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDV 389
++W+PCA+ C++ ML+DF KI I + +TIT ++Y ++ L+ K
Sbjct: 126 NYNTIFWSPCASSCVNSMLDDF-SKIDWVNRCICQAQTITRFVYNNKWVLDLMRKCIAGQ 184
Query: 390 DSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFW 449
+ + +T + +LTL L H+ +L +F S+++ +S A ++ FW
Sbjct: 185 ELVCSGITKCVSDFLTLQSLLRHRPKLKQMFHSSDYASSSYANRSLSSSCVEILDDDEFW 244
Query: 450 KDVLNCLRGAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKII 509
+ V + PL++V+ V KAA+GYIYE M + + IRT + I+
Sbjct: 245 RAVEEIAAVSEPLLRVMRDV-LGGKAAIGYIYESMTKVMDSIRTYYIMDEGKCKSFLDIV 303
Query: 510 DDRWDRQLHRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRK 563
+ +W +LH PLH+A FLNP ++Y P I Y L +++ ++R+
Sbjct: 304 EQKWQVELHSPLHSAAAFLNPSIQYNPEVKFFTSIKEEFYHVLDKVLTVPDQRQ 357
>UniRef100_O23355 Hypothetical protein AT4g15020 [Arabidopsis thaliana]
Length = 522
Score = 202 bits (513), Expect = 3e-50
Identities = 152/542 (28%), Positives = 249/542 (45%), Gaps = 56/542 (10%)
Query: 26 ARVRPKNAKGNKTDPAWEYAISTEPGSR-RVQCKFCQLAFTGSAYRVKHHLAGTSRDVAV 84
A + P K D AW++ + G R +++C +C+ F G + RD
Sbjct: 3 AELEPVALTPQKQDNAWKHCEIYKYGDRLQMRCLYCRKMFKGGGITRVKDILLVKRDKQC 62
Query: 85 CPSVPLDVKKLMQDKVNKLQKKLLEKANLIVVDAD----------GDKD-GAADADLEII 133
+D Q K +K + L A+L ++ D G K G++D ++
Sbjct: 63 -----IDGTVRRQRKRHKSSSEPLSVASLPPIEGDMMVVQPDVNDGFKSPGSSDVVVQNE 117
Query: 134 SGGRGKRKSAGEIAHKNMLKEDSLR*N------KAQSMVCGRRSTGKMLVR--------- 178
S G+ K + KN + S N +++ S+ K +V
Sbjct: 118 SLLSGRTKQRTYRSKKNAFENGSASNNVDLIGRDMDNLIPVAISSVKNIVHPSFRDRENT 177
Query: 179 ---LLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGK 235
+ F + F+ S + M + +A G G ++D++R L E+ K
Sbjct: 178 IHMAIGRFLFGIGADFDAVNSVNFQPMIDAIASGGFGVSAPTHDDLRGWILKNCVEEMAK 237
Query: 236 DLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIFK 295
+++ +A+W++ GC I+ + + +LNFLV+ P+ FLKS+D S++ +ADK+F+
Sbjct: 238 EIDECKAMWKRTGCSILVEELNSDKGFKVLNFLVYCPEKVVFLKSVDASEVLSSADKLFE 297
Query: 296 MIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKI 355
++ ++VEE+G NVVQV+T Y AG+ LM P LYW PCAAHCID MLE+F K+
Sbjct: 298 LLSELVEEVGSTNVVQVITKCDDYYVDAGKRLMLVYPSLYWVPCAAHCIDQMLEEF-GKL 356
Query: 356 PLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQ 415
++ I + + IT ++Y + A + AT++ TLG + E ++
Sbjct: 357 GWISETIEQAQAITRFVYNHS------------------AFSSSATNFATLGRIAELKSN 398
Query: 416 LIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKA 475
L + S EW ++ G +V N + + FWK V PL++ L +V E++
Sbjct: 399 LQAMVTSAEWNECSYSEEPSG-LVMNALTDEAFWKAVALVNHLTSPLLRALRIVCSEKRP 457
Query: 476 AMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYA 535
AMGY+Y + RAK+ I+T+ + Y WKIID G FLNP L Y
Sbjct: 458 AMGYVYAALYRAKDAIKTHLVNR-EDYIIYWKIIDHGGSNNSIFLCLLLGFFLNPKLFYN 516
Query: 536 PN 537
N
Sbjct: 517 TN 518
>UniRef100_Q5Z810 HAT dimerisation domain-containing protein-like [Oryza sativa]
Length = 657
Score = 194 bits (492), Expect = 9e-48
Identities = 114/336 (33%), Positives = 178/336 (52%), Gaps = 11/336 (3%)
Query: 235 KDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTADKIF 294
+DL+ H WE GC ++ D W R ++ ++ LVH KG F+KS+D SDI D++
Sbjct: 200 RDLKKH---WEMNGCSVILDTWESRCGKSFISVLVHCSKGMLFIKSMDVSDIIDDVDELA 256
Query: 295 KMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKR--PHLYWTPCAAHCIDLMLEDFE 352
M+ VVEE+G N+VQV+T++ + Y A E + KR ++T CA HCI+L+LE+
Sbjct: 257 VMLFRVVEEVGVLNIVQVITNDESPYMQAAEHAVLKRYGYSFFFTLCADHCINLLLENI- 315
Query: 353 KKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHFATSYLTLGCLNEH 412
+ + + K + IT +IY + L K+ + + + F ++TLG L
Sbjct: 316 AALDHVNEVLIKAREITRFIYSHAVPMELKGKYIQGGEILSSSNLKFVAMFITLGKLVSE 375
Query: 413 QNQLIDLFKSNEWKTSKLAKSKDGKIVQNVV-LSKVFWKDVLNCLRGAFPLVKVLCMVDW 471
+ L+++F S EW +S LA + V VV FW + L+ PL+ VL ++
Sbjct: 376 RINLVEMFSSPEWASSDLASRSSFRHVYEVVKTDNAFWSAAADILKLTDPLITVLYKLE- 434
Query: 472 EEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLNPM 531
+ +G +Y+ MD AKE I+ N + Y W ++D+ WD LH P+HAAG LNP
Sbjct: 435 ADNCPIGILYDAMDCAKEDIKCNLRDKHGDY---WPMVDEIWDHYLHTPVHAAGYILNPR 491
Query: 532 LRYAPNFTVDDEIVNGMYACLRRMVADAEKRKKKLI 567
+ Y F+ D EI +G AC+ R+ + KK I
Sbjct: 492 IFYTERFSYDTEIKSGTNACVTRLAKNHYDPKKVAI 527
>UniRef100_Q9SQN5 Hypothetical protein F19K16.28 [Arabidopsis thaliana]
Length = 518
Score = 191 bits (485), Expect = 6e-47
Identities = 108/362 (29%), Positives = 187/362 (50%), Gaps = 7/362 (1%)
Query: 202 MFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRR 261
M + VAK G GF S + ++L+ +I L+ W GC I+ + WTD +
Sbjct: 1 MLDAVAKCGPGFVAPSP---KTEWLDRVKSDISLQLKDTEKEWVTTGCTIIAEAWTDNKS 57
Query: 262 RTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYK 321
R ++NF V SP FF KS+D S K + + + D V+++IG+E++VQ++ DN+ Y
Sbjct: 58 RALINFSVSSPSRIFFHKSVDASSYFKNSKCLADLFDSVIQDIGQEHIVQIIMDNSFCYT 117
Query: 322 AAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISL 381
L++ ++ +PCA+ C++++LE+F K+ IS+ + I+ ++Y + ++ L
Sbjct: 118 GISNHLLQNYATIFVSPCASQCLNIILEEF-SKVDWVNQCISQAQVISKFVYNNSPVLDL 176
Query: 382 LHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQN 441
L K T D IR +T +++L+L + + + +L +F E+ T+ N
Sbjct: 177 LRKLTGGQDIIRSGVTRSVSNFLSLQSMMKQKARLKHMFNCPEYTTN--TNKPQSISCVN 234
Query: 442 VVLSKVFWKDVLNCLRGAFPLVKVLCMVDWEEKAAMGYIYEEMDRAKEKIRTNFSGQARS 501
++ FW+ V + + P++KVL V K A+G IYE M +AKE IRT +
Sbjct: 235 ILEDNDFWRAVEESVAISEPILKVLREVS-TGKPAVGSIYELMSKAKESIRTYYIMDENK 293
Query: 502 YNPIWKIIDDRWDRQLHRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEK 561
+ I+D W LH PLHAA FLNP ++Y P + + L +++ ++
Sbjct: 294 HKVFSDIVDTNWCEHLHSPLHAAAAFLNPSIQYNPEIKFLTSLKEDFFKVLEKLLPTSDL 353
Query: 562 RK 563
R+
Sbjct: 354 RR 355
>UniRef100_Q9LDK4 Transposase-like protein [Arabidopsis thaliana]
Length = 605
Score = 190 bits (482), Expect = 1e-46
Identities = 139/536 (25%), Positives = 249/536 (45%), Gaps = 45/536 (8%)
Query: 43 EYAISTEPGSRRVQCKFCQLAFTGSAYRVKHHLAGTSRDVAVCPSVPLDVKK-----LMQ 97
E+ I + RV+C +C S +R+KHHL DV C V L +++ LM+
Sbjct: 10 EHGICVDKKKSRVKCNYCGKEMN-SFHRLKHHLGAVGTDVTHCDQVSLTLRETFRTMLME 68
Query: 98 DKVNKLQKKLLEKANLIVVDADGDKDGAADADLEIISGGRGKRKSAGEIAHKNMLKEDSL 157
DK K + D+ + D+ + +S +G A E+ ++++L
Sbjct: 69 DKSGYTTPKTKRVGKFQMADSRKRRK-TEDSSSKSVSPEQGN--VAVEVDNQDLLS---- 121
Query: 158 R*NKAQSMVCGRRSTGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIGFKPLS 217
+KAQ + GR FFY + + + S + +M + G+G K
Sbjct: 122 --SKAQKCI-GR-------------FFYEHCVDLSAVDSPCFKEMMMAL---GVGQKIPD 162
Query: 218 YDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFF 277
++ + L E+ ++ + W+ GC I+ D W D + +++F+ P G +
Sbjct: 163 SHDLNGRLLQEAMKEVQDYVKNIKDSWKITGCSILLDAWIDPKGHDLVSFVADCPAGPVY 222
Query: 278 LKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAA-GELLMEKRPHLYW 336
LKSID S + + +++ +VEE+G NV Q++ + + + G+L ++W
Sbjct: 223 LKSIDVSVVKNDVTALLSLVNGLVEEVGVHNVTQIIACSTSGWVGELGKLFSGHDREVFW 282
Query: 337 TPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVD-SIRPA 395
+ +HC +LML K + D + K TI +I S + + ++ D ++ +
Sbjct: 283 SVSLSHCFELMLVKIGK-MRSFGDILDKVNTIWEFINNNPSALKIYRDQSHGKDITVSSS 341
Query: 396 MTHFATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNC 455
F YL L + + + L +F S+ WK ++GK V N+V FW+ V
Sbjct: 342 EFEFVKPYLILKSVFKAKKNLAAMFASSVWK------KEEGKSVSNLVNDSSFWEAVEEI 395
Query: 456 LRGAFPLVKVLCMV-DWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWD 514
L+ PL L + + + +GYIY+ +D K I+ F+ + + Y +W +IDD W+
Sbjct: 396 LKCTSPLTDGLRLFSNADNNQHVGYIYDTLDGIKLSIKKEFNDEKKHYLTLWDVIDDVWN 455
Query: 515 RQLHRPLHAAGLFLNPMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRKKKLISNL 570
+ LH PLHAAG +LNP Y+ +F +D E+ +G+ +V A++ + K+ S L
Sbjct: 456 KHLHNPLHAAGYYLNPTSFYSTDFHLDPEVSSGL---THSLVHVAKEGQIKIASQL 508
>UniRef100_Q9LDS0 Transposase-like protein [Arabidopsis thaliana]
Length = 544
Score = 184 bits (467), Expect = 7e-45
Identities = 100/335 (29%), Positives = 165/335 (48%), Gaps = 9/335 (2%)
Query: 232 EIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKTAD 291
E+ +E + W GC I+ D W D++ R ++ F+ P G +L S D SD
Sbjct: 104 EVQDRVEKIKESWAITGCSILLDAWVDQKGRDLVTFVADCPAGLVYLISFDVSDFKDDVT 163
Query: 292 KIFKMIDDVVEEIGEENVVQVVTDNAANYKAA-GELLMEKRPHLYWTPCAAHCIDLMLED 350
+ +++ +VEE+G NV Q++ + + + GEL ++W+ +HC +LML
Sbjct: 164 ALLSLVNGLVEEVGVRNVTQIIACSTSGWVGELGELFAGHDREVFWSVSVSHCFELMLVK 223
Query: 351 FEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVD-SIRPAMTHFATSYLTLGCL 409
K I D K I +I S++++ + +D ++ + F T YL L +
Sbjct: 224 ISK-IRSFGDIFDKVNNIWLFINNNPSVLNIFRDQCHGIDITVSSSEFEFVTPYLILESI 282
Query: 410 NEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGAFPLVKVLCMV 469
+ + L +F S+ W + + N+V FW+ V + L+ PL+ L +
Sbjct: 283 FKAKKNLTAMFASSNWNNEQCIA------ISNLVSDSSFWETVESVLKCTSPLIHGLLLF 336
Query: 470 DWEEKAAMGYIYEEMDRAKEKIRTNFSGQARSYNPIWKIIDDRWDRQLHRPLHAAGLFLN 529
+GY+Y+ MD KE I F+ + + Y P+W +IDD W++ LH PLHAAG FLN
Sbjct: 337 STANNQHLGYVYDTMDSIKESIAREFNHKPQFYKPLWDVIDDVWNKHLHNPLHAAGYFLN 396
Query: 530 PMLRYAPNFTVDDEIVNGMYACLRRMVADAEKRKK 564
P Y+ NF +D E+V G+ + L MV D + K
Sbjct: 397 PTAFYSTNFHLDIEVVTGLISSLIHMVEDCHVQFK 431
>UniRef100_Q6F2P4 Hypothetical protein OSJNBa0075J06.6 [Oryza sativa]
Length = 217
Score = 167 bits (424), Expect = 7e-40
Identities = 86/215 (40%), Positives = 128/215 (59%), Gaps = 2/215 (0%)
Query: 202 MFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRR 261
M E + + G K S E+ +L ++ + H+ WE GC IMTD WTDR+
Sbjct: 1 MLEAIGQFGRSLKGPSPYEMSGSFLQKRKEKVMDGFKEHKESWELTGCSIMTDAWTDRKG 60
Query: 262 RTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYK 321
R ++N +VHS G FL S++ S K IF+++D +EEIGE++VVQVVTDNA+
Sbjct: 61 RGVMNLVVHSAHGVLFLDSVECSGDRKDDKYIFELVDRYIEEIGEQHVVQVVTDNASVNT 120
Query: 322 AAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISL 381
A LL KRP ++W CAAHC+DLMLED K P+ + I+ + +T ++Y T ++ L
Sbjct: 121 TAASLLTAKRPSIFWNGCAAHCLDLMLEDIGKLGPVE-ETIANARQVTVFLYAHTRVLDL 179
Query: 382 LHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQL 416
+ KF N D +R +T FAT+YL L L +++ ++
Sbjct: 180 MRKFLNR-DLVRSGVTRFATAYLNLKSLLDNKKRV 213
>UniRef100_Q6F2S1 Hypothetical protein OSJNBa0060G17.6 [Oryza sativa]
Length = 217
Score = 164 bits (415), Expect = 8e-39
Identities = 85/215 (39%), Positives = 127/215 (58%), Gaps = 2/215 (0%)
Query: 202 MFELVAKHGIGFKPLSYDEIRNKYLNYFYGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRR 261
M E + + G K S E+ +L ++ + H+ WE GC IMTD WTDR+
Sbjct: 1 MLEAIGQFGRSLKGPSPYEMSGSFLQKRKEKVMDGFKEHKESWELTGCSIMTDAWTDRKG 60
Query: 262 RTILNFLVHSPKGTFFLKSIDPSDITKTADKIFKMIDDVVEEIGEENVVQVVTDNAANYK 321
R ++N +VHS G FL S++ S K IF+++D +EEIGE++VVQVVTDNA+
Sbjct: 61 RGVMNLVVHSAHGVLFLDSVECSGDRKDGKYIFELVDRYIEEIGEQHVVQVVTDNASVNT 120
Query: 322 AAGELLMEKRPHLYWTPCAAHCIDLMLEDFEKKIPLHTDAISKGKTITTYIYGRTSLISL 381
A LL KRP ++W AAHC+DLMLED K P+ + I+ + +T ++Y T ++ L
Sbjct: 121 TAASLLTAKRPSIFWNGYAAHCLDLMLEDIGKLGPVE-ETIANARQVTVFLYAHTRVLDL 179
Query: 382 LHKFTNDVDSIRPAMTHFATSYLTLGCLNEHQNQL 416
+ KF N D +R +T FAT+YL L L +++ ++
Sbjct: 180 MRKFLNR-DLVRSGVTRFATAYLNLKSLLDNKKRV 213
>UniRef100_Q75HZ7 Hypothetical protein OSJNBb0035N21.20 [Oryza sativa]
Length = 781
Score = 164 bits (414), Expect = 1e-38
Identities = 132/503 (26%), Positives = 222/503 (43%), Gaps = 61/503 (12%)
Query: 3 STSNVNETSSVASSAPVASSSA-----------AARVRPKNAKGNKTDPAWEYAISTEPG 51
S SN + + +S P + S + AARV P N K P W E
Sbjct: 112 SASNPSGNTGFDASRPSSGSGSTMGGHARSILNAARVIPPNDDEKK--PLWRERSRREC- 168
Query: 52 SRRVQCKFCQLAFTGSAYRVKHHLAGTSR-DVAVCPSVPLDVKKLMQDKVNKLQKKLLEK 110
+VQ +FC GS RVK HL VA C V +D+ +QD++ +
Sbjct: 169 --KVQMQFCNQVIHGSYSRVKAHLLKIGTIGVATCKKVTVDILGQLQDEMTR-------- 218
Query: 111 ANLIVVDADGDKDGAADADLEIISGGRGKRKSAGEIAHKNMLKEDSLR*NKAQSMVCGRR 170
+A ++ D L S RGKR++ I L +
Sbjct: 219 -----AEAISARNLPKDIPLPTESVSRGKRRAVSAIESSFNLDARA-------------- 259
Query: 171 STGKMLVRLLLSFFYNNAISFNVARSDEYLKMFELVAKHGIG-FKPLSYDEIRNKYLNYF 229
L L+ FY + I FNVAR+ + K F+ + +G + P +++++R L
Sbjct: 260 ----KLDALIARMFYTSGIPFNVARNPYFRKAFQFACNNQLGGYTPPNFNKLRTTLLVQE 315
Query: 230 YGEIGKDLEAHRAVWEKCGCIIMTDGWTDRRRRTILNFLVHSPKGTFFLKSIDPSDITKT 289
+ + L A ++ W G I++DGW+D +RR ILNFL + G FLK+I K
Sbjct: 316 RTNVERLLNAVKSTWSTKGVSIVSDGWSDAQRRPILNFLAVTEDGPMFLKAIYTEGEIKR 375
Query: 290 ADKIFKMIDDVVEEIGEENVVQVVTDNAANYKAAGELLMEKRPHLYWTPCAAHCIDLMLE 349
+ I + + ++EE+G NVVQV+TDNA+N +AAG ++ +K H++WTPC H ++L L+
Sbjct: 376 KEYIAEKMFAIIEEVGPNNVVQVITDNASNCRAAGLMVEQKYSHIFWTPCVVHTLNLALK 435
Query: 350 ----------DFEKKIPLHTDAISKGKTITTYIYGRTSLISLLHKFTNDVDSIRPAMTHF 399
D + ++ + I +I + +S+ + F+ + + A T F
Sbjct: 436 SICAAKNSSGDAFLEFQWISEVAADASAIKNFIMNHSMRLSMFNDFSK-LKFLAIADTRF 494
Query: 400 ATSYLTLGCLNEHQNQLIDLFKSNEWKTSKLAKSKDGKIVQNVVLSKVFWKDVLNCLRGA 459
A++ + L + LI + S +W + + V+ +L+ ++W V +
Sbjct: 495 ASTIVMLKRFRAIKENLILMVASEKWNAYREDNQVQAQHVKEKILNDLWWDKVKYIIDFC 554
Query: 460 FPLVKVLCMVDWEEKAAMGYIYE 482
P+ ++ D +K + IYE
Sbjct: 555 EPIYSMIRAAD-TDKPCLHLIYE 576
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,133,976,596
Number of Sequences: 2790947
Number of extensions: 49239219
Number of successful extensions: 250346
Number of sequences better than 10.0: 136
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 74
Number of HSP's that attempted gapping in prelim test: 249417
Number of HSP's gapped (non-prelim): 564
length of query: 677
length of database: 848,049,833
effective HSP length: 134
effective length of query: 543
effective length of database: 474,062,935
effective search space: 257416173705
effective search space used: 257416173705
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 78 (34.7 bits)
Medicago: description of AC138452.7


