

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138451.12 - phase: 0
(133 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
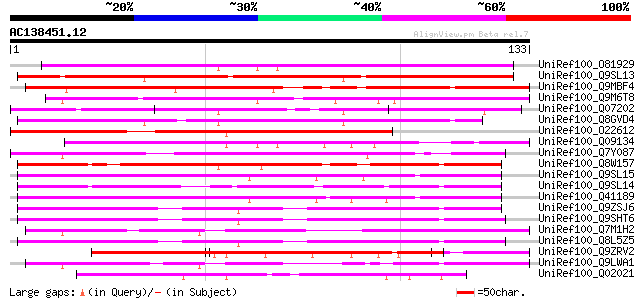
UniRef100_O81929 Glycine-rich protein 1 [Cicer arietinum] 139 1e-32
UniRef100_Q9SL13 Putative glycine-rich protein [Arabidopsis thal... 127 4e-29
UniRef100_Q9MBF4 Glycine-rich protein [Citrus unshiu] 119 1e-26
UniRef100_Q9M6T8 Glycine-rich protein [Nicotiana glauca] 114 6e-25
UniRef100_Q07202 Cold and drought-regulated protein CORA [Medica... 108 2e-23
UniRef100_Q8GVD4 Glycine-rich protein [Lilium hybrid cv. 'Star G... 102 1e-21
UniRef100_O22612 Dormancy-associated protein [Pisum sativum] 101 3e-21
UniRef100_Q09134 Abscisic acid and environmental stress inducibl... 99 2e-20
UniRef100_Q7Y087 GBR5 [Panax ginseng] 97 1e-19
UniRef100_Q8W157 Glycine-rich protein [Brassica oleracea] 97 1e-19
UniRef100_Q9SL15 Putative glycine-rich protein [Arabidopsis thal... 96 2e-19
UniRef100_Q9SL14 Putative glycine-rich protein [Arabidopsis thal... 96 2e-19
UniRef100_Q41189 Glycine-rich protein [Arabidopsis thaliana] 95 4e-19
UniRef100_Q9ZSJ6 Glycine-rich protein 3 short isoform [Arabidops... 94 9e-19
UniRef100_Q9SHT6 Expressed protein [Arabidopsis thaliana] 92 2e-18
UniRef100_Q7M1H2 Glycine-rich protein precursor [Petunia sp.] 91 4e-18
UniRef100_Q8L5Z5 Hypothetical protein At2g05380 [Arabidopsis tha... 91 6e-18
UniRef100_Q9ZRV2 Glycine-rich protein 2 [Cicer arietinum] 84 5e-16
UniRef100_Q9LWA1 Cell wall protein [Nicotiana tabacum] 77 6e-14
UniRef100_Q02021 Glycine-rich protein [Lycopersicon esculentum] 75 2e-13
>UniRef100_O81929 Glycine-rich protein 1 [Cicer arietinum]
Length = 154
Score = 139 bits (351), Expect = 1e-32
Identities = 78/154 (50%), Positives = 89/154 (57%), Gaps = 33/154 (21%)
Query: 9 ILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYG---------------- 52
ILGLLA++LLIS EVSARDLTE SSNT+KE +Q NE+ND KYG
Sbjct: 1 ILGLLAMVLLISFEVSARDLTETSSNTEKEVAEQKNEVNDVKYGGGNYGNGGGNYGQGGG 60
Query: 53 ---------SYGGGYPGRG------GGGNY--GGGYPRNRGGYPGNRGGNYGGGYPGNRG 95
+GGG G G GGGNY GGG + GG G+ GGNYG G G G
Sbjct: 61 NYGNGGGNYGHGGGNYGNGGGNYGHGGGNYGNGGGNYGHGGGNYGHGGGNYGNGGHGGHG 120
Query: 96 GNYCRYGCCGDRYYGSCRKCCSYAGEAVAMQTEN 129
G YC GCCG Y GSCR+CCSY GEA+ + T N
Sbjct: 121 GGYCGNGCCGGYYSGSCRRCCSYVGEAIDVNTLN 154
>UniRef100_Q9SL13 Putative glycine-rich protein [Arabidopsis thaliana]
Length = 135
Score = 127 bits (320), Expect = 4e-29
Identities = 68/132 (51%), Positives = 85/132 (63%), Gaps = 8/132 (6%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASS---NTQKEADDQTNELNDAKYGSYGGGYP 59
+ KA+L L L+ ++LLI+SEV ARDL E S+ N +++ QT + G YGG +P
Sbjct: 2 ASKALLFLSLI-VVLLIASEVVARDLAEKSAEQKNNERDEVKQTEQFGGFPGGGYGG-FP 59
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGG--NYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCS 117
G G GGN GGGY GGY GG N GGGY GNRGG YCRYGCC YYG C +CC+
Sbjct: 60 GGGYGGNPGGGYGNRGGGYRNRDGGYGNRGGGY-GNRGGGYCRYGCCYRGYYGGCFRCCA 118
Query: 118 YAGEAVAMQTEN 129
YAG+AV Q ++
Sbjct: 119 YAGQAVQTQPQS 130
>UniRef100_Q9MBF4 Glycine-rich protein [Citrus unshiu]
Length = 127
Score = 119 bits (299), Expect = 1e-26
Identities = 71/134 (52%), Positives = 85/134 (62%), Gaps = 15/134 (11%)
Query: 5 KAILILGLL-AIILLISSEVSARDLTEASSNTQKEADD--QTNELNDAKYGSYGGGYPGR 61
K L+LGLL AI LLISSEV+ARDL E S + ++AD +T+E+ D + G GG G
Sbjct: 4 KLFLLLGLLMAIALLISSEVAARDLAETSIDHNEKADKATETHEIEDGRGGYNRGGRGGY 63
Query: 62 GGGGN--YGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYA 119
GGG YGGG GGY G GG YGGG G GG +CRYGCCG Y+G CR CC+YA
Sbjct: 64 NGGGRGGYGGG-----GGYGG--GGGYGGG--GGHGGGHCRYGCCGHGYHGGCR-CCTYA 113
Query: 120 GEAVAMQTENNTRN 133
GEAV + E +N
Sbjct: 114 GEAVDTEPETEPQN 127
>UniRef100_Q9M6T8 Glycine-rich protein [Nicotiana glauca]
Length = 132
Score = 114 bits (284), Expect = 6e-25
Identities = 68/134 (50%), Positives = 81/134 (59%), Gaps = 13/134 (9%)
Query: 10 LGL-LAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRG--GGGN 66
LGL LAI ++ISS+V AR+L E S+ T KE +++ N+ GGGYPG G GGG
Sbjct: 2 LGLCLAIFVMISSDVFARELAETST-TAKEDSKKSDNKNEVHEDQLGGGYPGGGYPGGGY 60
Query: 67 YGGGYPRNRGGYPGNR--GGNYGGGYPGN-RGGN----YCRYGCCGDRYYGSCRKCCSYA 119
GGGYP GGYPG GG G GYPG RGG YCRYGCCG R Y C +CC Y
Sbjct: 61 PGGGYPG--GGYPGRGYPGGGRGRGYPGGGRGGGGRGGYCRYGCCGRRNYYGCSRCCYYK 118
Query: 120 GEAVAMQTENNTRN 133
GEA+ TE ++
Sbjct: 119 GEAMDKVTEGKPQH 132
>UniRef100_Q07202 Cold and drought-regulated protein CORA [Medicago sativa]
Length = 204
Score = 108 bits (271), Expect = 2e-23
Identities = 71/146 (48%), Positives = 88/146 (59%), Gaps = 18/146 (12%)
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYG---SYGGG 57
MDSKKAIL+L LLA+ L ISS +SARDLTE S++ +KE ++TNE+NDAKYG ++GGG
Sbjct: 1 MDSKKAILMLSLLAMAL-ISSVMSARDLTETSTDAKKEVVEKTNEVNDAKYGGGYNHGGG 59
Query: 58 YPGRGGGGNYGGGYPRNRGGYPGNRGG--------NYGGGYPGNRGGNYCRYGCCGDRYY 109
Y GGG N+GGGY GGY GG N GGG+ G+ GG Y G G
Sbjct: 60 Y--NGGGYNHGGGYNHGGGGYHNGGGGYNHGGGGYNGGGGHGGHGGGGYNGGGGHGGHGG 117
Query: 110 GSCRKCCSYAG----EAVAMQTENNT 131
G + G E+VA+QTE T
Sbjct: 118 GGYNGGGGHGGHGGAESVAVQTEEKT 143
Score = 52.4 bits (124), Expect = 2e-06
Identities = 28/60 (46%), Positives = 36/60 (59%), Gaps = 4/60 (6%)
Query: 38 EADDQTNELNDAKYGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGN 97
+ +++TNE+NDAKYG GRGG + GGGY N GG G GG+ G G G GG+
Sbjct: 138 QTEEKTNEVNDAKYGGGSNYNDGRGGYNHGGGGY--NHGG--GGHGGHGGHGGHGGHGGH 193
>UniRef100_Q8GVD4 Glycine-rich protein [Lilium hybrid cv. 'Star Gazer']
Length = 138
Score = 102 bits (255), Expect = 1e-21
Identities = 57/125 (45%), Positives = 73/125 (57%), Gaps = 9/125 (7%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASS-NTQKEADDQTNELNDAKYG---SYGGGY 58
+ KA+L+LG+L + L + + R+L E + NT+K A + + D KYG + GGGY
Sbjct: 2 ASKALLMLGVLIVAALFVTSDAGRELAEETKENTEKRATEAG--VADQKYGGGYNNGGGY 59
Query: 59 PGRGGGGNYGGGYPRNRGGYPGNRGG--NYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCC 116
PG GGG + GGGYP GGYPG GG GGGY GG C GCC YYG CR CC
Sbjct: 60 PGGGGGYHNGGGYPGGGGGYPGGGGGYPGGGGGYHNGGGGGRCYNGCCRRGYYGGCR-CC 118
Query: 117 SYAGE 121
++ E
Sbjct: 119 AHPDE 123
>UniRef100_O22612 Dormancy-associated protein [Pisum sativum]
Length = 129
Score = 101 bits (252), Expect = 3e-21
Identities = 58/106 (54%), Positives = 69/106 (64%), Gaps = 15/106 (14%)
Query: 1 MDSKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSY------ 54
MDS+KA+LILGLLA++LLISSEVSAR+LTE E ++++E+NDAKYG Y
Sbjct: 1 MDSRKAMLILGLLAMVLLISSEVSARELTE-------EVVEKSDEVNDAKYGGYGRGGGY 53
Query: 55 --GGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNY 98
GGGY GGG N GGGY GGY G G + GGG N GG Y
Sbjct: 54 HNGGGYHNGGGGYNGGGGYHNGGGGYNGGGGYHNGGGGYHNGGGGY 99
>UniRef100_Q09134 Abscisic acid and environmental stress inducible protein [Medicago
falcata]
Length = 159
Score = 99.4 bits (246), Expect = 2e-20
Identities = 71/168 (42%), Positives = 85/168 (50%), Gaps = 58/168 (34%)
Query: 15 IILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSY----GGGYPGRG---GGGNY 67
++LLISSEVSARDLTE +S+ +KE ++TNE+NDAKYG GGGY G G GGG Y
Sbjct: 1 MVLLISSEVSARDLTETTSDAKKEVVEKTNEVNDAKYGGGYNHGGGGYNGGGYNHGGGGY 60
Query: 68 ---------GGGYPRNRGGYP------GNRGGNY-----------------GGGYPG--- 92
GGGY GGY N GG Y GGGY G
Sbjct: 61 NNGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNNGGGGYNHGGGGYNGGGY 120
Query: 93 -------NRGGNYCRYGCCGDRYYGSCRKCCSYAGEAVAMQTENNTRN 133
N GG C+Y C G +CCS+A E VAMQ ++NT+N
Sbjct: 121 NHGGGGYNHGGGGCQYHCHG--------RCCSHA-EFVAMQAKDNTQN 159
>UniRef100_Q7Y087 GBR5 [Panax ginseng]
Length = 123
Score = 96.7 bits (239), Expect = 1e-19
Identities = 62/130 (47%), Positives = 75/130 (57%), Gaps = 17/130 (13%)
Query: 1 MDSKKAILILGL-LAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYP 59
M S KA L+LGL LAI+LLISSEV+AR+L EA + T TN+ +A GY
Sbjct: 1 MGSNKAFLLLGLSLAIVLLISSEVAARELAEAQTTT-------TNKNTEAATVDGRSGYN 53
Query: 60 GRGGGGNYGGGYPRNRGGYPGNRGGNYGGGY--PGNRGGNYCRYGCCGDRYYGSCRKCCS 117
G GGGG +GGG GGY G G + GGGY G GG C +G C RY +CC+
Sbjct: 54 GYGGGGYHGGGGYHGGGGYHGGGGYHGGGGYHGGGGHGGGGCSHGYC--RY-----RCCN 106
Query: 118 YAGEAVAMQT 127
YAGE V +T
Sbjct: 107 YAGEVVDAET 116
>UniRef100_Q8W157 Glycine-rich protein [Brassica oleracea]
Length = 124
Score = 96.7 bits (239), Expect = 1e-19
Identities = 59/130 (45%), Positives = 79/130 (60%), Gaps = 16/130 (12%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYG---SYGGGYP 59
+ KA+++LGL A++ +IS EV+A T + + E++D + G + GGGY
Sbjct: 2 ASKALVLLGLFALLFVIS-EVAA---TSEGQSLKSESEDTLQPDHSGGGGQGYNGGGGYN 57
Query: 60 GRGG---GGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCC 116
GRGG GG++GGG GGY G GG++GGG RGG YCR+GCC Y G C +CC
Sbjct: 58 GRGGYNGGGHHGGGGYNGGGGYNG--GGHHGGG---GRGGGYCRHGCCYRGYRG-CSRCC 111
Query: 117 SYAGEAVAMQ 126
SYAGEAV Q
Sbjct: 112 SYAGEAVQTQ 121
>UniRef100_Q9SL15 Putative glycine-rich protein [Arabidopsis thaliana]
Length = 145
Score = 95.9 bits (237), Expect = 2e-19
Identities = 57/142 (40%), Positives = 76/142 (53%), Gaps = 19/142 (13%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG-- 60
+ KA+++LGL A++L++S +A T S + + DQ ++ + GGGY G
Sbjct: 2 ASKALVLLGLFAVLLVVSEVAAASSATVNSESKETVKPDQRGYGDNGGNYNNGGGYQGGG 61
Query: 61 ----------RGGGGNYGGGYPRNRGG---YPGNRGGNYGGG---YPGNRGGNYCRYGCC 104
+GGGGNY GG R +GG Y G G GGG G GG+YCR+GCC
Sbjct: 62 GNYQGGGGNYQGGGGNYQGGGGRYQGGGGRYQGGGGRYQGGGGRQGGGGSGGSYCRHGCC 121
Query: 105 GDRYYGSCRKCCSYAGEAVAMQ 126
R Y C +CCSYAGEAV Q
Sbjct: 122 -YRGYNGCSRCCSYAGEAVQTQ 142
>UniRef100_Q9SL14 Putative glycine-rich protein [Arabidopsis thaliana]
Length = 115
Score = 95.9 bits (237), Expect = 2e-19
Identities = 57/124 (45%), Positives = 72/124 (57%), Gaps = 13/124 (10%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPGRG 62
+ KA+++ GL A++L+++ EV+A T S + + DQ N G +GG G
Sbjct: 2 ASKALVLFGLFAVLLVVT-EVAAASGTVKSESGETVQPDQYN-------GGHGGN-GGYN 52
Query: 63 GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
GGG Y GG N GGY N GG Y GG G R G YCRYGCC Y+G C +CCSYAGEA
Sbjct: 53 GGGGYNGGGGHNGGGY--NGGGGYNGGGHGGRHG-YCRYGCCYRGYHG-CSRCCSYAGEA 108
Query: 123 VAMQ 126
V Q
Sbjct: 109 VQTQ 112
>UniRef100_Q41189 Glycine-rich protein [Arabidopsis thaliana]
Length = 145
Score = 94.7 bits (234), Expect = 4e-19
Identities = 59/143 (41%), Positives = 78/143 (54%), Gaps = 21/143 (14%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGGYPG-- 60
+ KA+++LGL A++L++S +A T S + + DQ ++ + GGGY G
Sbjct: 2 ASKALVLLGLFAVLLVVSEVAAASSATVNSESKETVKPDQRGYGDNGGNYNNGGGYQGGG 61
Query: 61 ----------RGGGGNYGGGYPRNRGG---YPGNRGGNY--GGGYPGNRG--GNYCRYGC 103
+GGGGNY GG R +GG Y G GG Y GGG G G G+YCR+GC
Sbjct: 62 GNYQGGGGNYQGGGGNYQGGGGRYQGGGGRYQGG-GGRYQGGGGRQGGGGSRGSYCRHGC 120
Query: 104 CGDRYYGSCRKCCSYAGEAVAMQ 126
C R Y C +CCSYAGEAV Q
Sbjct: 121 C-YRGYNGCSRCCSYAGEAVQTQ 142
>UniRef100_Q9ZSJ6 Glycine-rich protein 3 short isoform [Arabidopsis thaliana]
Length = 116
Score = 93.6 bits (231), Expect = 9e-19
Identities = 53/125 (42%), Positives = 68/125 (54%), Gaps = 14/125 (11%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGG-YPGR 61
+ KA+L+LGL A + ++S +A + S T K E + +G GGG Y G
Sbjct: 2 ASKALLLLGLFAFLFIVSEMAAAGTVKSESEETVKP------EQHGGGFGDNGGGRYQGG 55
Query: 62 GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGE 121
GG G +GGG GY G G GGG GG+YCR+GCC Y+G C +CCSYAGE
Sbjct: 56 GGHGGHGGG------GYQGGGGRYQGGGGRQGGGGSYCRHGCCYKGYHG-CSRCCSYAGE 108
Query: 122 AVAMQ 126
AV Q
Sbjct: 109 AVQTQ 113
>UniRef100_Q9SHT6 Expressed protein [Arabidopsis thaliana]
Length = 116
Score = 92.4 bits (228), Expect = 2e-18
Identities = 52/126 (41%), Positives = 68/126 (53%), Gaps = 14/126 (11%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGG-YPGR 61
+ K +L+LGL A + ++S +A + S T K E + +G GGG Y G
Sbjct: 2 ASKTLLLLGLFAFLFIVSEMAAAGTVKSESEETVKP------EQHGGGFGDNGGGRYQGG 55
Query: 62 GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGE 121
GG G +GGG GY G G GGG GG+YCR+GCC Y+G C +CCSYAGE
Sbjct: 56 GGHGGHGGG------GYQGGGGRYQGGGGRQGGGGSYCRHGCCYKGYHG-CSRCCSYAGE 108
Query: 122 AVAMQT 127
AV Q+
Sbjct: 109 AVQTQS 114
>UniRef100_Q7M1H2 Glycine-rich protein precursor [Petunia sp.]
Length = 112
Score = 91.3 bits (225), Expect = 4e-18
Identities = 58/131 (44%), Positives = 67/131 (50%), Gaps = 24/131 (18%)
Query: 5 KAILILGL-LAIILLISSEVSARDLTEASSNTQKEADDQTNELN-DAKYGSYGGGYPGRG 62
KA LILGL L I+ LISSE AR+L E NT + NE D + GY G G
Sbjct: 4 KAFLILGLCLTILFLISSEAFARELAE---NTDQLKSANKNEAQVDGR-----SGYNGIG 55
Query: 63 GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
G YGG + RG G GG YCRYGCC YY C+KCCSYAG+A
Sbjct: 56 EDGYYGGRKGKGRG--------------KGKGGGGYCRYGCCRKGYYKGCKKCCSYAGQA 101
Query: 123 VAMQTENNTRN 133
+ TE N+ N
Sbjct: 102 MDKVTETNSHN 112
>UniRef100_Q8L5Z5 Hypothetical protein At2g05380 [Arabidopsis thaliana]
Length = 116
Score = 90.9 bits (224), Expect = 6e-18
Identities = 51/126 (40%), Positives = 68/126 (53%), Gaps = 14/126 (11%)
Query: 3 SKKAILILGLLAIILLISSEVSARDLTEASSNTQKEADDQTNELNDAKYGSYGGG-YPGR 61
+ K +L+LGL A + ++S +A + S T K E + +G GGG Y G
Sbjct: 2 ASKTLLLLGLFAFLFIVSEMAAAGTVKSESEETVKP------EQHGGGFGDNGGGRYQGG 55
Query: 62 GGGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGE 121
GG G +GGG GY G G GGG GG+YCR+GCC Y+G C +CCSYAG+
Sbjct: 56 GGHGGHGGG------GYQGGGGRYQGGGGRQGGGGSYCRHGCCYKGYHG-CSRCCSYAGK 108
Query: 122 AVAMQT 127
AV Q+
Sbjct: 109 AVQTQS 114
>UniRef100_Q9ZRV2 Glycine-rich protein 2 [Cicer arietinum]
Length = 208
Score = 84.3 bits (207), Expect = 5e-16
Identities = 53/104 (50%), Positives = 64/104 (60%), Gaps = 15/104 (14%)
Query: 22 EVSARDLTEASSNTQKEADDQTNELNDAKY------GSY--GGGYPGRGGG--GNYGGGY 71
EVSARDLTE SSNT+K+ ++ NE+ DA Y G+Y GGGYP GGG G+ GG Y
Sbjct: 1 EVSARDLTEISSNTEKDVAEKKNEVTDANYDGFNGGGNYGNGGGYPSNGGGNYGHGGGNY 60
Query: 72 PRNRG--GYPGNRGGNY--GGGYPGNRGGNYCRYGCCGDRYYGS 111
G G+ GN GGNY GGG G+ GGNY +G G YG+
Sbjct: 61 GNGGGNYGHGGNGGGNYGNGGGNYGHGGGNY-GHGSNGGGNYGN 103
Score = 61.2 bits (147), Expect = 5e-09
Identities = 39/87 (44%), Positives = 45/87 (50%), Gaps = 7/87 (8%)
Query: 51 YGSYGGGYPGRGGGGNYGGGYPRNRGGYPGNRGGNY--GGGYPGN--RGGNYCRYGCCGD 106
YG+ GG Y GG +GGG N GG G+ GGNY GGGYPGN GG Y G G
Sbjct: 125 YGNGGGSYGNGGGNYGHGGGNYGNGGGNYGHGGGNYGHGGGYPGNGGHGGGYPGNGGHGG 184
Query: 107 RYYGSCRKCCSYAGEAVAMQTENNTRN 133
+ G Y GEA +Q E+ T N
Sbjct: 185 GHGGHGG---GYVGEATDVQREDKTLN 208
Score = 51.2 bits (121), Expect = 5e-06
Identities = 32/65 (49%), Positives = 34/65 (52%), Gaps = 8/65 (12%)
Query: 52 GSYGGGYPGRGGGGNYGGGYPRNRG--GYPGNRGGNY--GGGYPGNRGGNY----CRYGC 103
G+YG G G G GN GG Y G G+ GN GGNY GGG GN GGNY YG
Sbjct: 89 GNYGHGSNGGGNYGNGGGNYGHGGGNYGHGGNGGGNYGNGGGSYGNGGGNYGHGGGNYGN 148
Query: 104 CGDRY 108
G Y
Sbjct: 149 GGGNY 153
>UniRef100_Q9LWA1 Cell wall protein [Nicotiana tabacum]
Length = 106
Score = 77.4 bits (189), Expect = 6e-14
Identities = 57/131 (43%), Positives = 68/131 (51%), Gaps = 30/131 (22%)
Query: 5 KAILILGL-LAIILLISSEVSARDLTEASSNTQKEADDQTNELN-DAKYGSYGGGYPGRG 62
KA L LGL LAI LIS EV A +L E S++ + D NE++ D + GY G G
Sbjct: 4 KAFLFLGLFLAIFFLISFEVVAAELAETSNSMK---SDNENEVHFDGR-----SGYNGIG 55
Query: 63 GGGNYGGGYPRNRGGYPGNRGGNYGGGYPGNRGGNYCRYGCCGDRYYGSCRKCCSYAGEA 122
G YGGG +GGGY RG CRYGCC Y G C++CCSYAGEA
Sbjct: 56 NDGYYGGG---------------HGGGYK-RRG---CRYGCCRKGYNG-CKRCCSYAGEA 95
Query: 123 VAMQTENNTRN 133
+ TE N
Sbjct: 96 MDKVTEVQPHN 106
>UniRef100_Q02021 Glycine-rich protein [Lycopersicon esculentum]
Length = 132
Score = 75.5 bits (184), Expect = 2e-13
Identities = 56/135 (41%), Positives = 67/135 (49%), Gaps = 38/135 (28%)
Query: 18 LISSEVSARDLTEASSNTQKEADDQT---NELNDAKYGSY---------GGGYPGRGGGG 65
+ISS+V AR+L E S+ T +E ++ NE+++A+YG Y GGGYPG G GG
Sbjct: 1 MISSDVLARELAETSTTTSEEDSKKSSNKNEVHEAEYGGYPGGGGGYPGGGGYPGGGRGG 60
Query: 66 NYGGGYPRNRGGYPGNRGGNYGGGYPGNRG--GNYCRY------GCCGDRYY-------- 109
GGGYP GG G GG GGGYPG G G RY G G RY
Sbjct: 61 G-GGGYP--GGGRSGGGGGYPGGGYPGGGGYRGGGGRYPGGGGGGRGGGRYSGGGGRGGG 117
Query: 110 -------GSCRKCCS 117
G RKCCS
Sbjct: 118 GGRGGRGGGGRKCCS 132
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 289,611,927
Number of Sequences: 2790947
Number of extensions: 17172822
Number of successful extensions: 243904
Number of sequences better than 10.0: 6675
Number of HSP's better than 10.0 without gapping: 3650
Number of HSP's successfully gapped in prelim test: 3347
Number of HSP's that attempted gapping in prelim test: 85193
Number of HSP's gapped (non-prelim): 70678
length of query: 133
length of database: 848,049,833
effective HSP length: 109
effective length of query: 24
effective length of database: 543,836,610
effective search space: 13052078640
effective search space used: 13052078640
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC138451.12


