

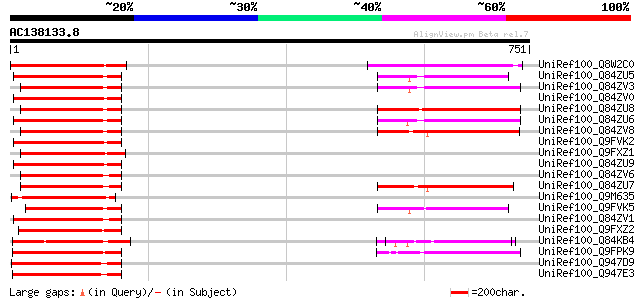
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.8 + phase: 0 /pseudo
(751 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Gl... 200 1e-49
UniRef100_Q84ZU5 R 8 protein [Glycine max] 195 4e-48
UniRef100_Q84ZV3 R 4 protein [Glycine max] 191 7e-47
UniRef100_Q84ZV0 R 14 protein [Glycine max] 191 9e-47
UniRef100_Q84ZU8 R 10 protein [Glycine max] 190 1e-46
UniRef100_Q84ZU6 R 1 protein [Glycine max] 189 3e-46
UniRef100_Q84ZV8 R 3 protein [Glycine max] 187 8e-46
UniRef100_Q9FVK2 Resistance protein MG13 [Glycine max] 187 1e-45
UniRef100_Q9FXZ1 Resistance protein MG23 [Glycine max] 185 4e-45
UniRef100_Q84ZU9 R 13 protein [Glycine max] 185 5e-45
UniRef100_Q84ZV6 R 6 protein [Glycine max] 180 1e-43
UniRef100_Q84ZU7 R 5 protein [Glycine max] 180 1e-43
UniRef100_Q9M635 Hypothetical protein [Glycine max] 180 1e-43
UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max] 179 4e-43
UniRef100_Q84ZV1 R 9 protein [Glycine max] 178 5e-43
UniRef100_Q9FXZ2 Resistance protein LM12 [Glycine max] 174 7e-42
UniRef100_Q84KB4 MRGH5 [Cucumis melo] 171 7e-41
UniRef100_Q9FPK9 Putative resistance protein [Glycine max] 166 2e-39
UniRef100_Q947D9 Resistance gene analog NBS9 [Helianthus annuus] 166 2e-39
UniRef100_Q947E3 Resistance gene analog NBS5 [Helianthus annuus] 164 9e-39
>UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Glycine max]
Length = 1124
Score = 200 bits (508), Expect = 1e-49
Identities = 102/168 (60%), Positives = 128/168 (75%), Gaps = 1/168 (0%)
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
M SSS+S SY + VFLSFRG DTR GFTGNLYKAL+D+GI+TF+DD + RG++IT
Sbjct: 1 MAKQSSSSSFSYRFSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQIT 60
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
L KAIEESRIFI V S NYASSSFCL+ELD+I+ K KG +LPVF+ VDPS VR+H
Sbjct: 61 SGLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNH 120
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPGQ 168
GS+G+AL HEK+F++ +ME+L+ WK AL++ ANLSGYH G+
Sbjct: 121 TGSFGKALTNHEKKFKS-TNDMEKLETWKMALNKVANLSGYHHFKHGE 167
Score = 161 bits (407), Expect = 8e-38
Identities = 93/223 (41%), Positives = 133/223 (58%), Gaps = 6/223 (2%)
Query: 519 YSSCFKRKHYKKGKAFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSI 578
YSS + + G AFKKM LKTLII +GH SKG K+ SL+ L+W S
Sbjct: 546 YSSFEEVEIQWDGDAFKKMKNLKTLIIRSGHFSKGPKHFPKSLRVLEWWRYPSHYFPYDF 605
Query: 579 LSKASEIKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFE 638
+ I + +C + + + KKF ++T L D C++LT IPDVS + +L+KLSF+
Sbjct: 606 QMEKLAIFNLPDCGFTSRELAAMLKKKFVNLTSLNFDSCQHLTLIPDVSCVPHLQKLSFK 665
Query: 639 YCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCK 698
C NL IH S+G L KL L A GC LK FPP+ L SL++LKL C+SL++FP++L K
Sbjct: 666 DCDNLYAIHPSVGFLEKLRILDAEGCSRLKNFPPIKLTSLEQLKLGFCHSLENFPEILGK 725
Query: 699 MTNIDKIWFWYTSIRELPSSFQNLSELDELSVREFGMLRFPKH 741
M NI ++ T +++ P SFQNL+ L+ + +L FP++
Sbjct: 726 MENITELDLEQTPVKKFPLSFQNLTRLETV------LLCFPRN 762
>UniRef100_Q84ZU5 R 8 protein [Glycine max]
Length = 892
Score = 195 bits (496), Expect = 4e-48
Identities = 101/157 (64%), Positives = 121/157 (76%), Gaps = 4/157 (2%)
Query: 6 SSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLK 65
++T+ S Y Y VFLSF G DTR GFTG LYKAL D+GI TFIDD L+RG+EI P+L
Sbjct: 2 AATTRSLAYNYDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSN 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYG 125
AI+ESRI I V S NYASSSFCLDEL I+HC K++G V+PVF+ VDPSHVRH KGSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYH 162
EA+A+H+KRF+ N E+LQ W+ AL Q A+LSGYH
Sbjct: 121 EAMAKHQKRFK---ANKEKLQKWRMALHQVADLSGYH 154
Score = 147 bits (371), Expect = 1e-33
Identities = 89/198 (44%), Positives = 115/198 (57%), Gaps = 17/198 (8%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSS------ILSKA--SE 584
AF KM LK LII N SKG Y L+ L+W S L S+ ++ K S
Sbjct: 554 AFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFDPINLVICKLPDSS 613
Query: 585 IKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLI 644
I SF S+KK +T+L D CE+LT IPDVS L NL++LSF +C++L+
Sbjct: 614 ITSFE---------FHGSSKKLGHLTVLNFDRCEFLTKIPDVSDLPNLKELSFNWCESLV 664
Query: 645 TIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDK 704
+ +SIG LNKL+ LSA+GCR L FPPL L SL+ L L C SL+ FP++L +M NI
Sbjct: 665 AVDDSIGFLNKLKTLSAYGCRKLTSFPPLNLTSLETLNLGGCSSLEYFPEILGEMKNITV 724
Query: 705 IWFWYTSIRELPSSFQNL 722
+ I+ELP SFQNL
Sbjct: 725 LALHDLPIKELPFSFQNL 742
>UniRef100_Q84ZV3 R 4 protein [Glycine max]
Length = 895
Score = 191 bits (485), Expect = 7e-47
Identities = 99/147 (67%), Positives = 116/147 (78%), Gaps = 4/147 (2%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLYKAL D+GI T IDD L RG+EITP+L KAI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRF 135
V S NYASSSFCLDEL I+HC K++G V+PVF+ VDPS VRH KGSYGEA+A+H+KRF
Sbjct: 72 VLSQNYASSSFCLDELVTILHC-KSEGLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 130
Query: 136 QNDPKNMERLQGWKDALSQAANLSGYH 162
+ E+LQ W+ AL Q A+LSGYH
Sbjct: 131 K---AKKEKLQKWRMALKQVADLSGYH 154
Score = 159 bits (401), Expect = 4e-37
Identities = 95/215 (44%), Positives = 129/215 (59%), Gaps = 17/215 (7%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSS------ILSKA--SE 584
AF KM LK LII NG SKG Y L+ L+W S L S+ ++ K S
Sbjct: 553 AFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSNFLPSNFDPINLVICKLPDSS 612
Query: 585 IKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLI 644
IKSF S+KK +T+L D C++LT IPDVS L NL +LSFE C++L+
Sbjct: 613 IKSFE---------FHGSSKKLGHLTVLKFDRCKFLTQIPDVSDLPNLRELSFEDCESLV 663
Query: 645 TIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDK 704
+ +SIG L KL++LSA+GCR L FPPL L SL+ L+LS C SL+ FP++L +M NI +
Sbjct: 664 AVDDSIGFLKKLKKLSAYGCRKLTSFPPLNLTSLETLQLSSCSSLEYFPEILGEMENIRE 723
Query: 705 IWFWYTSIRELPSSFQNLSELDELSVREFGMLRFP 739
+ I+ELP SFQNL+ L L++ G+++ P
Sbjct: 724 LRLTGLYIKELPFSFQNLTGLRLLALSGCGIVQLP 758
>UniRef100_Q84ZV0 R 14 protein [Glycine max]
Length = 641
Score = 191 bits (484), Expect = 9e-47
Identities = 96/157 (61%), Positives = 122/157 (77%), Gaps = 4/157 (2%)
Query: 6 SSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLK 65
++T+ S + Y VFLSFRG DTRYGFTGNLY+AL +KGI+TF D+ L G+EITP+L K
Sbjct: 2 AATTRSLPFIYDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYG 125
AI+ESRI I V S NYA SSFCLDEL I+HC K++G V+PVF+ VDPS +RH KGSYG
Sbjct: 62 AIQESRIAITVLSQNYAFSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDLRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYH 162
EA+ +H+KRF++ ME+LQ W+ AL Q A+LSG+H
Sbjct: 121 EAMIKHQKRFES---KMEKLQKWRMALKQVADLSGHH 154
>UniRef100_Q84ZU8 R 10 protein [Glycine max]
Length = 901
Score = 190 bits (483), Expect = 1e-46
Identities = 96/147 (65%), Positives = 114/147 (77%), Gaps = 4/147 (2%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIP 75
Y VFL+FRG DTRYGFTGNLY+AL DKGI+TF D+ L RG EITP+LLKAI+ESRI I
Sbjct: 12 YDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRF 135
V S NYASSSFCLDEL I+HC K++G V+PVF+ VDPS VRH KGSYG +A+H+KRF
Sbjct: 72 VLSKNYASSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDVRHQKGSYGVEMAKHQKRF 130
Query: 136 QNDPKNMERLQGWKDALSQAANLSGYH 162
+ E+LQ W+ AL Q A+L GYH
Sbjct: 131 K---AKKEKLQKWRIALKQVADLCGYH 154
Score = 157 bits (398), Expect = 8e-37
Identities = 92/209 (44%), Positives = 127/209 (60%), Gaps = 6/209 (2%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEI-KSFSNC 591
AF KM LK LII N SKG Y L+ L+W S L S+ I K +C
Sbjct: 556 AFMKMENLKILIIRNDKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFHPNNLVICKLPDSC 615
Query: 592 IYL*CTMISFSA-KKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSI 650
+ T F KF +T+L D+C++LT IPDVS L NL +LSFE C++L+ + +SI
Sbjct: 616 M----TSFEFHGPSKFGHLTVLKFDNCKFLTQIPDVSDLPNLRELSFEECESLVAVDDSI 671
Query: 651 GHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYT 710
G LNKL++LSA+GC LK FPPL L SL+ L+LS C SL+ FP+++ +M NI ++ +
Sbjct: 672 GFLNKLKKLSAYGCSKLKSFPPLNLTSLQTLELSQCSSLEYFPEIIGEMENIKHLFLYGL 731
Query: 711 SIRELPSSFQNLSELDELSVREFGMLRFP 739
I+EL SFQNL L L++R G+++ P
Sbjct: 732 PIKELSFSFQNLIGLRWLTLRSCGIVKLP 760
>UniRef100_Q84ZU6 R 1 protein [Glycine max]
Length = 902
Score = 189 bits (480), Expect = 3e-46
Identities = 99/157 (63%), Positives = 120/157 (76%), Gaps = 4/157 (2%)
Query: 6 SSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLK 65
++T+ S Y VFL+FRG DTRYGFTGNLYKAL DKGI+TF D++ L G++ITP+L K
Sbjct: 2 AATTRSLASIYDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYG 125
AI+ESRI I V S NYASSSFCLDEL I+HC K +G V+PVF VDPS VRH KGSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KREGLLVIPVFHNVDPSAVRHLKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYH 162
EA+A+H+KRF+ E+LQ W+ AL Q A+LSGYH
Sbjct: 121 EAMAKHQKRFK---AKKEKLQKWRMALHQVADLSGYH 154
Score = 157 bits (398), Expect = 8e-37
Identities = 94/215 (43%), Positives = 127/215 (58%), Gaps = 18/215 (8%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSL------SSSILSKA--SE 584
AF KM LK LII NG SKG Y L L+W S L ++ ++ K S
Sbjct: 556 AFMKMENLKILIIRNGKFSKGPNYFPEGLTVLEWHRYPSNCLPYNFHPNNLLICKLPDSS 615
Query: 585 IKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLI 644
I SF KF +T+L D CE+LT IPDVS L NL++LSF++C++LI
Sbjct: 616 ITSFE----------LHGPSKFWHLTVLNFDQCEFLTQIPDVSDLPNLKELSFDWCESLI 665
Query: 645 TIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDK 704
+ +SIG LNKL++LSA+GCR L+ FPPL L SL+ L+LS C SL+ FP++L +M NI
Sbjct: 666 AVDDSIGFLNKLKKLSAYGCRKLRSFPPLNLTSLETLQLSGCSSLEYFPEILGEMENIKA 725
Query: 705 IWFWYTSIRELPSSFQNLSELDELSVREFGMLRFP 739
+ I+ELP SFQNL L L++ G+++ P
Sbjct: 726 LDLDGLPIKELPFSFQNLIGLCRLTLNSCGIIQLP 760
>UniRef100_Q84ZV8 R 3 protein [Glycine max]
Length = 897
Score = 187 bits (476), Expect = 8e-46
Identities = 98/147 (66%), Positives = 114/147 (76%), Gaps = 4/147 (2%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLYKAL D+GI TFIDD L RG+EITP+L KAI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRF 135
V S NYASSSFCLDEL ++ C K KG V+PVF+ VDPS VR KGSYGEA+A+H+KRF
Sbjct: 72 VLSQNYASSSFCLDELVTVLLC-KRKGLLVIPVFYNVDPSDVRQQKGSYGEAMAKHQKRF 130
Query: 136 QNDPKNMERLQGWKDALSQAANLSGYH 162
+ E+LQ W+ AL Q A+LSGYH
Sbjct: 131 K---AKKEKLQKWRMALHQVADLSGYH 154
Score = 155 bits (392), Expect = 4e-36
Identities = 90/208 (43%), Positives = 128/208 (61%), Gaps = 8/208 (3%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSNCI 592
AF KM LK LII NG SKG Y L+ L+W S L S+ + + C
Sbjct: 553 AFMKMENLKILIIRNGKFSKGPNYFPQGLRVLEWHRYPSNCLPSNF-----DPINLVICK 607
Query: 593 YL*CTMISFS---AKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNS 649
+M SF + K +T+L D C++LT IPDVS L NL +LSF++C++L+ + +S
Sbjct: 608 LPDSSMTSFEFHGSSKLGHLTVLKFDWCKFLTQIPDVSDLPNLRELSFQWCESLVAVDDS 667
Query: 650 IGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWY 709
IG LNKL++L+A+GCR L FPPL L SL+ L+LS C SL+ FP++L +M NI+++
Sbjct: 668 IGFLNKLKKLNAYGCRKLTSFPPLHLTSLETLELSHCSSLEYFPEILGEMENIERLDLHG 727
Query: 710 TSIRELPSSFQNLSELDELSVREFGMLR 737
I+ELP SFQNL L +LS+ G+++
Sbjct: 728 LPIKELPFSFQNLIGLQQLSMFGCGIVQ 755
>UniRef100_Q9FVK2 Resistance protein MG13 [Glycine max]
Length = 344
Score = 187 bits (475), Expect = 1e-45
Identities = 96/157 (61%), Positives = 120/157 (76%), Gaps = 4/157 (2%)
Query: 6 SSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLK 65
++T+ S Y VFLSFRG+DTRYGFTGNLYKAL DKG +TF D++ L G EITP+LLK
Sbjct: 2 AATTRSLASIYDVFLSFRGTDTRYGFTGNLYKALCDKGFHTFFDEDKLHSGEEITPALLK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYG 125
AI++SR+ I V S NYA SSFCLDEL I HC K +G V+PVF+ VDPS+VRH KGSYG
Sbjct: 62 AIQDSRVAIIVLSENYAFSSFCLDELVTIFHC-KREGLLVIPVFYKVDPSYVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYH 162
EA+ +H++RF++ ME+LQ W+ AL Q A+LSG H
Sbjct: 121 EAMTKHQERFKD---KMEKLQEWRMALKQVADLSGSH 154
>UniRef100_Q9FXZ1 Resistance protein MG23 [Glycine max]
Length = 435
Score = 185 bits (470), Expect = 4e-45
Identities = 94/153 (61%), Positives = 119/153 (77%), Gaps = 3/153 (1%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTR+GFTGNLY L ++GI+TFIDD LQ+G+EIT +L +AIE+S+IFI
Sbjct: 8 YDVFLSFRGEDTRHGFTGNLYNVLRERGIDTFIDDEELQKGHEITKALEEAIEKSKIFII 67
Query: 76 VFSINYASSSFCLDELDHIIHCYKTKG-RPVLPVFFGVDPSHVRHHKGSYGEALAEHEKR 134
V S NYASSSFCL+EL HI++ K K R +LPVF+ VDPS VR+H+GS+GEALA HEK+
Sbjct: 68 VLSENYASSSFCLNELTHILNFTKGKSDRSILPVFYKVDPSDVRYHRGSFGEALANHEKK 127
Query: 135 FQNDPKNMERLQGWKDALSQAANLSGYHDSPPG 167
+++ ME+LQ WK AL Q +N SG+H P G
Sbjct: 128 LKSN--YMEKLQIWKMALQQVSNFSGHHFQPDG 158
>UniRef100_Q84ZU9 R 13 protein [Glycine max]
Length = 641
Score = 185 bits (469), Expect = 5e-45
Identities = 97/157 (61%), Positives = 118/157 (74%), Gaps = 4/157 (2%)
Query: 6 SSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLK 65
++T+ S Y VFLSFRG DTR GFTGNLYKAL D+GI TFIDD L RG++ITP+L
Sbjct: 2 AATTRSLASIYDVFLSFRGLDTRNGFTGNLYKALGDRGIYTFIDDQELPRGDKITPALSN 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYG 125
AI ESRI I V S NYA SSFCLDEL I+HC K++G V+PVF+ VDPS VRH KGSYG
Sbjct: 62 AINESRIAITVLSENYAFSSFCLDELVTILHC-KSEGLLVIPVFYKVDPSDVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYH 162
E + +H+KRF++ ME+L+ W+ AL Q A+LSGYH
Sbjct: 121 ETMTKHQKRFES---KMEKLREWRMALQQVADLSGYH 154
>UniRef100_Q84ZV6 R 6 protein [Glycine max]
Length = 264
Score = 180 bits (457), Expect = 1e-43
Identities = 89/147 (60%), Positives = 110/147 (74%), Gaps = 6/147 (4%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIP 75
Y VFL+FRG DTRYGFTGNLY+AL+DKGI TF D+ L G EITP+LLKAI++SRI I
Sbjct: 12 YDVFLNFRGEDTRYGFTGNLYRALSDKGIRTFFDEEKLHSGEEITPALLKAIKDSRIAIT 71
Query: 76 VFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRF 135
V S ++ASSSFCLDEL I+HC + G ++PVF+ V PS VRH KG+YGEALA+H+ RF
Sbjct: 72 VLSEDFASSSFCLDELTSIVHCAQYNGMMIIPVFYKVYPSDVRHQKGTYGEALAKHKIRF 131
Query: 136 QNDPKNMERLQGWKDALSQAANLSGYH 162
E+ Q W+ AL Q A+LSG+H
Sbjct: 132 P------EKFQNWEMALRQVADLSGFH 152
>UniRef100_Q84ZU7 R 5 protein [Glycine max]
Length = 907
Score = 180 bits (457), Expect = 1e-43
Identities = 92/147 (62%), Positives = 110/147 (74%), Gaps = 6/147 (4%)
Query: 16 YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIP 75
Y VFLSFRG DTRYGFTGNLYKAL DKGI+TF D++ L G EITP+LLKAI++SRI I
Sbjct: 12 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 71
Query: 76 VFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRF 135
V S ++ASSSFCLDEL I+ C + G V+PVF+ V P VRH KG+YGEALA+H+KRF
Sbjct: 72 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 131
Query: 136 QNDPKNMERLQGWKDALSQAANLSGYH 162
++LQ W+ AL Q ANLSG H
Sbjct: 132 P------DKLQKWERALRQVANLSGLH 152
Score = 154 bits (388), Expect = 1e-35
Identities = 91/199 (45%), Positives = 121/199 (60%), Gaps = 8/199 (4%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSNCI 592
AF KM LK LII NG SKG Y L+ L+W SK L S+ I C
Sbjct: 552 AFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSKCLPSNFHPNNLLI-----CK 606
Query: 593 YL*CTMISFS---AKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNS 649
+M SF + KF +T+L D+C++LT IPDVS L NL +LSF+ C++L+ + +S
Sbjct: 607 LPDSSMASFEFHGSSKFGHLTVLKFDNCKFLTQIPDVSDLPNLRELSFKGCESLVAVDDS 666
Query: 650 IGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWY 709
IG LNKL++L+A+GCR L FPPL L SL+ L+LS C SL+ FP++L +M NI ++
Sbjct: 667 IGFLNKLKKLNAYGCRKLTSFPPLNLTSLETLQLSGCSSLEYFPEILGEMENIKQLVLRD 726
Query: 710 TSIRELPSSFQNLSELDEL 728
I+ELP SFQNL L L
Sbjct: 727 LPIKELPFSFQNLIGLQVL 745
>UniRef100_Q9M635 Hypothetical protein [Glycine max]
Length = 148
Score = 180 bits (457), Expect = 1e-43
Identities = 96/150 (64%), Positives = 114/150 (76%), Gaps = 7/150 (4%)
Query: 3 SPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPS 62
S SSS+S +YD VFLSFRG DTR FTG+LY L KGI+TFIDD LQRG +ITP+
Sbjct: 5 SRSSSSSSNYD----VFLSFRGEDTRSAFTGHLYNTLQSKGIHTFIDDEKLQRGEQITPA 60
Query: 63 LLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKG 122
L+KAIE+SR+ I V S +YASSSFCLDEL I+HC + K V+PVF+ VDPS VRH KG
Sbjct: 61 LMKAIEDSRVAITVLSEHYASSSFCLDELATILHCDQRKRLLVIPVFYKVDPSDVRHQKG 120
Query: 123 SYGEALAEHEKRFQNDPKNMERLQGWKDAL 152
SYGEALA+ E+RFQ+DP E+LQ WK AL
Sbjct: 121 SYGEALAKLERRFQHDP---EKLQNWKMAL 147
>UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max]
Length = 863
Score = 179 bits (453), Expect = 4e-43
Identities = 92/139 (66%), Positives = 109/139 (78%), Gaps = 4/139 (2%)
Query: 24 GSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIPVFSINYAS 83
G DTR GFTG LYKAL D+GI TFIDD L+RG+EI P+L AI+ESRI I V S NYAS
Sbjct: 3 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 62
Query: 84 SSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYGEALAEHEKRFQNDPKNME 143
SSFCLDEL I+HC K++G V+PVF+ VDPSHVRH KGSYGEA+A+H+KRF+ N E
Sbjct: 63 SSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFK---ANKE 118
Query: 144 RLQGWKDALSQAANLSGYH 162
+LQ W+ AL Q A+LSGYH
Sbjct: 119 KLQKWRMALHQVADLSGYH 137
Score = 144 bits (363), Expect = 1e-32
Identities = 89/198 (44%), Positives = 117/198 (58%), Gaps = 9/198 (4%)
Query: 533 AFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSS------ILSKA--SE 584
AF KM LK LII N SKG Y L+ L+W S L S+ ++ K S
Sbjct: 530 AFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCLPSNFDPINLVICKLPDSS 589
Query: 585 IKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLI 644
I SF ++ S S +K +T+L D CE+LT IPDVS L NL++LSF +C++L+
Sbjct: 590 ITSFEFHGSSKASLKS-SLQKLGHLTVLNFDRCEFLTKIPDVSDLPNLKELSFNWCESLV 648
Query: 645 TIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDK 704
+ +SIG LNKL+ LSA+GCR L FPPL L SL+ L L C SL+ FP++L +M NI
Sbjct: 649 AVDDSIGFLNKLKTLSAYGCRKLTSFPPLNLTSLETLNLGGCSSLEYFPEILGEMKNITV 708
Query: 705 IWFWYTSIRELPSSFQNL 722
+ I+ELP SFQNL
Sbjct: 709 LALHDLPIKELPFSFQNL 726
>UniRef100_Q84ZV1 R 9 protein [Glycine max]
Length = 264
Score = 178 bits (452), Expect = 5e-43
Identities = 90/157 (57%), Positives = 114/157 (72%), Gaps = 6/157 (3%)
Query: 6 SSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLK 65
++T+ S Y VFL+FRG DTRYGFT NLY+AL+DKGI TF D+ L G EITP+LLK
Sbjct: 2 AATTSSRASIYDVFLNFRGEDTRYGFTSNLYRALSDKGIRTFFDEEKLHSGEEITPALLK 61
Query: 66 AIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSYG 125
AI++SRI I V S ++ASSSFCLDEL I+HC + G ++PVF+ V PS VRH KG+YG
Sbjct: 62 AIKDSRIAITVLSEDFASSSFCLDELTSIVHCAQYNGMMIIPVFYKVYPSDVRHQKGTYG 121
Query: 126 EALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYH 162
EALA+H+ RF E+ Q W+ AL Q A+LSG+H
Sbjct: 122 EALAKHKIRFP------EKFQNWEMALRQVADLSGFH 152
>UniRef100_Q9FXZ2 Resistance protein LM12 [Glycine max]
Length = 438
Score = 174 bits (442), Expect = 7e-42
Identities = 89/150 (59%), Positives = 113/150 (75%), Gaps = 3/150 (2%)
Query: 14 YKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIF 73
+ Y VFLSFRG DTRYGFTG LY L ++GI+TFIDD+ Q G+EIT +L AIE+S+IF
Sbjct: 6 FSYDVFLSFRGEDTRYGFTGYLYNVLRERGIHTFIDDDEPQEGDEITTALEAAIEKSKIF 65
Query: 74 IPVFSINYASSSFCLDELDHIIHCYKTKGRP-VLPVFFGVDPSHVRHHKGSYGEALAEHE 132
I V S NYASSSFCL+ L HI++ K VLPVF+ V+PS VRHH+GS+GEALA HE
Sbjct: 66 IIVLSENYASSSFCLNSLTHILNFTKENNDVLVLPVFYRVNPSDVRHHRGSFGEALANHE 125
Query: 133 KRFQNDPKNMERLQGWKDALSQAANLSGYH 162
K+ ++ NME+L+ WK AL Q +N+SG+H
Sbjct: 126 KK--SNSNNMEKLETWKMALHQVSNISGHH 153
>UniRef100_Q84KB4 MRGH5 [Cucumis melo]
Length = 1092
Score = 171 bits (433), Expect = 7e-41
Identities = 90/170 (52%), Positives = 117/170 (67%), Gaps = 7/170 (4%)
Query: 5 SSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLL 64
SSS+S +++ Y VFLSFRG DTR FTG+LY L KG+N FIDD GL+RG +I+ +L
Sbjct: 10 SSSSSPIFNWSYDVFLSFRGEDTRSNFTGHLYMFLRQKGVNVFIDD-GLERGEQISETLF 68
Query: 65 KAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSY 124
K I+ S I I +FS NYASS++CLDEL I+ C K+KG+ VLP+F+ VDPS VR G +
Sbjct: 69 KTIQNSLISIVIFSENYASSTWCLDELVEIMECKKSKGQKVLPIFYKVDPSDVRKQNGWF 128
Query: 125 GEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDSPPGQIHLQQD 174
E LA+HE F ME++ W+DAL+ AANLSG+H + HL QD
Sbjct: 129 REGLAKHEANF------MEKIPIWRDALTTAANLSGWHLGARKEAHLIQD 172
Score = 82.0 bits (201), Expect = 6e-14
Identities = 66/189 (34%), Positives = 96/189 (49%), Gaps = 12/189 (6%)
Query: 545 IENGHCSKGLKY----LRSSLKALKWEGCLSKSL---SSSILSKASEIKSFSNCIYL*CT 597
++ HC K K S+L++L +E C + + S L+K +K NC L
Sbjct: 704 LDLSHCKKLEKIPDISSASNLRSLSFEQCTNLVMIHDSIGSLTKLVTLK-LQNCSNLKKL 762
Query: 598 MISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLE 657
S QD+ L C+ L IPD S SNL+ LS E C +L +H+SIG L+KL
Sbjct: 763 PRYISWNFLQDLN---LSWCKKLEEIPDFSSTSNLKHLSLEQCTSLRVVHDSIGSLSKLV 819
Query: 658 RLSAFGCRTLKRFPP-LGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELP 716
L+ C L++ P L L SL+ L LS C L++FP++ M ++ + T+IRELP
Sbjct: 820 SLNLEKCSNLEKLPSYLKLKSLQNLTLSGCCKLETFPEIDENMKSLYILRLDSTAIRELP 879
Query: 717 SSFQNLSEL 725
S L+ L
Sbjct: 880 PSIGYLTHL 888
Score = 80.1 bits (196), Expect = 2e-13
Identities = 66/208 (31%), Positives = 101/208 (47%), Gaps = 9/208 (4%)
Query: 531 GKAFKKMTRLKTLIIENGHCSKGLKYLRSS--LKALKWEGCLS-KSLSSSILSKASEIK- 586
GK + RLK L + + K + ++ L+ L C + K++ S LS +
Sbjct: 621 GKGLQNCMRLKLLDLRHSVILKKISESSAAPNLEELYLSNCSNLKTIPKSFLSLRKLVTL 680
Query: 587 SFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITI 646
+C+ L I S ++ + L L HC+ L IPD+S SNL LSFE C NL+ I
Sbjct: 681 DLHHCVNL--KKIPRSYISWEALEDLDLSHCKKLEKIPDISSASNLRSLSFEQCTNLVMI 738
Query: 647 HNSIGHLNKLERLSAFGCRTLKRFPP-LGLASLKELKLSCCYSLKSFPKLLCKMTNIDKI 705
H+SIG L KL L C LK+ P + L++L LS C L+ P +N+ +
Sbjct: 739 HDSIGSLTKLVTLKLQNCSNLKKLPRYISWNFLQDLNLSWCKKLEEIPD-FSSTSNLKHL 797
Query: 706 WF-WYTSIRELPSSFQNLSELDELSVRE 732
TS+R + S +LS+L L++ +
Sbjct: 798 SLEQCTSLRVVHDSIGSLSKLVSLNLEK 825
>UniRef100_Q9FPK9 Putative resistance protein [Glycine max]
Length = 1093
Score = 166 bits (420), Expect = 2e-39
Identities = 90/158 (56%), Positives = 111/158 (69%), Gaps = 4/158 (2%)
Query: 5 SSSTSISYDYK-YQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSL 63
S + S S D + Y VFLSFRG DTR FTGNLY L +GI+TFI D + G EI SL
Sbjct: 2 SKAVSESTDIRVYDVFLSFRGEDTRRSFTGNLYNCLEKRGIHTFIGDYDFESGEEIKASL 61
Query: 64 LKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGS 123
+AIE SR+F+ VFS NYASSS+CLD L I+ + RPV+PVFF V+PSHVRH KG
Sbjct: 62 SEAIEHSRVFVIVFSENYASSSWCLDGLVRILDFTEDNHRPVIPVFFDVEPSHVRHQKGI 121
Query: 124 YGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGY 161
YGEALA HE+R +P++ ++ W++AL QAANLSGY
Sbjct: 122 YGEALAMHERRL--NPESY-KVMKWRNALRQAANLSGY 156
Score = 145 bits (367), Expect = 3e-33
Identities = 87/209 (41%), Positives = 120/209 (56%), Gaps = 7/209 (3%)
Query: 531 GKAFKKMTRLKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSN 590
G AF KM L+TLII SKG K + LK L+W GC SKSL S + I
Sbjct: 552 GMAFVKMISLRTLIIRKMF-SKGPKNFQI-LKMLEWWGCPSKSLPSDFKPEKLAILKLPY 609
Query: 591 CIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSI 650
++ S F M +L D CE+LT PD+SG L++L F +C+NL+ IH+S+
Sbjct: 610 SGFM-----SLELPNFLHMRVLNFDRCEFLTRTPDLSGFPILKELFFVFCENLVEIHDSV 664
Query: 651 GHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYT 710
G L+KLE ++ GC L+ FPP+ L SL+ + LS C SL SFP++L KM NI + YT
Sbjct: 665 GFLDKLEIMNFEGCSKLETFPPIKLTSLESINLSHCSSLVSFPEILGKMENITHLSLEYT 724
Query: 711 SIRELPSSFQNLSELDELSVREFGMLRFP 739
+I +LP+S + L L L + GM++ P
Sbjct: 725 AISKLPNSIRELVRLQSLELHNCGMVQLP 753
>UniRef100_Q947D9 Resistance gene analog NBS9 [Helianthus annuus]
Length = 304
Score = 166 bits (420), Expect = 2e-39
Identities = 82/159 (51%), Positives = 110/159 (68%), Gaps = 6/159 (3%)
Query: 3 SPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPS 62
S SSS+S S+ K++VFLSFRG DTR F +LYK L +GI T+ DD L RG I P+
Sbjct: 33 SSSSSSSSSHSLKHEVFLSFRGEDTRKNFVDHLYKDLVQQGIQTYKDDETLPRGERIGPA 92
Query: 63 LLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKG 122
LLKAI+ES + VFS NYA SS+CLDEL HI+ C T+G+ V+P+F+ VDPS VR G
Sbjct: 93 LLKAIQESHFAVVVFSENYADSSWCLDELAHIMECVDTRGQIVIPIFYHVDPSDVRKQNG 152
Query: 123 SYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGY 161
YG+A +HE+ KN ++++ W++AL +A NLSG+
Sbjct: 153 KYGKAFTKHER------KNKQKVESWRNALEKAGNLSGW 185
>UniRef100_Q947E3 Resistance gene analog NBS5 [Helianthus annuus]
Length = 285
Score = 164 bits (415), Expect = 9e-39
Identities = 82/157 (52%), Positives = 109/157 (69%), Gaps = 6/157 (3%)
Query: 5 SSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLL 64
SSS+S S+ K++VFLSFRG DTR F +LYK L +GI T+ DD L+RG I P+LL
Sbjct: 36 SSSSSSSHSLKHEVFLSFRGEDTRKNFVDHLYKDLVQQGIQTYKDDETLRRGESIRPALL 95
Query: 65 KAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHHKGSY 124
KAI+ESRI + VFS NYA SS+CLDEL II C T G+ V+P+F+ VDPS VR G Y
Sbjct: 96 KAIQESRIAVIVFSENYADSSWCLDELQQIIECMDTNGQIVIPIFYHVDPSDVRKQNGKY 155
Query: 125 GEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGY 161
G+A +H+K +N ++++ W+ AL +A NLSG+
Sbjct: 156 GKAFRKHKK------ENKQKVESWRKALEKAGNLSGW 186
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.347 0.151 0.545
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,138,099,440
Number of Sequences: 2790947
Number of extensions: 42625424
Number of successful extensions: 172915
Number of sequences better than 10.0: 657
Number of HSP's better than 10.0 without gapping: 362
Number of HSP's successfully gapped in prelim test: 308
Number of HSP's that attempted gapping in prelim test: 169515
Number of HSP's gapped (non-prelim): 2210
length of query: 751
length of database: 848,049,833
effective HSP length: 135
effective length of query: 616
effective length of database: 471,271,988
effective search space: 290303544608
effective search space used: 290303544608
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC138133.8


