

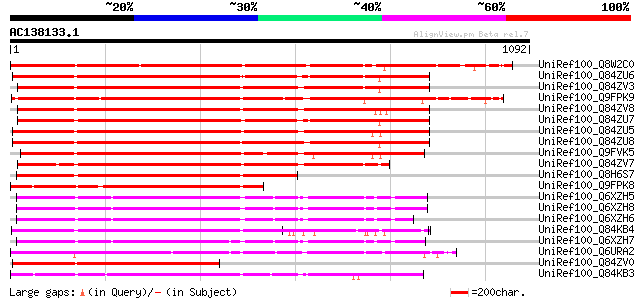
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.1 + phase: 0 /pseudo
(1092 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Gl... 935 0.0
UniRef100_Q84ZU6 R 1 protein [Glycine max] 834 0.0
UniRef100_Q84ZV3 R 4 protein [Glycine max] 832 0.0
UniRef100_Q9FPK9 Putative resistance protein [Glycine max] 826 0.0
UniRef100_Q84ZV8 R 3 protein [Glycine max] 819 0.0
UniRef100_Q84ZU7 R 5 protein [Glycine max] 818 0.0
UniRef100_Q84ZU5 R 8 protein [Glycine max] 816 0.0
UniRef100_Q84ZU8 R 10 protein [Glycine max] 815 0.0
UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max] 775 0.0
UniRef100_Q84ZV7 R 12 protein [Glycine max] 720 0.0
UniRef100_Q8H6S7 Resistance protein KR3 [Glycine max] 566 e-159
UniRef100_Q9FPK8 Putative resistance protein [Glycine max] 524 e-147
UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuber... 519 e-145
UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum] 516 e-144
UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuber... 510 e-142
UniRef100_Q84KB4 MRGH5 [Cucumis melo] 508 e-142
UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuber... 508 e-142
UniRef100_Q6URA2 TIR-NBS-LRR type R protein 7 [Malus baccata] 499 e-139
UniRef100_Q84ZV0 R 14 protein [Glycine max] 493 e-137
UniRef100_Q84KB3 MRGH63 [Cucumis melo] 482 e-134
>UniRef100_Q8W2C0 Functional candidate resistance protein KR1 [Glycine max]
Length = 1124
Score = 935 bits (2416), Expect = 0.0
Identities = 542/1113 (48%), Positives = 707/1113 (62%), Gaps = 93/1113 (8%)
Query: 1 MQSPSSSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEIT 60
M SSS+S SY + VFL+FRG DTR GFTGNLYKAL D+GIHTF+D+ ++ RGD+IT
Sbjct: 1 MAKQSSSSSFSYRFSNDVFLSFRGEDTRRGFTGNLYKALSDRGIHTFMDDKKIPRGDQIT 60
Query: 61 PSLLKAIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQ 120
L KAIEESRIFI V S NYASSSFCL+EL +I+ K KG L+LPVF+ V+P+ VR+
Sbjct: 61 SGLEKAIEESRIFIIVLSENYASSSFCLNELDYILKFIKGKGILILPVFYKVDPSDVRNH 120
Query: 121 KGSYGEALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYHDSPPG--YEYKLIGKIVK 178
GS+G+AL HEK+F++ ME+L+ WK AL++ ANLSGYH G YEY+ I +IV+
Sbjct: 121 TGSFGKALTNHEKKFKST-NDMEKLETWKMALNKVANLSGYHHFKHGEEYEYEFIQRIVE 179
Query: 179 YISNKISQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIY 238
+S KI++ PLHVA YPVGL+SR+Q++K+LLD GSD VHM+GI+G+GG+GK+TLA A+Y
Sbjct: 180 LVSKKINRAPLHVADYPVGLESRIQEVKALLDVGSDDVVHMLGIHGLGGVGKTTLAAAVY 239
Query: 239 NFIADQFECSCFLENVKESSASNNLKNLQQELLLKTLQLEIKLGSVSEGIPKIKERLHGK 298
N IAD FE CFL+NV+E+S + L++LQ+ LL + + E KL V +GI I+ RL K
Sbjct: 240 NSIADHFEALCFLQNVRETSKKHGLQHLQRNLLSE-MAGEDKLIGVKQGISIIEHRLRQK 298
Query: 299 KILLILDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEA 358
K+LLILDDVDK +QL+ALAGR D FGPGSRVIITTRDK LL CHG+E+TY V ELNE A
Sbjct: 299 KVLLILDDVDKREQLQALAGRPDLFGPGSRVIITTRDKQLLACHGVERTYEVNELNEEYA 358
Query: 359 LELLRWKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGR 418
LELL WKAFK EKV Y+D+L RA YASGLPLA+EV+GSNL GK+I + S LD+Y R
Sbjct: 359 LELLNWKAFKLEKVDPFYKDVLNRAATYASGLPLALEVIGSNLSGKNIEQWISALDRYKR 418
Query: 419 IPHKDIQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLV 478
IP+K+IQ+IL++SYDAL+E+EQS+FLDIACC K L EV+ ILH H+G+ +K H+ VLV
Sbjct: 419 IPNKEIQEILKVSYDALEEDEQSIFLDIACCFKKYDLAEVQDILHAHHGHCMKHHIGVLV 478
Query: 479 DKSLIKISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTG 538
+KSLIKIS + VTLH+LIE MGKE+VR+ESP+EPG+RSRLW DIV VL EN G
Sbjct: 479 EKSLIKISLDGY----VTLHDLIEDMGKEIVRKESPQEPGKRSRLWLPTDIVQVLEENKG 534
Query: 539 TGKTEMICMNLHSM--ESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWE 596
T +ICMN +S E I G AFKKM LKTLII +GH SKG KH P SL+ L+W
Sbjct: 535 TSHIGIICMNFYSSFEEVEIQWDGDAFKKMKNLKTLIIRSGHFSKGPKHFPKSLRVLEWW 594
Query: 597 GCLSKSLSSSILSKASEIKSFSNCIYL*CTMISFSPISSFVFMQKFQDMTILILDHCEYL 656
S + I + +C + S + + +KF ++T L D C++L
Sbjct: 595 RYPSHYFPYDFQMEKLAIFNLPDCGFT-------SRELAAMLKKKFVNLTSLNFDSCQHL 647
Query: 657 THIPDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKE 716
T IPDVS + +L+KLSF+ C NL IH S+G L KL L A GC +LK FPP+ L SL++
Sbjct: 648 TLIPDVSCVPHLQKLSFKDCDNLYAIHPSVGFLEKLRILDAEGCSRLKNFPPIKLTSLEQ 707
Query: 717 LDICCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFP 776
L + C SL++FPE+L KM NI E+DL+ + + P SFQNL+ L E +L FP
Sbjct: 708 LKLGFCHSLENFPEILGKMENITELDLE-QTPVKKFPLSFQNLTRL------ETVLLCFP 760
Query: 777 KHNDRMYSKVF-----------------------------------------SKVTKLRI 795
++ + +F S V L +
Sbjct: 761 RNQANGCTGIFLSNICPMQESPELINVIGVGWEGCLFRKEDEGAENVSLTTSSNVQFLDL 820
Query: 796 YECNLSDEYLQIVLKWCVNVELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGI 855
CNLSD++ I L NV L+LS NNF ++PEC+ EC L L L+YC L EIRGI
Sbjct: 821 RNCNLSDDFFPIALPCFANVMELNLSGNNFTVIPECIKECRFLTTLYLNYCERLREIRGI 880
Query: 856 PPNLKELSAYQCKSLSSSCRRMLMSQELHEARCTRFLFPNEKEGIPDWFEHQSRGDTISF 915
PPNLK A +C SL+SSCR ML+SQELHEA T F P K IP+WF+ Q+ ISF
Sbjct: 881 PPNLKYFYAEECLSLTSSCRSMLLSQELHEAGRTFFYLPGAK--IPEWFDFQTSEFPISF 938
Query: 916 WFRKEIPSMTYICIPPEGNNWSADTRVNYFVNGYEIEIDYCPRFSYVYFKHTNLFHTPKL 975
WFR + P++ I +S+ + N R + + NLF++ L
Sbjct: 939 WFRNKFPAIAICHIIKRVAEFSSSRGWTFRPN---------IRTKVIINGNANLFNSVVL 989
Query: 976 N-------ELGKRQFEYIMEKGLLKNEWIHVEFKVKDRDNIIFRNA----QMGIHVWKEK 1024
+L + +++ LL+NEW H E F A + G+HV K++
Sbjct: 990 GSDCTCLFDLRGERVTDNLDEALLENEWNHAEVTC---PGFTFTFAPTFIKTGLHVLKQE 1046
Query: 1025 SNTEEENVVFIDPYIRKTKSNEYLNASLSQKKR 1057
SN E+ + F DP RKTK + N+S + +R
Sbjct: 1047 SNMED--IRFSDP-CRKTKLDNDFNSSKPKNQR 1076
>UniRef100_Q84ZU6 R 1 protein [Glycine max]
Length = 902
Score = 834 bits (2154), Expect = 0.0
Identities = 469/910 (51%), Positives = 598/910 (65%), Gaps = 50/910 (5%)
Query: 6 SSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLK 65
++T+ S Y VFLNFRG DTRYGFTGNLYKAL DKGIHTF D +L GD+ITP+L K
Sbjct: 2 AATTRSLASIYDVFLNFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGDDITPALSK 61
Query: 66 AIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+HC K +G LV+PVF V+P+ VRH KGSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KREGLLVIPVFHNVDPSAVRHLKGSYG 120
Query: 126 EALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
EA+A+H+KRF+ E+LQ W+ AL Q A+LSGYH YEYK IG IV+ +S KI
Sbjct: 121 EAMAKHQKRFK---AKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIGNIVEEVSRKI 177
Query: 185 SQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQ 244
+ PLHVA YPVGL S+V ++ LLD GSD VH++GI+G+GGLGK+TLA A+YNFIA
Sbjct: 178 NCAPLHVADYPVGLGSQVIEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALAVYNFIALH 237
Query: 245 FECSCFLENVKESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLI 303
F+ SCFL+NV+E S + LK+ Q LL K L + +I L S EG I+ RL KK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHFQSILLSKLLGEKDITLTSWQEGASMIQHRLRRKKVLLI 297
Query: 304 LDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLR 363
LDDVDK +QLEA+ GR DWFGPGSRVIITTRDKHLL H +E+TY V+ LN AL+LL
Sbjct: 298 LDDVDKREQLEAIVGRSDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNHNAALQLLT 357
Query: 364 WKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKD 423
W AFK EK+ Y+D+L R V YASGLPLA+EV+GS+LFGK++AE ES ++ Y RIP +
Sbjct: 358 WNAFKREKIDPIYDDVLNRVVTYASGLPLALEVIGSDLFGKTVAEWESAVEHYKRIPSDE 417
Query: 424 IQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLI 483
I KIL++S+DAL EE+++VFLDIACC KG + EV+ IL YG K H+ VLV+KSLI
Sbjct: 418 ILKILKVSFDALGEEQKNVFLDIACCFKGYKWTEVDDILRAFYGNCKKHHIGVLVEKSLI 477
Query: 484 KISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTE 543
K++ C+ SG V +H+LI+ MG+E+ RQ SP+EP + RLWS DI VL NTGT K E
Sbjct: 478 KLN-CYDSG-TVEMHDLIQDMGREIERQRSPEEPWKCKRLWSPKDIFQVLKHNTGTSKIE 535
Query: 544 MICM--NLHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSK 601
+IC+ ++ E ++ AF KM LK LII NG SKG + P L L+W S
Sbjct: 536 IICLDFSISDKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPEGLTVLEWHRYPSN 595
Query: 602 SLSSSILSKASEIKSFSNCIYL*CTMISFSPISSFVF--MQKFQDMTILILDHCEYLTHI 659
L +F L C + S I+SF KF +T+L D CE+LT I
Sbjct: 596 CLP----------YNFHPNNLLICKLPD-SSITSFELHGPSKFWHLTVLNFDQCEFLTQI 644
Query: 660 PDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDI 719
PDVS L NL++LSF+ C +LI + +SIG LNKL++LSA+GCRKL+ FPPL L SL+ L +
Sbjct: 645 PDVSDLPNLKELSFDWCESLIAVDDSIGFLNKLKKLSAYGCRKLRSFPPLNLTSLETLQL 704
Query: 720 CCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFP--- 776
CSSL+ FPE+L +M NIK +DLD + I ELP SFQNL L L++ +++ P
Sbjct: 705 SGCSSLEYFPEILGEMENIKALDLD-GLPIKELPFSFQNLIGLCRLTLNSCGIIQLPCSL 763
Query: 777 -----------------------KHNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCV 813
+ +++ S + SK CNL D++ K
Sbjct: 764 AMMPELSVFRIENCNRWHWVESEEGEEKVGSMISSKELWFIAMNCNLCDDFFLTGSKRFT 823
Query: 814 NVELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSS 873
VE LDLS NNF ILPE E L+ L + C L+EIRG+PPNL+ A C SL+SS
Sbjct: 824 RVEYLDLSGNNFTILPEFFKELQFLRALMVSDCEHLQEIRGLPPNLEYFDARNCASLTSS 883
Query: 874 CRRMLMSQEL 883
+ ML++Q L
Sbjct: 884 TKSMLLNQVL 893
>UniRef100_Q84ZV3 R 4 protein [Glycine max]
Length = 895
Score = 832 bits (2149), Expect = 0.0
Identities = 460/901 (51%), Positives = 602/901 (66%), Gaps = 54/901 (5%)
Query: 16 YQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIA 75
Y VFL+FRG DTR+GFTGNLYKALDD+GI+T ID+ EL RGDEITP+L KAI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTSIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRF 135
V S NYASSSFCLDELV I+HC K++G LV+PVF+ V+P+ VRHQKGSYGEA+A+H+KRF
Sbjct: 72 VLSQNYASSSFCLDELVTILHC-KSEGLLVIPVFYKVDPSDVRHQKGSYGEAMAKHQKRF 130
Query: 136 QNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISQQPLHVATY 194
+ E+LQ W+ AL Q A+LSGYH + YEYK IG IV+ +S KIS+ LHVA Y
Sbjct: 131 K---AKKEKLQKWRMALKQVADLSGYHFEDGDAYEYKFIGSIVEEVSRKISRASLHVADY 187
Query: 195 PVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCFLENV 254
PVGL+S+V ++ LLD GSD VH++GI+G+GGLGK+TLA +YN IA F+ SCFL+NV
Sbjct: 188 PVGLESQVTEVMKLLDVGSDDLVHIIGIHGMGGLGKTTLALEVYNLIALHFDESCFLQNV 247
Query: 255 KESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLILDDVDKLDQL 313
+E S + LK+LQ LL K L + +I L S EG I+ RL KK+LLILDDV+K +QL
Sbjct: 248 REESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASTIQHRLQRKKVLLILDDVNKREQL 307
Query: 314 EALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKNEKVP 373
+A+ GR DWFGPGSRVIITTRDKHLL CH +E+TY V+ LN AL+LL W AFK EK+
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHLLKCHEVERTYEVKVLNHNAALQLLTWNAFKREKID 367
Query: 374 SSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILRLSYD 433
SYED+L R V YASGLPLA+E++GSN+FGKS+A ES ++ Y RIP+ +I +IL++S+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNMFGKSVAGWESAVEHYKRIPNDEILEILKVSFD 427
Query: 434 ALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCFFSGI 493
AL EE+++VFLDIA C+KGC+L EVE +L Y +K H+ VLVDKSLIK+ GI
Sbjct: 428 ALGEEQKNVFLDIAFCLKGCKLTEVEHMLCSLYDNCMKHHIDVLVDKSLIKVK----HGI 483
Query: 494 KVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICM--NLHS 551
V +H+LI+V+G+E+ RQ SP+EPG+R RLW DI+HVL +NTGT K E+IC+ ++
Sbjct: 484 -VEMHDLIQVVGREIERQRSPEEPGKRKRLWLPKDIIHVLKDNTGTSKIEIICLDFSISY 542
Query: 552 MESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILSKA 611
E ++ AF KM LK LII NG SKG + P L+ L+W S L S+
Sbjct: 543 KEETVEFNENAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSNFLPSNF---- 598
Query: 612 SEIKSFSNCIYL*CTMISFSPISSFVF---MQKFQDMTILILDHCEYLTHIPDVSGLSNL 668
+ I L + S I SF F +K +T+L D C++LT IPDVS L NL
Sbjct: 599 -------DPINLVICKLPDSSIKSFEFHGSSKKLGHLTVLKFDRCKFLTQIPDVSDLPNL 651
Query: 669 EKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDICCCSSLKSF 728
+LSFE C +L+ + +SIG L KL++LSA+GCRKL FPPL L SL+ L + CSSL+ F
Sbjct: 652 RELSFEDCESLVAVDDSIGFLKKLKKLSAYGCRKLTSFPPLNLTSLETLQLSSCSSLEYF 711
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFP------------ 776
PE+L +M NI+E+ L + I ELP SFQNL+ L L++ +++ P
Sbjct: 712 PEILGEMENIRELRLT-GLYIKELPFSFQNLTGLRLLALSGCGIVQLPCSLAMMPELSSF 770
Query: 777 --------------KHNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCVNVELLDLSH 822
+ +++ S + SK CNL D++ K +V L+LS
Sbjct: 771 YTDYCNRWQWIELEEGEEKLGSIISSKAQLFCATNCNLCDDFFLAGFKRFAHVGYLNLSG 830
Query: 823 NNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCRRMLMSQE 882
NNF ILPE E L+ L + C L+EIRG+PP L+ A C S +SS ML++Q
Sbjct: 831 NNFTILPEFFKELQFLRTLDVSDCEHLQEIRGLPPILEYFDARNCVSFTSSSTSMLLNQV 890
Query: 883 L 883
L
Sbjct: 891 L 891
>UniRef100_Q9FPK9 Putative resistance protein [Glycine max]
Length = 1093
Score = 826 bits (2134), Expect = 0.0
Identities = 495/1123 (44%), Positives = 677/1123 (60%), Gaps = 129/1123 (11%)
Query: 5 SSSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLL 64
S ST I Y VFL+FRG DTR FTGNLY L+ +GIHTFI +++ + G+EI SL
Sbjct: 6 SESTDIRV---YDVFLSFRGEDTRRSFTGNLYNCLEKRGIHTFIGDYDFESGEEIKASLS 62
Query: 65 KAIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSY 124
+AIE SR+F+ VFS NYASSS+CLD LV I+ + R V+PVFF VEP+ VRHQKG Y
Sbjct: 63 EAIEHSRVFVIVFSENYASSSWCLDGLVRILDFTEDNHRPVIPVFFDVEPSHVRHQKGIY 122
Query: 125 GEALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGY-HDSPPGYEYKLIGKIVKYISNK 183
GEALA HE+R +P+S + ++ W+ AL QAANLSGY GYEYKLI KIV+ ISNK
Sbjct: 123 GEALAMHERRL--NPESYKVMK-WRNALRQAANLSGYAFKHGDGYEYKLIEKIVEDISNK 179
Query: 184 IS-QQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIA 242
I +P V PVGL+ R+ ++ LLD S GVHM+GI GIGG+GK+TLA+A+Y+ A
Sbjct: 180 IKISRP--VVDRPVGLEYRMLEVDWLLDATSLAGVHMIGICGIGGIGKTTLARAVYHSAA 237
Query: 243 DQFECSCFLENVKESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKIL 301
F+ SCFL NV+E++ + L +LQQ LL + + I+L SV +GI IK+ L K++L
Sbjct: 238 GHFDTSCFLGNVRENAMKHGLVHLQQTLLAEIFRENNIRLTSVEQGISLIKKMLPRKRLL 297
Query: 302 LILDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALEL 361
L+LDDV +LD L AL G DWFGPGSRVIITTRD+HLL HG++K Y VE L EALEL
Sbjct: 298 LVLDDVCELDDLRALVGSPDWFGPGSRVIITTRDRHLLKAHGVDKVYEVEVLANGEALEL 357
Query: 362 LRWKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPH 421
L WKAF+ ++V + + L RA+ +ASG+PLA+E++GS+L+G+ I E ESTLD+Y + P
Sbjct: 358 LCWKAFRTDRVHPDFINKLNRAITFASGIPLALELIGSSLYGRGIEEWESTLDQYEKNPP 417
Query: 422 KDIQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKS 481
+DI L++S+DAL E+ VFLDIAC G L E+E IL H+G +K H+ LV+KS
Sbjct: 418 RDIHMALKISFDALGYLEKEVFLDIACFFNGFELAEIEHILGAHHGCCLKFHIGALVEKS 477
Query: 482 LIKISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGK 541
LI I +V +H+LI+ MG+E+VRQESP+ PG+RSRLWS +DIVHVL +NTGT K
Sbjct: 478 LIMID----EHGRVQMHDLIQQMGREIVRQESPEHPGKRSRLWSTEDIVHVLEDNTGTCK 533
Query: 542 TEMICMNLHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSK 601
+ I ++ E V+ G AF KM L+TLII SKG K+ LK L+W GC SK
Sbjct: 534 IQSIILDFSKSEKVVQWDGMAFVKMISLRTLIIRK-MFSKGPKNF-QILKMLEWWGCPSK 591
Query: 602 SLSSSILSKASEIKSFSNCIYL*CTMISFSPISSFVFMQ--KFQDMTILILDHCEYLTHI 659
SL S + I P S F+ ++ F M +L D CE+LT
Sbjct: 592 SLPSDFKPEKLAILKL--------------PYSGFMSLELPNFLHMRVLNFDRCEFLTRT 637
Query: 660 PDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDI 719
PD+SG L++L F C NL+ IH+S+G L+KLE ++ GC KL+ FPP+ L SL+ +++
Sbjct: 638 PDLSGFPILKELFFVFCENLVEIHDSVGFLDKLEIMNFEGCSKLETFPPIKLTSLESINL 697
Query: 720 CCCSSLKSFPELLCKMTNIKEIDLDY----------------------NISIGELPSSFQ 757
CSSL SFPE+L KM NI + L+Y N + +LPSS
Sbjct: 698 SHCSSLVSFPEILGKMENITHLSLEYTAISKLPNSIRELVRLQSLELHNCGMVQLPSSIV 757
Query: 758 NLSELDELSVREARMLRFPKHNDRMYSKVF----SKVTKLRIYECNLSDEYLQIVLKWCV 813
L EL+ LS+ + LRF K ++ + +K S + ++ ++ C++SDE++ L W
Sbjct: 758 TLRELEVLSICQCEGLRFSKQDEDVKNKSLLMPSSYLKQVNLWSCSISDEFIDTGLAWFA 817
Query: 814 NVELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQ------- 866
NV+ LDLS NNF ILP C+ EC L+ L L YC+ L EIRGIPPNL+ LSA +
Sbjct: 818 NVKSLDLSANNFTILPSCIQECRLLRKLYLDYCTHLHEIRGIPPNLETLSAIRCTSLKDL 877
Query: 867 -------------------------------------------CKSLSSSCRRMLMSQEL 883
C+SL++SCRRML+ QEL
Sbjct: 878 DLAVPLESTKEGCCLRQLILDDCENLQEIRGIPPSIEFLSATNCRSLTASCRRMLLKQEL 937
Query: 884 HEARCTRFLFPNEKEGIPDWFEHQSRGDTISFWFRKEIPSMTYICIPPEGNNWSADTRVN 943
HEA R+ P + IP+WFEH SRG +ISFWFR + P ++ +C+ + +
Sbjct: 938 HEAGNKRYSLPGTR--IPEWFEHCSRGQSISFWFRNKFPVIS-LCLAGLMHKHPFGLKPI 994
Query: 944 YFVNGYEIEIDYCPRFSYVYFKHTNLFHTPKLNELGKRQ--FEYIMEKGLLKNEWIH--- 998
+NG +++ ++ R+ Y F T + G+RQ FE +++ + +N+W H
Sbjct: 995 VSINGNKMKTEFQRRWFYFEFP----VLTDHILIFGERQIKFEDNVDEVVSENDWNHVVV 1050
Query: 999 ---VEFKVKDRDNIIFRNAQMGIHVWKEKSNTEEENVVFIDPY 1038
V+FK + ++ R G+HV K KS+ E+ + FIDPY
Sbjct: 1051 SVDVDFKWNPTEPLVVRT---GLHVIKPKSSVED--IRFIDPY 1088
>UniRef100_Q84ZV8 R 3 protein [Glycine max]
Length = 897
Score = 819 bits (2115), Expect = 0.0
Identities = 457/904 (50%), Positives = 600/904 (65%), Gaps = 57/904 (6%)
Query: 16 YQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIA 75
Y VFL+FRG DTR+GFTGNLYKALDD+GI+TFID+ EL RGDEITP+L KAI+ESRI I
Sbjct: 12 YDVFLSFRGLDTRHGFTGNLYKALDDRGIYTFIDDQELPRGDEITPALSKAIQESRIAIT 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRF 135
V S NYASSSFCLDELV ++ C K KG LV+PVF+ V+P+ VR QKGSYGEA+A+H+KRF
Sbjct: 72 VLSQNYASSSFCLDELVTVLLC-KRKGLLVIPVFYNVDPSDVRQQKGSYGEAMAKHQKRF 130
Query: 136 QNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISQQPLHVATY 194
+ E+LQ W+ AL Q A+LSGYH YEYK I IV+ +S +I++ PLHVA Y
Sbjct: 131 K---AKKEKLQKWRMALHQVADLSGYHFKDGDAYEYKFIQSIVEQVSREINRTPLHVADY 187
Query: 195 PVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCFLENV 254
PVGL S+V +++ LLD GS VH++GI+G+GGLGK+TLA A+YN IA F+ SCFL+NV
Sbjct: 188 PVGLGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALHFDESCFLQNV 247
Query: 255 KESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLILDDVDKLDQL 313
+E S + LK+LQ +L K L + +I L S EG I+ RL KK+LLILDDVDK QL
Sbjct: 248 REESNKHGLKHLQSIILSKLLGEKDINLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQL 307
Query: 314 EALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKNEKVP 373
+A+ GR DWFGPGSRVIITTRDKH+L H +E+TY V+ LN++ AL+LL+W AFK EK
Sbjct: 308 KAIVGRPDWFGPGSRVIITTRDKHILKYHEVERTYEVKVLNQSAALQLLKWNAFKREKND 367
Query: 374 SSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILRLSYD 433
SYED+L R V YASGLPLA+E++GSNLFGK++AE ES ++ Y RIP +I +IL++S+D
Sbjct: 368 PSYEDVLNRVVTYASGLPLALEIIGSNLFGKTVAEWESAMEHYKRIPSDEILEILKVSFD 427
Query: 434 ALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCFFSGI 493
AL EE+++VFLDIACC+KGC+L EVE +L Y +K H+ VLVDKSL K+ GI
Sbjct: 428 ALGEEQKNVFLDIACCLKGCKLTEVEHMLRGLYDNCMKHHIDVLVDKSLTKVR----HGI 483
Query: 494 KVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICM--NLHS 551
V +H+LI+ MG+E+ RQ SP+EPG+R RLWS DI+ VL NTGT K E+I + ++
Sbjct: 484 -VEMHDLIQDMGREIERQRSPEEPGKRKRLWSPKDIIQVLKHNTGTSKIEIIYVDFSISD 542
Query: 552 MESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILSKA 611
E ++ AF KM LK LII NG SKG + P L+ L+W S L S+
Sbjct: 543 KEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPQGLRVLEWHRYPSNCLPSNF---- 598
Query: 612 SEIKSFSNCIYL*CTMISFSPISSFVF--MQKFQDMTILILDHCEYLTHIPDVSGLSNLE 669
+ I L + S ++SF F K +T+L D C++LT IPDVS L NL
Sbjct: 599 -------DPINLVICKLPDSSMTSFEFHGSSKLGHLTVLKFDWCKFLTQIPDVSDLPNLR 651
Query: 670 KLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDICCCSSLKSFP 729
+LSF+ C +L+ + +SIG LNKL++L+A+GCRKL FPPL L SL+ L++ CSSL+ FP
Sbjct: 652 ELSFQWCESLVAVDDSIGFLNKLKKLNAYGCRKLTSFPPLHLTSLETLELSHCSSLEYFP 711
Query: 730 ELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSV-------REARMLRFPK----- 777
E+L +M NI+ +DL + + I ELP SFQNL L +LS+ + PK
Sbjct: 712 EILGEMENIERLDL-HGLPIKELPFSFQNLIGLQQLSMFGCGIVQLRCSLAMMPKLSAFK 770
Query: 778 ----------HNDRMYSKVFSKVT--------KLRIYECNLSDEYLQIVLKWCVNVELLD 819
++ KV S ++ CNL D++ K +V L+
Sbjct: 771 FVNCNRWQWVESEEAEEKVGSIISSEARFWTHSFSAKNCNLCDDFFLTGFKKFAHVGYLN 830
Query: 820 LSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCRRMLM 879
LS NNF ILPE E L L + +C L+EIRGIP NL+ +A C SL+SS + ML+
Sbjct: 831 LSRNNFTILPEFFKELQFLGSLNVSHCKHLQEIRGIPQNLRLFNARNCASLTSSSKSMLL 890
Query: 880 SQEL 883
+Q L
Sbjct: 891 NQVL 894
>UniRef100_Q84ZU7 R 5 protein [Glycine max]
Length = 907
Score = 818 bits (2114), Expect = 0.0
Identities = 451/901 (50%), Positives = 595/901 (65%), Gaps = 56/901 (6%)
Query: 16 YQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIA 75
Y VFL+FRG DTRYGFTGNLYKAL DKGIHTF D +L G+EITP+LLKAI++SRI I
Sbjct: 12 YDVFLSFRGEDTRYGFTGNLYKALCDKGIHTFFDEDKLHSGEEITPALLKAIQDSRIAIT 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRF 135
V S ++ASSSFCLDEL I+ C + G +V+PVF+ V P VRHQKG+YGEALA+H+KRF
Sbjct: 72 VLSEDFASSSFCLDELATILFCAQYNGMMVIPVFYKVYPCDVRHQKGTYGEALAKHKKRF 131
Query: 136 QNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISQQPLHVATY 194
++LQ W+ AL Q ANLSG H YEYK IG+IV +S KI+ LHVA
Sbjct: 132 P------DKLQKWERALRQVANLSGLHFKDRDEYEYKFIGRIVASVSEKINPASLHVADL 185
Query: 195 PVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYN--FIADQFECSCFLE 252
PVGL+S+VQ+++ LLD G+ GV M+GI+G+GG+GKSTLA+A+YN I + F+ CFLE
Sbjct: 186 PVGLESKVQEVRKLLDVGNHDGVCMIGIHGMGGIGKSTLARAVYNDLIITENFDGLCFLE 245
Query: 253 NVKESSASNNLKNLQQELLLKTLQLEIKLGSVSEGIPKIKERLHGKKILLILDDVDKLDQ 312
NV+ESS ++ L++LQ LL + L +IK+ S +GI KI+ L GKK+LLILDDVDK Q
Sbjct: 246 NVRESSNNHGLQHLQSILLSEILGEDIKVRSKQQGISKIQSMLKGKKVLLILDDVDKPQQ 305
Query: 313 LEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKNEKV 372
L+ +AGR DWFGPGS +IITTRDK LL HG++K Y VE LN+ AL+LL W AFK EK+
Sbjct: 306 LQTIAGRRDWFGPGSIIIITTRDKQLLAPHGVKKRYEVEVLNQNAALQLLTWNAFKREKI 365
Query: 373 PSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILRLSY 432
SYED+L R V YASGLPLA+EV+GSN+FGK +AE +S ++ Y RIP+ +I +IL++S+
Sbjct: 366 DPSYEDVLNRVVTYASGLPLALEVIGSNMFGKRVAEWKSAVEHYKRIPNDEILEILKVSF 425
Query: 433 DALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCFFSG 492
DAL EE+++VFLDIACC KGC+L EVE +L Y +K H+ VLVDKSLIK+
Sbjct: 426 DALGEEQKNVFLDIACCFKGCKLTEVEHMLRGLYNNCMKHHIDVLVDKSLIKVRHG---- 481
Query: 493 IKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICM--NLH 550
V +H+LI+V+G+E+ RQ SP+EPG+ RLW DI+ VL NTGT K E+IC+ ++
Sbjct: 482 -TVNMHDLIQVVGREIERQISPEEPGKCKRLWLPKDIIQVLKHNTGTSKIEIICLDFSIS 540
Query: 551 SMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILSK 610
E ++ AF KM LK LII NG SKG + P L+ L+W SK L S
Sbjct: 541 DKEQTVEWNQNAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSKCLPS----- 595
Query: 611 ASEIKSFSNCIYL*CTMISFSPISSFVF--MQKFQDMTILILDHCEYLTHIPDVSGLSNL 668
+F L C + S ++SF F KF +T+L D+C++LT IPDVS L NL
Sbjct: 596 -----NFHPNNLLICKLPD-SSMASFEFHGSSKFGHLTVLKFDNCKFLTQIPDVSDLPNL 649
Query: 669 EKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDICCCSSLKSF 728
+LSF+ C +L+ + +SIG LNKL++L+A+GCRKL FPPL L SL+ L + CSSL+ F
Sbjct: 650 RELSFKGCESLVAVDDSIGFLNKLKKLNAYGCRKLTSFPPLNLTSLETLQLSGCSSLEYF 709
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFP------------ 776
PE+L +M NIK++ L ++ I ELP SFQNL L L + ++ P
Sbjct: 710 PEILGEMENIKQLVL-RDLPIKELPFSFQNLIGLQVLYLWSCLIVELPCRLVMMPELFQL 768
Query: 777 --------------KHNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCVNVELLDLSH 822
+ +++ S + SK R CNL D++ K +VE LDLS
Sbjct: 769 HIEYCNRWQWVESEEGEEKVGSILSSKARWFRAMNCNLCDDFFLTGSKRFTHVEYLDLSG 828
Query: 823 NNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCRRMLMSQE 882
NNF ILPE E L+ L + C L++IRG+PPNLK+ A C SL+SS + ML++Q
Sbjct: 829 NNFTILPEFFKELKFLRTLDVSDCEHLQKIRGLPPNLKDFRAINCASLTSSSKSMLLNQV 888
Query: 883 L 883
L
Sbjct: 889 L 889
>UniRef100_Q84ZU5 R 8 protein [Glycine max]
Length = 892
Score = 816 bits (2108), Expect = 0.0
Identities = 458/908 (50%), Positives = 597/908 (65%), Gaps = 50/908 (5%)
Query: 6 SSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLK 65
++T+ S Y Y VFL+F G DTR GFTG LYKAL D+GI+TFID+ EL+RGDEI P+L
Sbjct: 2 AATTRSLAYNYDVFLSFTGQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSN 61
Query: 66 AIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+HC K++G LV+PVF+ V+P+ VRHQKGSYG
Sbjct: 62 AIQESRIAITVLSQNYASSSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
EA+A+H+KRF+ + E+LQ W+ AL Q A+LSGYH YEY+ IG IV+ IS K
Sbjct: 121 EAMAKHQKRFK---ANKEKLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKF 177
Query: 185 SQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQ 244
S+ LHVA YPVGL+S V ++ LLD GS VH++GI+G+GGLGK+TLA A++NFIA
Sbjct: 178 SRASLHVADYPVGLESEVTEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALH 237
Query: 245 FECSCFLENVKESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLI 303
F+ SCFL+NV+E S + LK+LQ LL K L + +I L S EG I+ RL KK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLI 297
Query: 304 LDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLR 363
LDDVDK QL+A+ GR DWFGPGSRVIITTRDKHLL H +E+TY V+ LN++ AL+LL
Sbjct: 298 LDDVDKRQQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLT 357
Query: 364 WKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKD 423
W AFK EK+ SYED+L R V YASGLPLA+EV+GSNLF K++AE ES ++ Y RIP +
Sbjct: 358 WNAFKREKIDPSYEDVLNRVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDE 417
Query: 424 IQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLI 483
IQ+IL++S+DAL EE+++VFLDIACC KG EV+ IL YG K H+ VLV+KSL+
Sbjct: 418 IQEILKVSFDALGEEQKNVFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLV 477
Query: 484 KISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTE 543
K+S C V +H++I+ MG+E+ RQ SP+EPG+ RL DI+ VL +NTGT K E
Sbjct: 478 KVSCC----DTVEMHDMIQDMGREIERQRSPEEPGKCKRLLLPKDIIQVLKDNTGTSKIE 533
Query: 544 MICM--NLHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSK 601
+IC+ ++ E ++ AF KM LK LII N SKG + P L+ L+W S
Sbjct: 534 IICLDFSISDKEETVEWNENAFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSN 593
Query: 602 SLSSSILSKASEIKSFSNCIYL*CTMISFSPISSFVF---MQKFQDMTILILDHCEYLTH 658
L S+ + I L + S I+SF F +K +T+L D CE+LT
Sbjct: 594 CLPSNF-----------DPINLVICKLPDSSITSFEFHGSSKKLGHLTVLNFDRCEFLTK 642
Query: 659 IPDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELD 718
IPDVS L NL++LSF C +L+ + +SIG LNKL+ LSA+GCRKL FPPL L SL+ L+
Sbjct: 643 IPDVSDLPNLKELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSFPPLNLTSLETLN 702
Query: 719 ICCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSE-----LDELSVREAR-- 771
+ CSSL+ FPE+L +M NI + L +++ I ELP SFQNL LD + + R
Sbjct: 703 LGGCSSLEYFPEILGEMKNITVLAL-HDLPIKELPFSFQNLIGLLFLWLDSCGIVQLRCS 761
Query: 772 MLRFPK----------------HNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCVNV 815
+ PK ++ KV + +CNL D++ I K +V
Sbjct: 762 LATMPKLCEFCITDSCNRWQWVESEEGEEKVVGSILSFEATDCNLCDDFFFIGSKRFAHV 821
Query: 816 ELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCR 875
L+L NNF ILPE E L L +H C L+EIRG+PPNLK A C SL+SS +
Sbjct: 822 GYLNLPGNNFTILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNLKHFDARNCASLTSSSK 881
Query: 876 RMLMSQEL 883
ML++Q L
Sbjct: 882 SMLLNQVL 889
>UniRef100_Q84ZU8 R 10 protein [Glycine max]
Length = 901
Score = 815 bits (2105), Expect = 0.0
Identities = 453/910 (49%), Positives = 601/910 (65%), Gaps = 50/910 (5%)
Query: 6 SSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLK 65
++T+ S Y VFLNFRG DTRYGFTGNLY+AL DKGIHTF D +L RG+EITP+LLK
Sbjct: 2 AATTRSRASIYDVFLNFRGGDTRYGFTGNLYRALCDKGIHTFFDEKKLHRGEEITPALLK 61
Query: 66 AIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYG 125
AI+ESRI I V S NYASSSFCLDELV I+HC K++G LV+PVF+ V+P+ VRHQKGSYG
Sbjct: 62 AIQESRIAITVLSKNYASSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDVRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
+A+H+KRF+ E+LQ W+ AL Q A+L GYH YEYK I IV+ +S +I
Sbjct: 121 VEMAKHQKRFK---AKKEKLQKWRIALKQVADLCGYHFKDGDAYEYKFIQSIVEQVSREI 177
Query: 185 SQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQ 244
++ PLHVA YPVGL S+V +++ LLD GS VH++GI+G+GGLGK+TLA A+YN IA
Sbjct: 178 NRAPLHVADYPVGLGSQVIEVRKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVYNLIALH 237
Query: 245 FECSCFLENVKESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLI 303
F+ SCFL+NV+E S + LK+LQ LL K L + +I L S EG I+ RL KK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLI 297
Query: 304 LDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLR 363
LDDVDK +QL+A+ GR DWFGPGSRVIITTRDKHLL H +E+TY V+ LN++ AL+LL+
Sbjct: 298 LDDVDKREQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLK 357
Query: 364 WKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKD 423
W AFK EK+ SYED+L R V YASGLPLA+EV+GSNLFGK++AE ES ++ Y RIP +
Sbjct: 358 WNAFKREKIDPSYEDVLNRVVTYASGLPLALEVIGSNLFGKTVAEWESAMEHYKRIPSDE 417
Query: 424 IQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLI 483
I +IL++S+DAL EE+++VFLDIACC +G + EV+ IL YG K H+ VLV+KSLI
Sbjct: 418 ILEILKVSFDALGEEQKNVFLDIACCFRGYKWTEVDDILRALYGNCKKHHIGVLVEKSLI 477
Query: 484 KISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTE 543
K++ C+ + V +H+LI+ M +E+ R+ SP+EPG+ RLW DI+ V +NTGT K E
Sbjct: 478 KLN-CYGTDT-VEMHDLIQDMAREIERKRSPQEPGKCKRLWLPKDIIQVFKDNTGTSKIE 535
Query: 544 MICMN--LHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSK 601
+IC++ + E ++ AF KM LK LII N SKG + P L+ L+W S
Sbjct: 536 IICLDSSISDKEETVEWNENAFMKMENLKILIIRNDKFSKGPNYFPEGLRVLEWHRYPSN 595
Query: 602 SLSSSILSKASEIKSFSNCIYL*CTMISFSPISSFVFM--QKFQDMTILILDHCEYLTHI 659
L S+ I + S ++SF F KF +T+L D+C++LT I
Sbjct: 596 CLPSNFHPNNLVICKLPD-----------SCMTSFEFHGPSKFGHLTVLKFDNCKFLTQI 644
Query: 660 PDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDI 719
PDVS L NL +LSFE C +L+ + +SIG LNKL++LSA+GC KLK FPPL L SL+ L++
Sbjct: 645 PDVSDLPNLRELSFEECESLVAVDDSIGFLNKLKKLSAYGCSKLKSFPPLNLTSLQTLEL 704
Query: 720 CCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFP--- 776
CSSL+ FPE++ +M NIK + L Y + I EL SFQNL L L++R +++ P
Sbjct: 705 SQCSSLEYFPEIIGEMENIKHLFL-YGLPIKELSFSFQNLIGLRWLTLRSCGIVKLPCSL 763
Query: 777 -----------------------KHNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCV 813
+ ++ S SK + +CNL D++ K
Sbjct: 764 AMMPELFEFHMEYCNRWQWVESEEGEKKVGSIPSSKAHRFSAKDCNLCDDFFLTGFKTFA 823
Query: 814 NVELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSS 873
V L+LS NNF ILPE E L+ L + C L+EIRG+PPNL+ A C SL+SS
Sbjct: 824 RVGHLNLSGNNFTILPEFFKELQLLRSLMVSDCEHLQEIRGLPPNLEYFDARNCASLTSS 883
Query: 874 CRRMLMSQEL 883
+ ML++Q L
Sbjct: 884 SKNMLLNQVL 893
>UniRef100_Q9FVK5 Resistance protein LM6 [Glycine max]
Length = 863
Score = 775 bits (2002), Expect = 0.0
Identities = 442/888 (49%), Positives = 573/888 (63%), Gaps = 65/888 (7%)
Query: 24 GSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIAVFSINYAS 83
G DTR GFTG LYKAL D+GI+TFID+ EL+RGDEI P+L AI+ESRI I V S NYAS
Sbjct: 3 GQDTRQGFTGYLYKALCDRGIYTFIDDQELRRGDEIKPALSNAIQESRIAITVLSQNYAS 62
Query: 84 SSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRFQNDPKSME 143
SSFCLDELV I+HC K++G LV+PVF+ V+P+ VRHQKGSYGEA+A+H+KRF+ + E
Sbjct: 63 SSFCLDELVTILHC-KSQGLLVIPVFYKVDPSHVRHQKGSYGEAMAKHQKRFK---ANKE 118
Query: 144 RLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKISQQPLHVATYPVGLQSRV 202
+LQ W+ AL Q A+LSGYH YEY+ IG IV+ IS K S+ LHVA YPVGL+S V
Sbjct: 119 KLQKWRMALHQVADLSGYHFKDGDSYEYEFIGSIVEEISRKFSRASLHVADYPVGLESEV 178
Query: 203 QQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCFLENVKESSASNN 262
++ LLD GS VH++GI+G+GGLGK+TLA A++NFIA F+ SCFL+NV+E S +
Sbjct: 179 TEVMKLLDVGSHDVVHIIGIHGMGGLGKTTLALAVHNFIALHFDESCFLQNVREESNKHG 238
Query: 263 LKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLILDDVDKLDQLEALAGRLD 321
LK+LQ LL K L + +I L S EG I+ RL KK+LLILDDVDK QL+A+ GR D
Sbjct: 239 LKHLQSILLSKLLGEKDITLTSWQEGASMIQHRLQRKKVLLILDDVDKRQQLKAIVGRPD 298
Query: 322 WFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKNEKVPSSYEDILK 381
WFGPGSRVIITTRDKHLL H +E+TY V+ LN++ AL+LL W AFK EK+ SYED+L
Sbjct: 299 WFGPGSRVIITTRDKHLLKYHEVERTYEVKVLNQSAALQLLTWNAFKREKIDPSYEDVLN 358
Query: 382 RAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILRLSYDALDEEEQS 441
R V YASGLPLA+EV+GSNLF K++AE ES ++ Y RIP +IQ+IL++S+DAL EE+++
Sbjct: 359 RVVTYASGLPLALEVIGSNLFEKTVAEWESAMEHYKRIPSDEIQEILKVSFDALGEEQKN 418
Query: 442 VFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCFFSGIKVTLHELI 501
VFLDIACC KG EV+ IL YG K H+ VLV+KSL+K+S C V +H++I
Sbjct: 419 VFLDIACCFKGYEWTEVDNILRDLYGNCTKHHIGVLVEKSLVKVSCC----DTVEMHDMI 474
Query: 502 EVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICM--NLHSMESVIDKK 559
+ MG+E+ RQ SP+EPG+ RL DI+ V K E+IC+ ++ E ++
Sbjct: 475 QDMGREIERQRSPEEPGKCKRLLLPKDIIQVF-------KIEIICLDFSISDKEETVEWN 527
Query: 560 GKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILSKASEIKSFSN 619
AF KM LK LII N SKG + P L+ L+W S L S+ +
Sbjct: 528 ENAFMKMKNLKILIIRNCKFSKGPNYFPEGLRVLEWHRYPSNCLPSNF-----------D 576
Query: 620 CIYL*CTMISFSPISSFVF-----------MQKFQDMTILILDHCEYLTHIPDVSGLSNL 668
I L + S I+SF F +QK +T+L D CE+LT IPDVS L NL
Sbjct: 577 PINLVICKLPDSSITSFEFHGSSKASLKSSLQKLGHLTVLNFDRCEFLTKIPDVSDLPNL 636
Query: 669 EKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDICCCSSLKSF 728
++LSF C +L+ + +SIG LNKL+ LSA+GCRKL FPPL L SL+ L++ CSSL+ F
Sbjct: 637 KELSFNWCESLVAVDDSIGFLNKLKTLSAYGCRKLTSFPPLNLTSLETLNLGGCSSLEYF 696
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSE-----LDELSVREAR--MLRFPK---- 777
PE+L +M NI + L +++ I ELP SFQNL LD + + R + PK
Sbjct: 697 PEILGEMKNITVLAL-HDLPIKELPFSFQNLIGLLFLWLDSCGIVQLRCSLATMPKLCEF 755
Query: 778 ------------HNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCVNVELLDLSHNNF 825
++ KV + +CNL D++ I K +V L+L NNF
Sbjct: 756 CITDSCNRWQWVESEEGEEKVVGSILSFEATDCNLCDDFFFIGSKRFAHVGYLNLPGNNF 815
Query: 826 KILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSS 873
ILPE E L L +H C L+EIRG+PPNLK A C SL+SS
Sbjct: 816 TILPEFFKELQFLTTLVVHDCKHLQEIRGLPPNLKHFDARNCASLTSS 863
>UniRef100_Q84ZV7 R 12 protein [Glycine max]
Length = 893
Score = 720 bits (1858), Expect = 0.0
Identities = 404/791 (51%), Positives = 520/791 (65%), Gaps = 37/791 (4%)
Query: 16 YQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFIA 75
Y VFL+F DT GFT LYKAL+D+GI+TF + EL R E+TP L KAI SR+ I
Sbjct: 12 YDVFLSFIREDTHRGFTFYLYKALNDRGIYTFFYDQELPRETEVTPGLYKAILASRVAII 71
Query: 76 VFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKRF 135
V S NYA SSFCLDELV I+HC R V+PVF V+P+ VRHQKGSYGEA+A+H+KRF
Sbjct: 72 VLSENYAFSSFCLDELVTILHCE----REVIPVFHNVDPSDVRHQKGSYGEAMAKHQKRF 127
Query: 136 QNDPKSMERLQGWKEALSQAANLSGYHDSPPG-YEYKLIGKIVKYISNKISQQPLHVATY 194
+ ++LQ W+ AL Q ANL GYH G YEY LIG+IVK +S LHVA Y
Sbjct: 128 K-----AKKLQKWRMALKQVANLCGYHFKDGGSYEYMLIGRIVKQVSRMFGLASLHVADY 182
Query: 195 PVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCFLENV 254
PVGL+S+V ++ LLD GSD VH++GI+G+GGLGK+TLA A+YNFIA F+ SCFL+NV
Sbjct: 183 PVGLESQVTEVMKLLDVGSDDVVHIIGIHGMGGLGKTTLAMAVYNFIAPHFDESCFLQNV 242
Query: 255 KESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLILDDVDKLDQL 313
+E S + LK+LQ LL K L + +I L S EG I+ RL KKILLILDDVDK +QL
Sbjct: 243 REESNKHGLKHLQSVLLSKLLGEKDITLTSWQEGASMIQHRLRLKKILLILDDVDKREQL 302
Query: 314 EALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKNEKVP 373
+A+ G+ DWFGPGSRVIITTRDKHLL H +E+TY V LN +A +LL W AFK EK+
Sbjct: 303 KAIVGKPDWFGPGSRVIITTRDKHLLKYHEVERTYEVNVLNHDDAFQLLTWNAFKREKID 362
Query: 374 SSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILRLSYD 433
SY+D+L R V YASGLPLA+EV+GSNL+GK++AE ES L+ Y RIP +I KIL +S+D
Sbjct: 363 PSYKDVLNRVVTYASGLPLALEVIGSNLYGKTVAEWESALETYKRIPSNEILKILEVSFD 422
Query: 434 ALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKS-LIKISWCFFSG 492
AL+EE+++VFLDIACC KG + EV I Y H+ VLV+KS L+K+SW
Sbjct: 423 ALEEEQKNVFLDIACCFKGYKWTEVYDIFRALYSNCKMHHIGVLVEKSLLLKVSW----R 478
Query: 493 IKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICM--NLH 550
V +H+LI+ MG+++ RQ SP+EPG+ RLWS DI+ VL NTGT K E+IC+ ++
Sbjct: 479 DNVEMHDLIQDMGRDIERQRSPEEPGKCKRLWSPKDIIQVLKHNTGTSKLEIICLDSSIS 538
Query: 551 SMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILSK 610
E ++ AF KM LK LII NG SKG + P L+ L+W S L S+
Sbjct: 539 DKEETVEWNENAFMKMENLKILIIRNGKFSKGPNYFPEGLRVLEWHRYPSNCLPSNF--- 595
Query: 611 ASEIKSFSNCIYL*CTMISFSPISSFVF--MQKFQDMTILILDHCEYLTHIPDVSGLSNL 668
+ I L + S I+S F K +T+L D C++LT IPDVS L NL
Sbjct: 596 --------DPINLVICKLPDSSITSLEFHGSSKLGHLTVLKFDKCKFLTQIPDVSDLPNL 647
Query: 669 EKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPPLGLASLKELDICCCSSLKSF 728
+LSF C +L+ I +SIG LNKLE L+A GCRKL FPPL L SL+ L++ CSSL+ F
Sbjct: 648 RELSFVGCESLVAIDDSIGFLNKLEILNAAGCRKLTSFPPLNLTSLETLELSHCSSLEYF 707
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFPKHNDRMYSKVFS 788
PE+L +M NI + L+ + I ELP SFQNL L E+++R R++R R +
Sbjct: 708 PEILGEMENITALHLE-RLPIKELPFSFQNLIGLREITLRRCRIVRL-----RCSLAMMP 761
Query: 789 KVTKLRIYECN 799
+ + +I CN
Sbjct: 762 NLFRFQIRNCN 772
>UniRef100_Q8H6S7 Resistance protein KR3 [Glycine max]
Length = 636
Score = 566 bits (1458), Expect = e-159
Identities = 301/601 (50%), Positives = 419/601 (69%), Gaps = 21/601 (3%)
Query: 15 KYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIFI 74
+Y VF+NFRG DTR+ FTG+L+KAL +KGI F+D ++++RGDEI +L +AI+ SRI I
Sbjct: 34 RYDVFINFRGEDTRFAFTGHLHKALCNKGIRAFMDENDIKRGDEIRATLEEAIKGSRIAI 93
Query: 75 AVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEKR 134
VFS +YASSSFCLDEL I+ CY+ K LV+PVF+ V+P+ VR +GSY E LA E+R
Sbjct: 94 TVFSKDYASSSFCLDELATILGCYREKTLLVIPVFYKVDPSDVRRLQGSYAEGLARLEER 153
Query: 135 FQNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI--SQQPLHV 191
F ++ WK+AL + A L+G+H GYE+K I KIV + +KI ++ ++V
Sbjct: 154 FH------PNMENWKKALQKVAELAGHHFKDGAGYEFKFIRKIVDDVFDKINKAEASIYV 207
Query: 192 ATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCFL 251
A +PVGL V++++ LL+ GS + M+GI+G+GG+GKSTLA+A+YN D F+ SCFL
Sbjct: 208 ADHPVGLHLEVEKIRKLLEAGSSDAISMIGIHGMGGVGKSTLARAVYNLHTDHFDDSCFL 267
Query: 252 ENVKESSASNNLKNLQQELLLKTLQLEIKLGSVSEGIPKIKERLHGKKILLILDDVDKLD 311
+NV+E S + LK LQ LL + L+ EI L S +G IK +L GKK+LL+LDDVD+
Sbjct: 268 QNVREESNRHGLKRLQSILLSQILKKEINLASEQQGTSMIKNKLKGKKVLLVLDDVDEHK 327
Query: 312 QLEALAGRLDW----FGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAF 367
QL+A+ G+ W FG +IITTRDK LL +G+++T+ V+EL++ +A++LL+ KAF
Sbjct: 328 QLQAIVGKSVWSESEFGTRLVLIITTRDKQLLTSYGVKRTHEVKELSKKDAIQLLKRKAF 387
Query: 368 KN-EKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQK 426
K ++V SY +L V + SGLPLA+EV+GSNLFGKSI E ES + +Y RIP+K+I K
Sbjct: 388 KTYDEVDQSYNQVLNDVVTWTSGLPLALEVIGSNLFGKSIKEWESAIKQYQRIPNKEILK 447
Query: 427 ILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKIS 486
IL++S+DAL+EEE+SVFLDI CC+KG + E+E ILH Y +K H+ VLVDKSLI+I
Sbjct: 448 ILKVSFDALEEEEKSVFLDITCCLKGYKCREIEDILHSLYDNCMKYHIGVLVDKSLIQI- 506
Query: 487 WCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMIC 546
S +VTLH+LIE MGKE+ RQ+SPKE G+R RLW DI+ VL +N+GT + ++IC
Sbjct: 507 ----SDDRVTLHDLIENMGKEIDRQKSPKETGKRRRLWLLKDIIQVLKDNSGTSEVKIIC 562
Query: 547 MN--LHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLS 604
++ + + I+ G AFK+M LK LII NG S+G +LP SL+ L+W S L
Sbjct: 563 LDFPISDKQETIEWNGNAFKEMKNLKALIIRNGILSQGPNYLPESLRILEWHRHPSHCLP 622
Query: 605 S 605
S
Sbjct: 623 S 623
>UniRef100_Q9FPK8 Putative resistance protein [Glycine max]
Length = 522
Score = 524 bits (1350), Expect = e-147
Identities = 279/538 (51%), Positives = 387/538 (71%), Gaps = 21/538 (3%)
Query: 1 MQSPSSSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEI- 59
M S+S + Y++ VFL+FRG DTR GF GNLYKAL +KG HTF +L RG+EI
Sbjct: 1 MAGSERSSSRVWHYEFDVFLSFRGEDTRLGFVGNLYKALTEKGFHTFF-REKLVRGEEIA 59
Query: 60 -TPSLL-KAIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIV 117
+PS++ KAI+ SR+F+ VFS NYASS+ CL+EL+ I+ + R VLPVF+ V+P+ V
Sbjct: 60 ASPSVVEKAIQHSRVFVVVFSQNYASSTRCLEELLSILRFSQDNRRPVLPVFYYVDPSDV 119
Query: 118 RHQKGSYGEALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGY-HDSPPGYEYKLIGKI 176
Q G YGEALA HEKRF ++ +++ W++AL +AA LSG+ GYEY+LI KI
Sbjct: 120 GLQTGMYGEALAMHEKRFNSES---DKVMKWRKALCEAAALSGWPFKHGDGYEYELIEKI 176
Query: 177 VKYISNKISQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKA 236
V+ +S KI++ PVGLQ R+ ++ LLD S GVH++GIYG+GG+GK+TLA+A
Sbjct: 177 VEGVSKKINR--------PVGLQYRMLELNGLLDAASLSGVHLIGIYGVGGIGKTTLARA 228
Query: 237 IYNFIADQFECSCFLENVKESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERL 295
+Y+ +A QF+ CFL+ V+E++ + L +LQQ +L +T+ + +I+L SV +GI +K+RL
Sbjct: 229 LYDSVAVQFDALCFLDEVRENAMKHGLVHLQQTILAETVGEKDIRLPSVKQGITLLKQRL 288
Query: 296 HGKKILLILDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNE 355
K++LL+LDD+++ +QL+AL G WFGPGSRVIITTRD+ LL+ HG+EK Y VE L +
Sbjct: 289 QEKRVLLVLDDINESEQLKALVGSPGWFGPGSRVIITTRDRQLLESHGVEKIYEVENLAD 348
Query: 356 TEALELLRWKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDK 415
EALELL WKAFK +KV + + + RA+ YASGLPLA+EV+GSNLFG+ I E + TLD
Sbjct: 349 GEALELLCWKAFKTDKVYPDFINKIYRALTYASGLPLALEVIGSNLFGREIVEWQYTLDL 408
Query: 416 YGRIPHKDIQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLR 475
Y +I KDIQKIL++S+DALDE E+ +FLDIAC KGC+L +VE I+ YG S+K+ +
Sbjct: 409 YEKIHDKDIQKILKISFDALDEHEKDLFLDIACFFKGCKLAQVESIVSGRYGDSLKAIID 468
Query: 476 VLVDKSLIKISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVL 533
VL++K+LIKI +V +H+LI+ MG+E+VRQESPK PG SRLWS +D+ VL
Sbjct: 469 VLLEKTLIKID----EHGRVKMHDLIQQMGREIVRQESPKHPGNCSRLWSPEDVADVL 522
>UniRef100_Q6XZH5 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 519 bits (1336), Expect = e-145
Identities = 327/877 (37%), Positives = 509/877 (57%), Gaps = 46/877 (5%)
Query: 14 YKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIF 73
+ Y VFL+FRG D R F +LY AL K I+TF D+ +L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELVSSIEESRIA 75
Query: 74 IAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL I+ C KG++V+PVF+ V+P+ VR QK +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 134 RFQNDPKSMERLQGWKEALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SQQPLH 190
RFQ D ++Q W+ AL +AAN+SG+ ++ G+E +++ KI + I ++ SQ+
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCF 250
A VG++S + Q+ +L GS GVH +GI G+ G+GK+TLA+ IY+ I QF+ +CF
Sbjct: 191 NARNLVGMESHMHQVYKMLGIGSG-GVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 251 LENVKESSASNNLKNLQQELLLKTLQLE-IKLGSVSEGIPKIKERLHGKKILLILDDVDK 309
L V++ SA L+ LQ+ LL + L ++ +++ EG K+RL KK+LL+LDDVD
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKN 369
+DQL ALAG +WFG GSR+IITT+DKHLL + EK Y ++ LN E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 370 EKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILR 429
+ +ED+ + + + GLPLA++V+GS L+G+ + E S +++ +IP +I K L
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 430 LSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCF 489
S+ L EQ +FLDIAC G + + V +IL + + ++VL++K LI
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIG-IKVLMEKCLITT---- 484
Query: 490 FSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICMNL 549
G ++T+H+LI+ MG +VR+E+ +P SRLW ++DI VL N GT K E + ++L
Sbjct: 485 LQG-RITIHQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKIEGMSLHL 543
Query: 550 HSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILS 609
+ E V + GKAF +MTRL+ L +N + +G + LP L+ L W G SKSL +S
Sbjct: 544 TNEEEV-NFGGKAFMQMTRLRFLKFQNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSF-- 600
Query: 610 KASEIKSFSNCIYL*CTMISFSPISSFVFMQKFQDMTILILDHCEYLTHIPDVSGLSNLE 669
K ++ S + I + + + + L H + L +PD S NLE
Sbjct: 601 KGDQLVSLK--------LKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLE 652
Query: 670 KLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPP-LGLASLKELDICCCSSLKSF 728
+L E C +L+ I+ SI +L KL L+ CR LK P + L L+ L + CS L++F
Sbjct: 653 RLVLEECTSLVEINFSIENLGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTF 712
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFPKHNDRMYSKVF- 787
PE+ KM + E+ LD S+ ELP+S +NLS + ++ L + KH + + S +F
Sbjct: 713 PEIEEKMNCLAELYLDAT-SLSELPASVENLSGVGVIN------LSYCKHLESLPSSIFR 765
Query: 788 -SKVTKLRIYEC----NLSDEYLQIVLKWCVNVELLDLSHNNFKILPECLSECHHLKHLG 842
+ L + C NL D+ L V +E L +H + +P +S +LK L
Sbjct: 766 LKCLKTLDVSGCSKLKNLPDD-----LGLLVGLEQLHCTHTAIQTIPSSMSLLKNLKRLS 820
Query: 843 LHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCRRMLM 879
L C++L + ++ ++LS C +++
Sbjct: 821 LSGCNALSSQVSSSSHGQKSMGVNFQNLSGLCSLIML 857
>UniRef100_Q6XZH8 Nematode resistance protein [Solanum tuberosum]
Length = 1136
Score = 516 bits (1330), Expect = e-144
Identities = 325/877 (37%), Positives = 508/877 (57%), Gaps = 46/877 (5%)
Query: 14 YKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIF 73
+ Y VFL+FRG D R F +LY AL+ K I+TF D+ +L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELVSSIEESRIA 75
Query: 74 IAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL I+ C KG++V+PVF+ V+P+ VR QK +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 134 RFQNDPKSMERLQGWKEALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SQQPLH 190
RFQ D ++Q W+ AL +AAN+SG+ ++ G+E +++ KI + I ++ SQ+
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTANGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCF 250
A VG++S + ++ +L GS GVH +GI G+ G+GK+TLA+ IY+ I QF+ +CF
Sbjct: 191 NARNLVGMESHMHKVYKMLGIGSG-GVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 251 LENVKESSASNNLKNLQQELLLKTLQLE-IKLGSVSEGIPKIKERLHGKKILLILDDVDK 309
L V++ SA L+ LQ+ LL + L ++ +++ EG K+RL KK+LL+LDDVD
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKN 369
+DQL ALAG +WFG GSR+IITT+DKHLL + EK Y ++ LN E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 370 EKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILR 429
+ +ED+ + + + GLPLA++V+GS L+G+ + E S +++ +IP +I K L
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 430 LSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCF 489
S+ L EQ +FLDIAC G + + V +IL + + ++VL++K LI I
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIG-IKVLMEKCLITI---- 484
Query: 490 FSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICMNL 549
G ++T+H+LI+ MG +VR+E+ +P SR+W ++DI VL N GT K E + ++L
Sbjct: 485 LQG-RITIHQLIQDMGWHIVRREATDDPRMCSRMWKREDICPVLERNLGTDKNEGMSLHL 543
Query: 550 HSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILS 609
+ E V + GKAF +MTRL+ L N + +G + LP L+ L W G SKSL +S
Sbjct: 544 TNEEEV-NFGGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSF-- 600
Query: 610 KASEIKSFSNCIYL*CTMISFSPISSFVFMQKFQDMTILILDHCEYLTHIPDVSGLSNLE 669
K ++ + I + + + + L H + L PD S NLE
Sbjct: 601 KGDQLVGLK--------LKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLE 652
Query: 670 KLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPP-LGLASLKELDICCCSSLKSF 728
+L E C +L+ I+ SI +L KL L+ CR LK P + L L+ L + CS L++F
Sbjct: 653 RLVLEECTSLVEINFSIENLGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTF 712
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFPKHNDRMYSKVF- 787
PE+ KM + E+ L S+ ELP+S +NLS + ++ L + KH + + S +F
Sbjct: 713 PEIEEKMNCLAELYLGAT-SLSELPASVENLSGVGVIN------LSYCKHLESLPSSIFR 765
Query: 788 -SKVTKLRIYEC----NLSDEYLQIVLKWCVNVELLDLSHNNFKILPECLSECHHLKHLG 842
+ L + C NL D+ L V +E L +H + +P +S +LKHL
Sbjct: 766 LKCLKTLDVSGCSKLKNLPDD-----LGLLVGLEELHCTHTAIQTIPSSMSLLKNLKHLS 820
Query: 843 LHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCRRMLM 879
L C++L + ++ ++LS C +++
Sbjct: 821 LSGCNALSSQVSSSSHGQKSMGVNFQNLSGLCSLIML 857
>UniRef100_Q6XZH6 Nematode resistance-like protein [Solanum tuberosum]
Length = 1136
Score = 510 bits (1313), Expect = e-142
Identities = 321/847 (37%), Positives = 494/847 (57%), Gaps = 46/847 (5%)
Query: 14 YKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIF 73
+ Y VFL+FRG D R F +LY AL K I+TF D+ +L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGEDVRKTFVDHLYLALQQKCINTFKDDEKLEKGKFISPELMSSIEESRIA 75
Query: 74 IAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL I+ C KG++V+PVF+ V+P+ VR QK +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIMECKNVKGQIVVPVFYDVDPSTVRKQKSIFGEAFSKHEA 135
Query: 134 RFQNDPKSMERLQGWKEALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SQQPLH 190
RFQ D ++Q W+ AL +AAN+SG+ ++ G+E +++ KI + I ++ SQ+
Sbjct: 136 RFQED-----KVQKWRAALEEAANISGWDLPNTSNGHEARVMEKIAEDIMARLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCF 250
A VG++S + ++ +L GS GVH +GI G+ G+GK+TLA+ IY+ I QF+ +CF
Sbjct: 191 NARNLVGMESHMLKVYKMLGIGSG-GVHFLGILGMSGVGKTTLARVIYDNIRSQFQGACF 249
Query: 251 LENVKESSASNNLKNLQQELLLKTLQLE-IKLGSVSEGIPKIKERLHGKKILLILDDVDK 309
L V++ SA L+ LQ+ LL + L ++ +++ + EG K+RL KK+LL+LDDVD
Sbjct: 250 LHEVRDRSAKQGLERLQEILLSEILVVKKLRINNSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKN 369
+DQL ALAG +WFG GSR+IITT+DKHLL + EK Y ++ LN E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMKTLNNYESLQLFKQHAFKK 369
Query: 370 EKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILR 429
+ +ED+ + + + GLPLA++V+GS L+G+ + E S +++ +IP +I K L
Sbjct: 370 NRPTKEFEDLSAQVIKHTDGLPLALKVLGSFLYGRGLDEWISEVERLKQIPENEILKKLE 429
Query: 430 LSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCF 489
S+ L EQ +FLDIAC G + + V +IL + + ++VL++K LI I
Sbjct: 430 QSFTGLHNTEQKIFLDIACFFSGKKKDSVTRILESFHFCPVIG-IKVLMEKCLITI---- 484
Query: 490 FSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICMNL 549
G ++T+H+LI+ MG +VR+E+ +P SRLW ++DI VL N GT K E + ++L
Sbjct: 485 LQG-RITIHQLIQDMGWHIVRREATDDPRMCSRLWKREDICPVLERNLGTDKNEGMSLHL 543
Query: 550 HSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILS 609
+ E V + GKAF +MTRL+ L N + +G + LP L+ L W G SKSL +S
Sbjct: 544 TNEEEV-NFGGKAFMQMTRLRFLKFRNAYVCQGPEFLPDELRWLDWHGYPSKSLPNSF-- 600
Query: 610 KASEIKSFSNCIYL*CTMISFSPISSFVFMQKFQDMTILILDHCEYLTHIPDVSGLSNLE 669
K ++ + I + + + + L H + L PD S NLE
Sbjct: 601 KGDQLVGLK--------LKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRTPDFSVTPNLE 652
Query: 670 KLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPP-LGLASLKELDICCCSSLKSF 728
+L E C +L+ I+ SI +L KL L+ CR LK P + L L+ L + CS L++F
Sbjct: 653 RLVLEECTSLVEINFSIENLGKLVLLNLKNCRNLKTLPKRIRLEKLEILVLTGCSKLRTF 712
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFPKHNDRMYSKVF- 787
PE+ KM + E+ L S+ LP+S +NLS + ++ L + KH + + S +F
Sbjct: 713 PEIEEKMNCLAELYLGAT-SLSGLPASVENLSGVGVIN------LSYCKHLESLPSSIFR 765
Query: 788 -SKVTKLRIYEC----NLSDEYLQIVLKWCVNVELLDLSHNNFKILPECLSECHHLKHLG 842
+ L + C NL D+ L V +E L +H +P +S +LK L
Sbjct: 766 LKCLKTLDVSGCSKLKNLPDD-----LGLLVGLEKLHCTHTAIHTIPSSMSLLKNLKRLS 820
Query: 843 LHYCSSL 849
L C++L
Sbjct: 821 LRGCNAL 827
>UniRef100_Q84KB4 MRGH5 [Cucumis melo]
Length = 1092
Score = 508 bits (1308), Expect = e-142
Identities = 342/943 (36%), Positives = 524/943 (55%), Gaps = 92/943 (9%)
Query: 5 SSSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLL 64
SSS+S + + Y VFL+FRG DTR FTG+LY L KG++ FID+ L+RG++I+ +L
Sbjct: 10 SSSSSPIFNWSYDVFLSFRGEDTRSNFTGHLYMFLRQKGVNVFIDDG-LERGEQISETLF 68
Query: 65 KAIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSY 124
K I+ S I I +FS NYASS++CLDELV I+ C K+KG+ VLP+F+ V+P+ VR Q G +
Sbjct: 69 KTIQNSLISIVIFSENYASSTWCLDELVEIMECKKSKGQKVLPIFYKVDPSDVRKQNGWF 128
Query: 125 GEALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYHDSPPGYEYKLIGKIVKYISNKI 184
E LA+HE F ME++ W++AL+ AANLSG+H E LI IVK + + +
Sbjct: 129 REGLAKHEANF------MEKIPIWRDALTTAANLSGWHLGARK-EAHLIQDIVKEVLSIL 181
Query: 185 SQ-QPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIAD 243
+ +PL+ + VG+ S+++ + + V+M+GIYGIGG+GK+TLAKA+Y+ +A
Sbjct: 182 NHTKPLNANEHLVGIDSKIEFLYRKEEMYKSECVNMLGIYGIGGIGKTTLAKALYDKMAS 241
Query: 244 QFECSCFLENVKESSAS-NNLKNLQQELLLKTLQLEIKLGSVSEGIPKIKERLHGKKILL 302
QFE C+L +V+E+S + L LQ++LL + L+ ++++ + GI IK RL KK+L+
Sbjct: 242 QFEGCCYLRDVREASKLFDGLTQLQKKLLFQILKYDLEVVDLDWGINIIKNRLRSKKVLI 301
Query: 303 ILDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELL 362
+LDDVDKL+QL+AL G DWFG G+++I+TTR+K LL HG +K Y V+ L++ EA+EL
Sbjct: 302 LLDDVDKLEQLQALVGGHDWFGQGTKIIVTTRNKQLLVSHGFDKMYEVQGLSKHEAIELF 361
Query: 363 RWKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKS-IAECESTLDKYGRIPH 421
R AFKN + S+Y D+ +RA Y +G PLA+ V+GS L +S +AE LD +
Sbjct: 362 RRHAFKNLQPSSNYLDLSERATRYCTGHPLALIVLGSFLCDRSDLAEWSGILDGFENSLR 421
Query: 422 KDIQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKS 481
KDI+ IL+LS+D L++E + +FLDI+C + G R+ V+++L + + + L D S
Sbjct: 422 KDIKDILQLSFDGLEDEVKEIFLDISCLLVGKRVSYVKKMLSECHSI-LDFGITKLKDLS 480
Query: 482 LIKISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGK 541
LI+ F +V +H+LI+ MG ++V ES +PG+RSRLW + DI+ V + N+G+
Sbjct: 481 LIR-----FEDDRVQMHDLIKQMGHKIVHDESHDQPGKRSRLWLEKDILEVFSNNSGSDA 535
Query: 542 TEMICMNLHSMESVIDKKGKAFKKMTRLKTLIIE-NGHCSKGLKHLPSSLKALKW----- 595
+ I + L + VID +AF+ M L+ L+++ N K +K+LP+ LK +KW
Sbjct: 536 VKAIKLVLTDPKRVIDLDPEAFRSMKNLRILMVDGNVRFCKKIKYLPNGLKWIKWHRFAH 595
Query: 596 -----------------------------EGCLSKSL----SSSILSKASEIKS------ 616
+ C+ L S IL K SE +
Sbjct: 596 PSLPSCFITKDLVGLDLQHSFITNFGKGLQNCMRLKLLDLRHSVILKKISESSAAPNLEE 655
Query: 617 --FSNCIYL*CTMISFSPISSFVFMQ---------------KFQDMTILILDHCEYLTHI 659
SNC L SF + V + ++ + L L HC+ L I
Sbjct: 656 LYLSNCSNLKTIPKSFLSLRKLVTLDLHHCVNLKKIPRSYISWEALEDLDLSHCKKLEKI 715
Query: 660 PDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPP-LGLASLKELD 718
PD+S SNL LSFE C NL+ IH+SIG L KL L C LK+ P + L++L+
Sbjct: 716 PDISSASNLRSLSFEQCTNLVMIHDSIGSLTKLVTLKLQNCSNLKKLPRYISWNFLQDLN 775
Query: 719 ICCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARML-RFPK 777
+ C L+ P+ +N+K + L+ S+ + S +LS+L L++ + L + P
Sbjct: 776 LSWCKKLEEIPD-FSSTSNLKHLSLEQCTSLRVVHDSIGSLSKLVSLNLEKCSNLEKLPS 834
Query: 778 HNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCVNVELLDLSHNNFKILPECLSECHH 837
Y K+ S + L + C + + +I + ++ +L L + LP + H
Sbjct: 835 -----YLKLKS-LQNLTLSGCCKLETFPEID-ENMKSLYILRLDSTAIRELPPSIGYLTH 887
Query: 838 LKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSSCRRMLMS 880
L L C++L + LK L LS S R + S
Sbjct: 888 LYMFDLKGCTNLISLPCTTHLLKSLGELH---LSGSSRFEMFS 927
Score = 105 bits (263), Expect = 6e-21
Identities = 104/374 (27%), Positives = 168/374 (44%), Gaps = 75/374 (20%)
Query: 574 IENGHCSKGLKHLP-----SSLKALKWEGCLSKSL---SSSILSKASEIKSFSNCIYL*C 625
++ HC K L+ +P S+L++L +E C + + S L+K +K NC L
Sbjct: 704 LDLSHCKK-LEKIPDISSASNLRSLSFEQCTNLVMIHDSIGSLTKLVTLK-LQNCSNL-- 759
Query: 626 TMISFSPISSFVFMQKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFECCYNLITIHNS 685
+ ++ QD+ L C+ L IPD S SNL+ LS E C +L +H+S
Sbjct: 760 -----KKLPRYISWNFLQDLN---LSWCKKLEEIPDFSSTSNLKHLSLEQCTSLRVVHDS 811
Query: 686 IGHLNKLERLSAFGCRKLKRFPP-LGLASLKELDICCCSSLKSFPELLCKMTNIKEIDLD 744
IG L+KL L+ C L++ P L L SL+ L + C L++FPE+ M ++ + LD
Sbjct: 812 IGSLSKLVSLNLEKCSNLEKLPSYLKLKSLQNLTLSGCCKLETFPEIDENMKSLYILRLD 871
Query: 745 YNI------SIG-----------------ELPSSFQNLSELDELSVR------------- 768
SIG LP + L L EL +
Sbjct: 872 STAIRELPPSIGYLTHLYMFDLKGCTNLISLPCTTHLLKSLGELHLSGSSRFEMFSYIWD 931
Query: 769 --------EARMLRFPKHNDRMYSKV------FSKVTKLRIYECNLSD-EYLQIVLKWCV 813
++++ ++ +S+V F T L + CN+S+ ++L+I+
Sbjct: 932 PTINPVCSSSKIMETSLTSEFFHSRVPKESLCFKHFTLLDLEGCNISNVDFLEILCNVAS 991
Query: 814 NVELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLSSS 873
++ + LS NNF LP CL + L++L L C L+EI +P ++ + A C SLS S
Sbjct: 992 SLSSILLSENNFSSLPSCLHKFMSLRNLELRNCKFLQEIPNLPLCIQRVDATGCVSLSRS 1051
Query: 874 CRRML---MSQELH 884
+L SQ++H
Sbjct: 1052 PNNILDIISSQQVH 1065
>UniRef100_Q6XZH7 Nematode resistance-like protein [Solanum tuberosum]
Length = 1121
Score = 508 bits (1308), Expect = e-142
Identities = 324/873 (37%), Positives = 505/873 (57%), Gaps = 47/873 (5%)
Query: 14 YKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLKAIEESRIF 73
+ Y VFL+FRG + R F +LY AL+ K I+TF D+ +L++G I+P L+ +IEESRI
Sbjct: 16 WSYDVFLSFRGENVRKTFVDHLYLALEQKCINTFKDDEKLEKGKFISPELMSSIEESRIA 75
Query: 74 IAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYGEALAEHEK 133
+ +FS NYA+S++CLDEL II C KG++V+PVF+ V+P+ VR QK +GEA ++HE
Sbjct: 76 LIIFSKNYANSTWCLDELTKIIECKNVKGQIVVPVFYDVDPSTVRRQKNIFGEAFSKHEA 135
Query: 134 RFQNDPKSMERLQGWKEALSQAANLSGYH--DSPPGYEYKLIGKIVKYISNKI-SQQPLH 190
RF+ D +++ W+ AL +AAN+SG+ ++ G+E ++I KI + I ++ SQ+
Sbjct: 136 RFEED-----KVKKWRAALEEAANISGWDLPNTSNGHEARVIEKITEDIMVRLGSQRHAS 190
Query: 191 VATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQFECSCF 250
A VG++S + Q+ +L GS GV +GI G+ G+GK+TLA+ IY+ I QFE +CF
Sbjct: 191 NARNVVGMESHMHQVYKMLGIGSG-GVRFLGILGMSGVGKTTLARVIYDNIQSQFEGACF 249
Query: 251 LENVKESSASNNLKNLQQELLLKTLQLE-IKLGSVSEGIPKIKERLHGKKILLILDDVDK 309
L V++ SA L++LQ+ LL + L ++ +++ EG K+RL KK+LL+LDDVD
Sbjct: 250 LHEVRDRSAKQGLEHLQEILLSEILVVKKLRINDSFEGANMQKQRLQYKKVLLVLDDVDH 309
Query: 310 LDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLRWKAFKN 369
+DQL ALAG +WFG GSR+IITT+DKHLL + EK Y + L++ E+L+L + AFK
Sbjct: 310 IDQLNALAGEREWFGDGSRIIITTKDKHLLVKYETEKIYRMGTLDKYESLQLFKQHAFKK 369
Query: 370 EKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKDIQKILR 429
+ED+ + + + GLPLA++V+GS L+G+ + E S +++ +IP +I K L
Sbjct: 370 NHSTKEFEDLSAQVIEHTGGLPLALKVLGSFLYGRGLDEWISEVERLKQIPQNEILKKLE 429
Query: 430 LSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLRVLVDKSLIKISWCF 489
S+ L+ EQ +FLDIAC G + + V +IL + +S ++VL++K LI I
Sbjct: 430 PSFTGLNNIEQKIFLDIACFFSGKKKDSVTRIL-ESFHFSPVIGIKVLMEKCLITI---- 484
Query: 490 FSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTENTGTGKTEMICMNL 549
G ++T+H+LI+ MG +VR+E+ P SRLW ++DI VL +N T K E + ++L
Sbjct: 485 LKG-RITIHQLIQEMGWHIVRREASYNPRICSRLWKREDICPVLEQNLCTDKIEGMSLHL 543
Query: 550 HSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSSLKALKWEGCLSKSLSSSILS 609
+ E V + GKA +MT L+ L N + +G + LP L+ L W G SK+L +S
Sbjct: 544 TNEEEV-NFGGKALMQMTSLRFLKFRNAYVYQGPEFLPDELRWLDWHGYPSKNLPNSF-- 600
Query: 610 KASEIKSFSNCIYL*CTMISFSPISSFVFMQKFQDMTILILDHCEYLTHIPDVSGLSNLE 669
K ++ S + I + + + + L H + L +PD S NLE
Sbjct: 601 KGDQLVSLK--------LKKSRIIQLWKTSKDLGKLKYMNLSHSQKLIRMPDFSVTPNLE 652
Query: 670 KLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPP-LGLASLKELDICCCSSLKSF 728
+L E C +L+ I+ SIG L KL L+ CR LK P + L L+ L + CS L++F
Sbjct: 653 RLVLEECTSLVEINFSIGDLGKLVLLNLKNCRNLKTIPKRIRLEKLEVLVLSGCSKLRTF 712
Query: 729 PELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSVREARMLRFPKHNDRMYSKVF- 787
PE+ KM + E+ L S+ ELP+S +N S + ++ L + KH + + S +F
Sbjct: 713 PEIEEKMNRLAELYLGAT-SLSELPASVENFSGVGVIN------LSYCKHLESLPSSIFR 765
Query: 788 -SKVTKLRIYEC----NLSDEYLQIVLKWCVNVELLDLSHNNFKILPECLSECHHLKHLG 842
+ L + C NL D+ L V +E L +H + +P +S +LKHL
Sbjct: 766 LKCLKTLDVSGCSKLKNLPDD-----LGLLVGIEKLHCTHTAIQTIPSSMSLLKNLKHLS 820
Query: 843 LHYCSSL-EEIRGIPPNLKELSAYQCKSLSSSC 874
L C++L ++ K + ++LS C
Sbjct: 821 LSGCNALSSQVSSSSHGQKSMGINFFQNLSGLC 853
>UniRef100_Q6URA2 TIR-NBS-LRR type R protein 7 [Malus baccata]
Length = 1095
Score = 499 bits (1286), Expect = e-139
Identities = 338/976 (34%), Positives = 522/976 (52%), Gaps = 77/976 (7%)
Query: 2 QSPSSSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITP 61
++ SSS+S+S + Y +FL+FRG DTR GFTG+L+ AL D+G ++D +L RG+EI
Sbjct: 9 EASSSSSSMSKLWNYDLFLSFRGEDTRNGFTGHLHAALKDRGYQAYMDQDDLNRGEEIKE 68
Query: 62 SLLKAIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQK 121
L +AIE SRI I VFS YA SS+CLDELV I+ C GR VLP+F+ V+P+ VR Q
Sbjct: 69 ELFRAIEGSRISIIVFSKRYADSSWCLDELVKIMECRSKLGRHVLPIFYHVDPSHVRKQD 128
Query: 122 GSYGEALAEHEKRF------QNDPKSMERLQGWKEALSQAANLSGYHD---SPPGYEYKL 172
G EA +HE+ + ER++ WK+AL++AANLSG HD + G E L
Sbjct: 129 GDLAEAFLKHEEGIGEGTDGKKREAKQERVKQWKKALTEAANLSG-HDLRITDNGREANL 187
Query: 173 IGK-IVKYISNK--ISQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLG 229
+ IV I K +S L VA + VG+ SR+Q + S L G + V MVGI+G+GGLG
Sbjct: 188 CPREIVDNIITKWLMSTNKLRVAKHQVGINSRIQDIISRLSSGGSN-VIMVGIWGMGGLG 246
Query: 230 KSTLAKAIYNFIADQFECSCFLENVKESSASNNLKNLQQELLLKTLQLEIKLGSVSEGIP 289
K+T AKAIYN I +F+ FL +V +++ + L LQ+EL+ L+ + K+ SV EGI
Sbjct: 247 KTTAAKAIYNQIHHEFQFKSFLPDVGNAASKHGLVYLQKELIYDILKTKSKISSVDEGIG 306
Query: 290 KIKERLHGKKILLILDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYA 349
I+++ +++L+I+D++D++ QL+A+ G DWFGPGSR+IITTRD+HLL ++KTY
Sbjct: 307 LIEDQFRHRRVLVIMDNIDEVGQLDAIVGNPDWFGPGSRIIITTRDEHLL--KQVDKTYV 364
Query: 350 VEELNETEALELLRWKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAEC 409
++L+E EALEL W AF N Y ++ ++ V Y GLPLA+EV+GS LF + IAE
Sbjct: 365 AQKLDEREALELFSWHAFGNNWPNEEYLELSEKVVSYCGGLPLALEVLGSFLFKRPIAEW 424
Query: 410 ESTLDKYGRIPHKDIQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYS 469
+S L+K R P I K LR+S++ LD+ ++++FLDI+C G + V ++L G+
Sbjct: 425 KSQLEKLKRTPEGKIIKSLRISFEGLDDAQKAIFLDISCFFIGEDKDYVAKVL-DGCGFY 483
Query: 470 IKSHLRVLVDKSLIKISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDI 529
+ VL ++ L+ + K+ +H+L+ M K ++ ++SP +PG+ SRLW + ++
Sbjct: 484 ATIGISVLRERCLVTV-----EHNKLNMHDLLREMAKVIISEKSPGDPGKWSRLWDKREV 538
Query: 530 VHVLTENTGTGKTEMICMNLHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKGLKHLPSS 589
++VLT +GT + E + + +AF + +L+ L + + KHLP
Sbjct: 539 INVLTNKSGTEEVEGLALPWGYRHDTAFST-EAFANLKKLRLLQLCRVELNGEYKHLPKE 597
Query: 590 LKALKWEGCLSKSLSSSILSKASEIKSFSNCIYL*CTMISFSPISSFVFMQKFQDMTILI 649
L L W C KS+ ++ + M + + + ++ L
Sbjct: 598 LIWLHWFECPLKSIPDDFFNQDKLVV---------LEMQWSKLVQVWEGSKSLHNLKTLD 648
Query: 650 LDHCEYLTHIPDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLSAFGCRKLKRFPP- 708
L L PD S + NLE+L C L IH SIGHL +L ++ C KL P
Sbjct: 649 LSESRSLQKSPDFSQVPNLEELILYNCKELSEIHPSIGHLKRLSLVNLEWCDKLISLPGD 708
Query: 709 -LGLASLKELDICCCSSLKSFPELLCKMTNIKEIDLDYNISIGELPSSFQNLSELDELSV 767
S++ L + C L+ E + +M +++ ++ +Y I E+P S L L LS+
Sbjct: 709 FYKSKSVEALLLNGCLILRELHEDIGEMISLRTLEAEYT-DIREVPPSIVRLKNLTRLSL 767
Query: 768 REARMLRFPKHNDRMYSKVFSKVTKLRIYECNLSDEYLQIVLKWCVNVELLDLSHNNFKI 827
+ P + S + +L + L+D+ + L ++++ L+L N+F
Sbjct: 768 SSVESIHLPHSLHGLNS-----LRELNLSSFELADDEIPKDLGSLISLQDLNLQRNDFHT 822
Query: 828 LPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAYQCKSLS--------SSCRRMLM 879
LP LS L+ L LH+C L I +P NLK L A C +L S+ R + +
Sbjct: 823 LPS-LSGLSKLETLRLHHCEQLRTITDLPTNLKFLLANGCPALETMPNFSEMSNIRELKV 881
Query: 880 SQELHEARCTRFLFPNEKEG---------------IPDWFEHQSRGDTISFWFRKEIPSM 924
S + + L N +G +PDWFE + G ++F +IP
Sbjct: 882 SDSPN--NLSTHLRKNILQGWTSCGFGGIFLHANYVPDWFEFVNEGTKVTF----DIP-- 933
Query: 925 TYICIPPEGNNWSADT 940
P +G N+ T
Sbjct: 934 -----PSDGRNFEGLT 944
>UniRef100_Q84ZV0 R 14 protein [Glycine max]
Length = 641
Score = 493 bits (1268), Expect = e-137
Identities = 254/437 (58%), Positives = 327/437 (74%), Gaps = 6/437 (1%)
Query: 6 SSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITPSLLK 65
++T+ S + Y VFL+FRG DTRYGFTGNLY+AL +KGIHTF D +L GDEITP+L K
Sbjct: 2 AATTRSLPFIYDVFLSFRGEDTRYGFTGNLYRALCEKGIHTFFDEEKLHGGDEITPALSK 61
Query: 66 AIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQKGSYG 125
AI+ESRI I V S NYA SSFCLDELV I+HC K++G LV+PVF+ V+P+ +RHQKGSYG
Sbjct: 62 AIQESRIAITVLSQNYAFSSFCLDELVTILHC-KSEGLLVIPVFYNVDPSDLRHQKGSYG 120
Query: 126 EALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYH-DSPPGYEYKLIGKIVKYISNKI 184
EA+ +H+KRF++ ME+LQ W+ AL Q A+LSG+H YEYK IG IV+ +S KI
Sbjct: 121 EAMIKHQKRFES---KMEKLQKWRMALKQVADLSGHHFKDGDAYEYKFIGSIVEEVSRKI 177
Query: 185 SQQPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYNFIADQ 244
++ LHV YPVGL+S+V + LLD GSD VH++GI+G+ GLGK+TL+ A+YN IA
Sbjct: 178 NRASLHVLDYPVGLESQVTDLMKLLDVGSDDVVHIIGIHGMRGLGKTTLSLAVYNLIALH 237
Query: 245 FECSCFLENVKESSASNNLKNLQQELLLKTL-QLEIKLGSVSEGIPKIKERLHGKKILLI 303
F+ SCFL+NV+E S + LK+LQ LLLK L + +I L S EG I+ RL KK+LLI
Sbjct: 238 FDESCFLQNVREESNKHGLKHLQSILLLKLLGEKDINLTSWQEGASMIQHRLRRKKVLLI 297
Query: 304 LDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETEALELLR 363
LDD D+ +QL+A+ GR DWFGPGSRVIITTRDKHLL HGIE+TY V+ LN+ AL+LL
Sbjct: 298 LDDADRHEQLKAIVGRPDWFGPGSRVIITTRDKHLLKYHGIERTYEVKVLNDNAALQLLT 357
Query: 364 WKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFGKSIAECESTLDKYGRIPHKD 423
W AF+ EK+ SYE +L R V YASGLPLA+EV+GS+LF K++AE E ++ Y RIP +
Sbjct: 358 WNAFRREKIDPSYEHVLNRVVAYASGLPLALEVIGSHLFEKTVAEWEYAVEHYSRIPIDE 417
Query: 424 IQKILRLSYDALDEEEQ 440
I IL++S+DA +E Q
Sbjct: 418 IVDILKVSFDATKQETQ 434
>UniRef100_Q84KB3 MRGH63 [Cucumis melo]
Length = 943
Score = 482 bits (1241), Expect = e-134
Identities = 334/906 (36%), Positives = 500/906 (54%), Gaps = 62/906 (6%)
Query: 2 QSPSSSTSISYEYKYQVFLNFRGSDTRYGFTGNLYKALDDKGIHTFIDNHELQRGDEITP 61
Q+ SS+S + + + VFL+FRG DTR FT +L L +GI+ FID +L RG+EI+
Sbjct: 3 QAGRSSSSSCFRWSFDVFLSFRGEDTRSNFTSHLNMTLRQRGINVFIDK-KLSRGEEISS 61
Query: 62 SLLKAIEESRIFIAVFSINYASSSFCLDELVHIIHCYKTKGRLVLPVFFAVEPTIVRHQK 121
SLL+AIEES++ I V S +YASSS+CL+ELV II C K +G++VLP+F+ V+P+ V +Q
Sbjct: 62 SLLEAIEESKVSIIVISESYASSSWCLNELVKIIMCNKLRGQVVLPIFYKVDPSEVGNQS 121
Query: 122 GSYGEALAEHEKRFQNDPKSMERLQGWKEALSQAANLSGYHDSPPGYEYKLIGKIVKYIS 181
G +GE A+ E RF +D +++ WKEAL +++SG+ E LI IV+ +
Sbjct: 122 GRFGEEFAKLEVRFSSD-----KMEAWKEALITVSHMSGWPVLQRDDEANLIQNIVQEVW 176
Query: 182 NKISQ--QPLHVATYPVGLQSRVQQMKSLLDEGSDHGVHMVGIYGIGGLGKSTLAKAIYN 239
++ + L VA YPVG+ Q+++LL +G MVG+YGIGG+GK+TLAKA+YN
Sbjct: 177 KELDRATMQLDVAKYPVGIDI---QVRNLLPHVMSNGTTMVGLYGIGGMGKTTLAKALYN 233
Query: 240 FIADQFECSCFLENVKESSAS-NNLKNLQQELLLKTLQLE-IKLGSVSEGIPKIKERLHG 297
IAD FE CFL N++E+S L LQ+ELL + L + IK+ ++ G+ I+ RL+
Sbjct: 234 KIADDFEGCCFLPNIREASNQYGGLVQLQRELLREILVDDSIKVSNLPRGVTIIRNRLYS 293
Query: 298 KKILLILDDVDKLDQLEALAGRLDWFGPGSRVIITTRDKHLLDCHGIEKTYAVEELNETE 357
KKILLILDDVD +QL+AL G DWFG GS+VI TTR+K LL HG +K +V L+ E
Sbjct: 294 KKILLILDDVDTREQLQALVGGHDWFGHGSKVIATTRNKQLLVTHGFDKMQSVVGLDYDE 353
Query: 358 ALELLRWKAFKNEKVPSSYEDILKRAVVYASGLPLAIEVVGSNLFG-KSIAECESTLDKY 416
ALEL W F+N + Y ++ KRAV Y GLPLA+EV+GS L + LD+Y
Sbjct: 354 ALELFSWHCFRNSHPLNDYLELSKRAVDYCKGLPLALEVLGSFLHSIDDPFNFKRILDEY 413
Query: 417 GR-IPHKDIQKILRLSYDALDEEEQSVFLDIACCIKGCRLEEVEQILHHHYGYSIKSHLR 475
+ K+IQ LR+SYD L++E + +F I+CC + +V+ +L ++ +
Sbjct: 414 EKYYLDKEIQDSLRISYDGLEDEVKEIFCYISCCFVREDINKVKMMLEACGCICLEKGIT 473
Query: 476 VLVDKSLIKISWCFFSGIKVTLHELIEVMGKEVVRQESPKEPGERSRLWSQDDIVHVLTE 535
L++ SL+ I +V +H++I+ MG+ + E+ K +R RL +DD ++VL
Sbjct: 474 KLMNLSLLTIG----RFNRVEMHDIIQQMGRTIHLSETSKS-HKRKRLLIKDDAMNVLKG 528
Query: 536 NTGTGKTEMICMNLHSMESVIDKKGKAFKKMTRLKTLIIENGHCSKG--LKHLPSSLKAL 593
N ++I N + +D +AF+K+ L L + N SK L++LPSSL+ +
Sbjct: 529 NKEARAVKVIKFNF-PKPTELDIDSRAFEKVKNLVVLEVGNATSSKSTTLEYLPSSLRWM 587
Query: 594 KWEGCLSKSLSSSILSKASEIKSFSNCIYL*CTMISFSPISSF-VFMQKFQDMTILILDH 652
W SL + + N + L + +S I F + + + L
Sbjct: 588 NWPQFPFSSLPPTY--------TMENLVEL---KLPYSSIKHFGQGYMSCERLKEINLTD 636
Query: 653 CEYLTHIPDVSGLSNLEKLSFECCYNLITIHNSIGHLNKLERLS-AFGCRKLKRFPP-LG 710
+L IPD+S NL+ L C NL+ +H SIG LNKL L + + ++FP L
Sbjct: 637 SNFLVEIPDLSTAINLKYLDLVGCENLVKVHESIGSLNKLVALHLSSSVKGFEQFPSHLK 696
Query: 711 LASLKELDICCC----------SSLKSFPEL--------------LCKMTNIKEIDLDYN 746
L SLK L + C +KS L + +T++K + L Y
Sbjct: 697 LKSLKFLSMKNCRIDEWCPQFSEEMKSIEYLSIGYSIVTHQLSPTIGYLTSLKHLTLYYC 756
Query: 747 ISIGELPSSFQNLSELDELSVREARMLRFPKHNDRMYSKVFSKVTKLRIYECNLSD-EYL 805
+ LPS+ LS L L V ++ + FP N +TKLR+ C +++ ++L
Sbjct: 757 KELTTLPSTIYRLSNLTSLIVLDSDLSTFPSLNHPSLPSSLFYLTKLRLVGCKITNLDFL 816
Query: 806 QIVLKWCVNVELLDLSHNNFKILPECLSECHHLKHLGLHYCSSLEEIRGIPPNLKELSAY 865
+ ++ +++ LDLS NNF LP C+ LK+L C LEEI +P + SA
Sbjct: 817 ETIVYVAPSLKELDLSENNFCRLPSCIINFKSLKYLYTMDCELLEEISKVPEGVICTSAA 876
Query: 866 QCKSLS 871
CKSL+
Sbjct: 877 GCKSLA 882
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.137 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,830,165,950
Number of Sequences: 2790947
Number of extensions: 79591320
Number of successful extensions: 247389
Number of sequences better than 10.0: 3090
Number of HSP's better than 10.0 without gapping: 1153
Number of HSP's successfully gapped in prelim test: 1983
Number of HSP's that attempted gapping in prelim test: 233497
Number of HSP's gapped (non-prelim): 8804
length of query: 1092
length of database: 848,049,833
effective HSP length: 138
effective length of query: 954
effective length of database: 462,899,147
effective search space: 441605786238
effective search space used: 441605786238
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC138133.1


