

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138130.16 + phase: 0
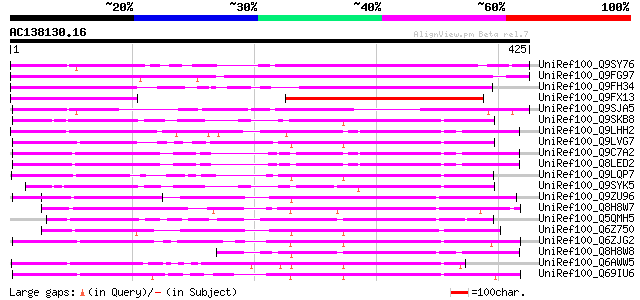
(425 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SY76 F14N23.22 [Arabidopsis thaliana] 246 1e-63
UniRef100_Q9FG97 Ankyrin-like protein [Arabidopsis thaliana] 234 3e-60
UniRef100_Q9FH34 Similarity to ankyrin-like protein [Arabidopsis... 228 2e-58
UniRef100_Q9FX13 F12G12.13 protein [Arabidopsis thaliana] 182 2e-44
UniRef100_Q9SJA5 Expressed protein [Arabidopsis thaliana] 139 2e-31
UniRef100_Q9SKB8 Ankyrin-like protein [Arabidopsis thaliana] 112 2e-23
UniRef100_Q9LHH2 Ankyrin-like protein [Arabidopsis thaliana] 112 2e-23
UniRef100_Q9LVG7 Ankyrin-like protein [Arabidopsis thaliana] 111 3e-23
UniRef100_Q9C7A2 Ankyrin-like protein; 93648-91299 [Arabidopsis ... 110 6e-23
UniRef100_Q8LED2 Ankyrin-like protein [Arabidopsis thaliana] 110 6e-23
UniRef100_Q9LQP7 F24B9.19 protein [Arabidopsis thaliana] 109 2e-22
UniRef100_Q9SYK5 F3F20.9 protein [Arabidopsis thaliana] 108 4e-22
UniRef100_Q9ZU96 Expressed protein [Arabidopsis thaliana] 105 2e-21
UniRef100_Q8H8W7 Putative ankyrin repeat containing protein [Ory... 105 3e-21
UniRef100_Q5QMH5 Ankyrin-like protein [Oryza sativa] 103 1e-20
UniRef100_Q6Z750 Ankyrin-like protein [Oryza sativa] 100 1e-19
UniRef100_Q6ZJG2 Ankyrin-like protein [Oryza sativa] 98 4e-19
UniRef100_Q8H8W8 Hypothetical protein [Oryza sativa] 97 7e-19
UniRef100_Q6AWW5 At5g02620 [Arabidopsis thaliana] 97 9e-19
UniRef100_Q69IU6 Ankyrin-like protein [Oryza sativa] 94 1e-17
>UniRef100_Q9SY76 F14N23.22 [Arabidopsis thaliana]
Length = 578
Score = 246 bits (627), Expect = 1e-63
Identities = 160/429 (37%), Positives = 229/429 (53%), Gaps = 68/429 (15%)
Query: 1 MNGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTN---LLHC 57
+ G V IL++F+ SF +T +ETVFHLA R DA VF+ + S G N LL
Sbjct: 214 LRGSVVILEEFLDKVPLSFSSITPSKETVFHLAARNKNMDAFVFMAE-SLGINSQILLQQ 272
Query: 58 QDRYGNSVLHLAVSGGRHK-MTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQ 116
D GN+VLH+A S + +++ K +DI ++N G A +L + E L
Sbjct: 273 TDESGNTVLHIAASVSFDAPLIRYIVGKNIVDITSKNKMGFEAFQLLPREAQDFE---LL 329
Query: 117 AIFIRAGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPP 176
+ ++R G + S ELD N + + E+EV+ +
Sbjct: 330 SRWLRFG-------TETSQELDSEN--------NVEQHEGSQEVEVIRLLRIIG------ 368
Query: 177 VSKSSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMH 236
++ + + ++K + K + V ++ ++MH
Sbjct: 369 ---------------------------INTSEIAERK----RSKEQEVERGRQNLEYQMH 397
Query: 237 QEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNI 296
EAL NARNTI +VAVLIA+V +A GI+PPGGVYQ+GP RGKS+VG+T+AFKVFAI NNI
Sbjct: 398 IEALQNARNTIAIVAVLIASVAYAGGINPPGGVYQDGPWRGKSLVGKTTAFKVFAICNNI 457
Query: 297 ALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQ 356
ALFTSL +VI+LVSI+P++RKP LL+ H++MWV+V FMAT Y+AA+WV +PH G Q
Sbjct: 458 ALFTSLGIVILLVSIIPYKRKPLKRLLVATHRMMWVSVGFMATAYIAASWVTIPHYHGTQ 517
Query: 357 WLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGAAESDKESENSDFQS 416
WL ++A+ GG+L +F L V + H W KK +GD + S SD
Sbjct: 518 WLFPAIVAVAGGALTVLFFYLGVETIGH------WFKKMNRVGDNIPSFARTS--SDLAV 569
Query: 417 SYLQGYHSY 425
S GY +Y
Sbjct: 570 SGKSGYFTY 578
>UniRef100_Q9FG97 Ankyrin-like protein [Arabidopsis thaliana]
Length = 535
Score = 234 bits (598), Expect = 3e-60
Identities = 149/434 (34%), Positives = 227/434 (51%), Gaps = 25/434 (5%)
Query: 1 MNGKVSILDDFVSGSAASFHYLTRE-EETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQD 59
MNG V L F++ + SF +T + ETVFHLA R+ +A +F+ + +N LL+ D
Sbjct: 118 MNGSVETLTAFINKAPLSFDSVTLQTSETVFHLAARHKKMEAFIFMAKNANLRRLLYELD 177
Query: 60 RYGNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQ------AMDSVESR 113
GN+VLH A S G + +++++ K+++ T+N +G A+D+L++ M +
Sbjct: 178 GEGNTVLHAAASVGFLSLVSYIVHEIKIEVTTQNDKGFEAVDLLNKDDEDFKMMSMILGH 237
Query: 114 QLQAIFIRAGGKRSIQSSSFSLELDKNNSPSPAYRLSPS--RRYIPNEMEVLTEMVSYDC 171
+ + A R + S E++ + ++P + NE + E +
Sbjct: 238 DSEIVQRAASSPRDAYTPSTQTEVENSEIHHEQGLVAPEIKEENVTNENNKVFEAIDL-- 295
Query: 172 ISPPPVSKSSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKH 231
P + D + SE F+ + ++P + + I + R
Sbjct: 296 ----PTKEDGDLKMLAGTDSETFQLPSSRTGILTPETETEMVISNTLHGIRHGLRESRIK 351
Query: 232 YHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFA 291
EM EAL NARNTI +VAVLIA+VTF G++PPGGVYQ+G GK+ G T AFKVF+
Sbjct: 352 EKEMQSEALQNARNTITVVAVLIASVTFTCGLNPPGGVYQDGHFIGKATAGGTVAFKVFS 411
Query: 292 ISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPH 351
+SN+IALFTSL +VI+L+SI+PFR K LII HK++W+AV MA+ YVA T V LPH
Sbjct: 412 VSNSIALFTSLCIVILLLSIIPFRTKSLKTFLIITHKMIWLAVIAMASAYVAGTCVTLPH 471
Query: 352 NQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGAAESDKESEN 411
++G +W+ L + LG +FI L L H RK K RK E
Sbjct: 472 SRGNKWVLKATLVIACVMLGGMFIFLWFKLANHMSRKKKMRKNMM----------SRIET 521
Query: 412 SDFQSSYLQGYHSY 425
+ ++++ GY+SY
Sbjct: 522 LNKAAAFVDGYYSY 535
>UniRef100_Q9FH34 Similarity to ankyrin-like protein [Arabidopsis thaliana]
Length = 652
Score = 228 bits (582), Expect = 2e-58
Identities = 138/396 (34%), Positives = 216/396 (53%), Gaps = 63/396 (15%)
Query: 1 MNGKVSILDDFVSGSAASFHYLTREE-ETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQD 59
+NG V IL +F+ + +SF+ T+ ETVFHLA +Y A +F+ Q +N LL+ D
Sbjct: 242 INGSVEILKEFLCKAPSSFNITTQGTIETVFHLAAKYQKTKAFIFMAQSANIRQLLYSLD 301
Query: 60 RYGNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIF 119
N+VLH+A S + ++++T +D+ +N +G A+D++D
Sbjct: 302 AEDNTVLHVAASVDSTSLVRHILSETTIDVTLKNKKGFAAVDLID--------------- 346
Query: 120 IRAGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSK 179
+ G + S F E +K P+ RY+ E + E++ +
Sbjct: 347 -KEGVDFPLLSLWFRDEAEKIQRPA---------RYVKFAHEPV-ELIRN--------TN 387
Query: 180 SSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQEA 239
+ + S + +A + G P N ++ H E+
Sbjct: 388 NGEKLSSESRAMDLLREGR------DPRNKEREMH----------------------SES 419
Query: 240 LLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALF 299
L NARNTI +VAVLIA+V F GI+PPGGV+Q+GP GK+ GRT AFK+F+++NNIALF
Sbjct: 420 LQNARNTITIVAVLIASVAFTCGINPPGGVHQDGPFIGKATAGRTLAFKIFSVANNIALF 479
Query: 300 TSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLS 359
TSLS+V +LVSI+ +R K + ++IAHK+MW+AVA MAT Y A+ W+ +PHN+G +WL
Sbjct: 480 TSLSIVTLLVSIISYRTKALKMCVVIAHKMMWLAVASMATAYAASAWITVPHNEGSKWLV 539
Query: 360 VLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKR 395
A+ +LG++F+ +S M+V+H L+K K R+ +
Sbjct: 540 YTTSAIASVALGSMFVYVSFMMVKHILKKDKLRRNQ 575
Score = 40.4 bits (93), Expect = 0.097
Identities = 17/32 (53%), Positives = 24/32 (74%)
Query: 320 TLLLIIAHKVMWVAVAFMATGYVAATWVILPH 351
T LL+I H++MWV VA M + Y+AA+ + LPH
Sbjct: 598 TKLLVITHRLMWVDVAAMESAYLAASSITLPH 629
>UniRef100_Q9FX13 F12G12.13 protein [Arabidopsis thaliana]
Length = 573
Score = 182 bits (461), Expect = 2e-44
Identities = 89/162 (54%), Positives = 119/162 (72%)
Query: 227 TKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSA 286
T K +MH EALLNARNTI +VAVLIA+V F GI+PPGGVYQEGP +GKS GRT A
Sbjct: 379 TPSKRESKMHAEALLNARNTITIVAVLIASVAFTCGINPPGGVYQEGPYKGKSTAGRTLA 438
Query: 287 FKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATW 346
F+VF+ISNNIALFTSL +VI+LVSI+P+R +P L + H+++WVAVA MA YV+A
Sbjct: 439 FQVFSISNNIALFTSLCIVILLVSIIPYRTRPLKNFLKLTHRILWVAVASMALAYVSAAS 498
Query: 347 VILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRK 388
+I+PH +G +WL +L++ LG +F ++ ++ H L+K
Sbjct: 499 IIIPHVEGKRWLFTTVLSISTLMLGGLFAFMTYKVIRHWLKK 540
Score = 70.5 bits (171), Expect = 9e-11
Identities = 36/104 (34%), Positives = 59/104 (56%)
Query: 1 MNGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDR 60
M + IL +F + F LT +ETVFHLA + A F+ + + NLLH DR
Sbjct: 227 MKCSIPILKEFSDKAPRYFDILTPAKETVFHLAAEHKNILAFYFMAESPDRNNLLHQVDR 286
Query: 61 YGNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILD 104
YGN+VLH AV + + + +T +D++ +N+ G+ A+D+++
Sbjct: 287 YGNTVLHTAVMSSCYSVIVSITYETTIDLSAKNNRGLKAVDLIN 330
>UniRef100_Q9SJA5 Expressed protein [Arabidopsis thaliana]
Length = 548
Score = 139 bits (350), Expect = 2e-31
Identities = 128/443 (28%), Positives = 185/443 (40%), Gaps = 130/443 (29%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTN---LLHCQD 59
G V IL++F+ S SF T +ETVFHLA R DA VF+ + + GT+ LL +D
Sbjct: 216 GSVIILEEFMDKSPLSFCVRTPSKETVFHLAARNKNTDAFVFMAE-NLGTSSPILLKKKD 274
Query: 60 RYGNSVLHLAVSGG-RHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAI 118
+ GN+VLH+A S + +++ K +DI
Sbjct: 275 QQGNTVLHIAASVSCGSPLIRYIVGKKIIDIR---------------------------- 306
Query: 119 FIRAGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPV- 177
D+NN AY L P + + E ++ + D + V
Sbjct: 307 -------------------DRNNMGYRAYHLLPRQA---QDYEFISSYLRCDTKTSEEVD 344
Query: 178 SKSSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQ 237
SK ++ P SE +S + + ++K K K +V + HEMH
Sbjct: 345 SKKAERNEPHIGHSEVIR--LLKLIEISTSEIAERK----KSKKHHVKRGHKSLEHEMHI 398
Query: 238 EALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIA 297
EAL NARNTI +VAVLIA+V++A GI+PPGGVYQ+GP +GKS+VG
Sbjct: 399 EALQNARNTIAIVAVLIASVSYAGGINPPGGVYQDGPWKGKSLVG--------------- 443
Query: 298 LFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQW 357
FMAT YVAA+ V +PH G +W
Sbjct: 444 --------------------------------------FMATAYVAASLVTIPHFPGTRW 465
Query: 358 LSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSK--------WRKKRKEIGDGAAESDKES 409
L +++++ GGSL +F L V + H +K + K + D A + E
Sbjct: 466 LFPVIISVAGGSLTVLFSYLGVETISHWFKKMNRVGRGLPIYFIKNNRVEDIPAIAKNEG 525
Query: 410 E-------NSDFQSSYLQGYHSY 425
E NSD +S GY +Y
Sbjct: 526 EMPSLARTNSDLAASEGSGYFTY 548
>UniRef100_Q9SKB8 Ankyrin-like protein [Arabidopsis thaliana]
Length = 662
Score = 112 bits (280), Expect = 2e-23
Identities = 96/398 (24%), Positives = 179/398 (44%), Gaps = 71/398 (17%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G V ++ + + ++ +T H+AV+ G D +V + V +L +D G
Sbjct: 303 GHVEVVKSLIGKDPSIGFRTDKKGQTALHMAVK-GQNDGIVVEL-VKPDVAVLSVEDNKG 360
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRA 122
N+ LH+A + GR K+ L++ +++N N G T LD+ ++ + + +L ++ A
Sbjct: 361 NTPLHIATNKGRIKIVRCLVSFEGINLNPINKAGDTPLDVSEK----IGNAELVSVLKEA 416
Query: 123 GGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSD 182
G + D +PA +L + I +E+
Sbjct: 417 GAATA---------KDLGKPQNPAKQLKQTVSDIKHEV---------------------- 445
Query: 183 SRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQEALLN 242
Q Q + + G +++ + K ++H L N
Sbjct: 446 ----QSQLQQSRQTGV---------------------RVQKI----AKRLKKLHISGLNN 476
Query: 243 ARNTIVLVAVLIATVTFAAGISPPGGVYQE---GPMRGKSMVGRTSAFKVFAISNNIALF 299
A N+ +VAVLIATV FAA + PG ++ G + G++ + + F VF I +++ALF
Sbjct: 477 AINSATVVAVLIATVAFAAIFTIPGQYEEDRSKGELLGQAHIANKAPFLVFFIFDSLALF 536
Query: 300 TSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLS 359
SL+VV+V S+V +K + L+ + +K+MW A F++ +V+ +++++ + WL+
Sbjct: 537 ISLAVVVVQTSVVVIEQKAKKKLVFVINKLMWCACLFISIAFVSLSYIVVGKEE--MWLA 594
Query: 360 VLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKE 397
V +GG + T + +V H + +SK R RKE
Sbjct: 595 VCATVIGGTIMLTTIGAMCYCVVMHRMEESKLRSIRKE 632
>UniRef100_Q9LHH2 Ankyrin-like protein [Arabidopsis thaliana]
Length = 1100
Score = 112 bits (280), Expect = 2e-23
Identities = 103/429 (24%), Positives = 191/429 (44%), Gaps = 40/429 (9%)
Query: 1 MNGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDR 60
M G +++ +S + + HLA R G + + L+ S L D+
Sbjct: 692 MRGHTEVVNQLLSKAGNLLEISRSNNKNALHLAARQGHVEVIKALL--SKDPQLARRIDK 749
Query: 61 YGNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFI 120
G + LH+AV G ++ L++ + + TAL + + + L FI
Sbjct: 750 KGQTALHMAVKGQSSEVVKLLLDADPAIVMQPDKSCNTALHVATRKKRAEVCITLIVWFI 809
Query: 121 RAGGKRSIQSSSFSL-----ELDKNNSPSPAYRLSPSRRYIPNEME--VLTEMVSY--DC 171
+ I SS F + EL + + A L+ + + E L+E SY +C
Sbjct: 810 L---RFLIGSSHFGIYLQIVELLLSLPDTNANTLTRDHKTALDIAEGLPLSEESSYIKEC 866
Query: 172 ISPPPVSKSSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVN---HTK 228
++ ++++ P+ + S +K H + + N H
Sbjct: 867 LARSGALRANELNQPRDELR-------------STVTQIKNDVHIQLEQTKRTNKNVHNI 913
Query: 229 RKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFK 288
K ++H+E + NA N++ +VAVL ATV FAA + PGG +G +VGR S FK
Sbjct: 914 SKELRKLHREGINNATNSVTVVAVLFATVAFAAIFTVPGGDNNDG---SAVVVGRAS-FK 969
Query: 289 VFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVI 348
+F I N +ALFTSL+VV+V +++V K + ++ + +K+MW+A + ++A+++++
Sbjct: 970 IFFIFNALALFTSLAVVVVQITLVRGETKAEKRVVEVINKLMWLASMCTSVAFLASSYIV 1029
Query: 349 LPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGAAESDKE 408
+ +W + L+ +G G I G+ + + ++ + R RK++
Sbjct: 1030 VGRKN--EWAAELVTVVG----GVIMAGVLGTMTYYVVKSKRTRSMRKKVKSARRSGSNS 1083
Query: 409 SENSDFQSS 417
+SDF +S
Sbjct: 1084 WHHSDFSNS 1092
>UniRef100_Q9LVG7 Ankyrin-like protein [Arabidopsis thaliana]
Length = 548
Score = 111 bits (278), Expect = 3e-23
Identities = 104/409 (25%), Positives = 183/409 (44%), Gaps = 51/409 (12%)
Query: 3 GKVSILDDFVSGSAASFHYLTREE-ETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRY 61
G V +++ + + +S + + +T H A R G + + +V V T D+
Sbjct: 153 GHVEVVEYLLEAAGSSLAAIAKSNGKTALHSAARNGHAEVVKAIVAVEPDTATR--TDKK 210
Query: 62 GNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIR 121
G + LH+AV G + L+ + +N +S+G TAL + R
Sbjct: 211 GQTPLHMAVKGQSIDVVVELMKGHRSSLNMADSKGNTALHVAT----------------R 254
Query: 122 AGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSS 181
G + ++ L LD N + SPS + I E T + + + P ++
Sbjct: 255 KGRIKIVE-----LLLDNNET-------SPSTKAINRAGE--TPLDTAEKTGHPQIAAVL 300
Query: 182 DSRS-PQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKR------KHYHE 234
+R P +A + +K + HH ++E+ T++ K ++
Sbjct: 301 KTRGVPSAKAINNTTRPNAARELKQTVSDIKHEVHH---QLEHARETRKRVQGIAKRINK 357
Query: 235 MHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE------GPMRGKSMVGRTSAFK 288
MH E L NA N+ +VAVLIATV FAA + PG E G G++ + AF
Sbjct: 358 MHVEGLDNAINSTTVVAVLIATVAFAAIFTVPGQYADELSSLLPGQSLGEANIADRPAFA 417
Query: 289 VFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVI 348
+F I ++IALF SL+VV+V S+V K + ++ + +K+MW+A ++ ++A +V+
Sbjct: 418 IFFIFDSIALFISLAVVVVQTSVVAIEHKAKKNMMAVINKLMWLACVLISVAFLALAFVV 477
Query: 349 LPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKE 397
+ + +WL+V + G + T + ++ H + S RK RKE
Sbjct: 478 VGEEE--RWLAVGVTVFGATIMLTTLGTMCYWVIMHRIEASNVRKSRKE 524
Score = 40.0 bits (92), Expect = 0.13
Identities = 27/106 (25%), Positives = 52/106 (48%), Gaps = 4/106 (3%)
Query: 2 NGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRY 61
NG ++ V+ + ++ +T H+AV+ D +V L++ + L+ D
Sbjct: 187 NGHAEVVKAIVAVEPDTATRTDKKGQTPLHMAVKGQSIDVVVELMKGHRSS--LNMADSK 244
Query: 62 GNSVLHLAVSGGRHKMTDFLINKTKLDINTR--NSEGMTALDILDQ 105
GN+ LH+A GR K+ + L++ + +T+ N G T LD ++
Sbjct: 245 GNTALHVATRKGRIKIVELLLDNNETSPSTKAINRAGETPLDTAEK 290
>UniRef100_Q9C7A2 Ankyrin-like protein; 93648-91299 [Arabidopsis thaliana]
Length = 590
Score = 110 bits (276), Expect = 6e-23
Identities = 97/415 (23%), Positives = 183/415 (43%), Gaps = 75/415 (18%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G V ++ +S + ++ +T H+AV+ + + L+ ++ D+
Sbjct: 243 GHVEVIKALLSKDPQLARRIDKKGQTALHMAVKGQSSEVVKLLLDADPA--IVMQPDKSC 300
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRA 122
N+ LH+A R ++ + L++ + NT + TALDI + S ES ++ R+
Sbjct: 301 NTALHVATRKKRAEIVELLLSLPDTNANTLTRDHKTALDIAEGLPLSEESSYIKECLARS 360
Query: 123 GGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSD 182
G +L ++ N P R + ++ I N++ + E
Sbjct: 361 G----------ALRANELNQPRDELRSTVTQ--IKNDVHIQLE----------------- 391
Query: 183 SRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQEALLN 242
Q NK + N++ RK +H+E + N
Sbjct: 392 -----------------------------QTKRTNKN-VHNISKELRK----LHREGINN 417
Query: 243 ARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSL 302
A N++ +VAVL ATV FAA + PGG +G +VGR S FK+F I N +ALFTSL
Sbjct: 418 ATNSVTVVAVLFATVAFAAIFTVPGGDNNDG---SAVVVGRAS-FKIFFIFNALALFTSL 473
Query: 303 SVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLL 362
+VV+V +++V K + ++ + +K+MW+A + ++A++++++ +W + L+
Sbjct: 474 AVVVVQITLVRGETKAEKRVVEVINKLMWLASMCTSVAFLASSYIVVGRKN--EWAAELV 531
Query: 363 LALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGAAESDKESENSDFQSS 417
+G G I G+ + + ++ + R RK++ +SDF +S
Sbjct: 532 TVVG----GVIMAGVLGTMTYYVVKSKRTRSMRKKVKSARRSGSNSWHHSDFSNS 582
>UniRef100_Q8LED2 Ankyrin-like protein [Arabidopsis thaliana]
Length = 534
Score = 110 bits (276), Expect = 6e-23
Identities = 97/415 (23%), Positives = 183/415 (43%), Gaps = 75/415 (18%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G V ++ +S + ++ +T H+AV+ + + L+ ++ D+
Sbjct: 187 GHVEVIKALLSKDPQLARRIDKKGQTALHMAVKGQSSEVVKLLLDADPA--IVMQPDKSC 244
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRA 122
N+ LH+A R ++ + L++ + NT + TALDI + S ES ++ R+
Sbjct: 245 NTALHVATRKKRAEIVELLLSLPDTNANTLTRDHKTALDIAEGLPLSEESSYIKECLARS 304
Query: 123 GGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSD 182
G +L ++ N P R + ++ I N++ + E
Sbjct: 305 G----------ALRANELNQPRDELRSTVTQ--IKNDVHIQLE----------------- 335
Query: 183 SRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQEALLN 242
Q NK + N++ RK +H+E + N
Sbjct: 336 -----------------------------QTKRTNKN-VHNISKELRK----LHREGINN 361
Query: 243 ARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSL 302
A N++ +VAVL ATV FAA + PGG +G +VGR S FK+F I N +ALFTSL
Sbjct: 362 ATNSVTVVAVLFATVAFAAIFTVPGGDNNDG---SAVVVGRAS-FKIFFIFNALALFTSL 417
Query: 303 SVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLL 362
+VV+V +++V K + ++ + +K+MW+A + ++A++++++ +W + L+
Sbjct: 418 AVVVVQITLVRGETKAEKRVVEVINKLMWLASMCTSVAFLASSYIVVGRKN--EWAAELV 475
Query: 363 LALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGAAESDKESENSDFQSS 417
+G G I G+ + + ++ + R RK++ +SDF +S
Sbjct: 476 TVVG----GVIMAGVLGTMTYYVVKSKRTRSMRKKVKSARRSGSNSWHHSDFSNS 526
>UniRef100_Q9LQP7 F24B9.19 protein [Arabidopsis thaliana]
Length = 543
Score = 109 bits (272), Expect = 2e-22
Identities = 96/407 (23%), Positives = 179/407 (43%), Gaps = 85/407 (20%)
Query: 2 NGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRY 61
NG V ++ ++ A + ++ +T H+AV+ + + L++ + ++ D
Sbjct: 179 NGHVKVIKALLASEPAIAIRMDKKGQTALHMAVKGTNVEVVEELIKADRSS--INIADTK 236
Query: 62 GNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIR 121
GN+ LH+A GR ++ L+ D N G TA +D+ E
Sbjct: 237 GNTALHIAARKGRSQIVKLLLANNMTDTKAVNRSGETA-------LDTAEK--------- 280
Query: 122 AGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSS 181
I + +L L K+ PS A + PS PN L + VS
Sbjct: 281 ------IGNPEVALILQKHGVPS-AKTIKPSG---PNPARELKQTVSD------------ 318
Query: 182 DSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKR------KHYHEM 235
+K + H+ ++E+ T++ K ++M
Sbjct: 319 ----------------------------IKHEVHN---QLEHTRLTRKRVQGIAKQLNKM 347
Query: 236 HQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE------GPMRGKSMVGRTSAFKV 289
H E L NA N+ +VAVLIATV FAA + PG ++ G G++ + T+ F +
Sbjct: 348 HTEGLNNAINSTTVVAVLIATVAFAAIFTVPGQYVEDTSKIPDGHSLGEANIASTTPFII 407
Query: 290 FAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVIL 349
F I ++IALF SL+VV+V S+V K + ++ + +K+MW+A ++ ++A ++V++
Sbjct: 408 FFIFDSIALFISLAVVVVQTSVVVIESKAKKQMMAVINKLMWLACVLISVAFLALSFVVV 467
Query: 350 PHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRK 396
+ +WL++ + A+G + T + +++H + + R R+
Sbjct: 468 GEEE--KWLAIWVTAIGATIMITTLGTMCYWIIQHKIEAANLRNIRR 512
>UniRef100_Q9SYK5 F3F20.9 protein [Arabidopsis thaliana]
Length = 627
Score = 108 bits (269), Expect = 4e-22
Identities = 100/390 (25%), Positives = 179/390 (45%), Gaps = 75/390 (19%)
Query: 14 GSAASFHYLT-REEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSG 72
G+ AS + T ++ +T H+AV+ G + +V L V +L +D GN+ LH A +
Sbjct: 277 GNDASIGFRTDKKGQTALHMAVK-GQNEGIV-LELVKPDPAILSVEDSKGNTPLHTATNK 334
Query: 73 GRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRAGGKRSIQSSS 132
GR K+ L++ +++N N G TALDI ++ + + +L ++ AG
Sbjct: 335 GRIKIVRCLVSFDGINLNAMNKAGDTALDIAEK----IGNPELVSVLKEAGA-------- 382
Query: 133 FSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASE 192
+ D +PA +L+ + I +E+ Q Q +
Sbjct: 383 -ATAKDLGKPRNPAKQLNQTVSDIKHEV--------------------------QSQLQQ 415
Query: 193 RFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKRKHYHEMHQEALLNARNTIVLVAV 252
+ G ++ + +K +H L NA N+ +VAV
Sbjct: 416 SRQTGV---------------------RVRRIAKRLKK----LHINGLNNAINSATVVAV 450
Query: 253 LIATVTFAAGISPPGGVYQEGPMRGKSMVGRT-----SAFKVFAISNNIALFTSLSVVIV 307
LIATV FAA + P G Y+E +G ++G + F VF I +++ALF SL+VV+V
Sbjct: 451 LIATVAFAAIFTIP-GQYEEDRTKGLLLLGEARIAGKAPFLVFFIFDSLALFISLAVVVV 509
Query: 308 LVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGG 367
S+V +K + L+ + +K+MW+A F++ +V+ +++++ WL++ +GG
Sbjct: 510 QTSVVVIEQKAKKNLVFVINKLMWLACLFISVAFVSLSFIVVGKED--IWLAICATIIGG 567
Query: 368 GSLGTIFIGLSVMLVEHSLRKSKWRKKRKE 397
+ T + +V H + +SK + RKE
Sbjct: 568 TIMLTTIGAMCYCVVMHRIEESKLKSLRKE 597
>UniRef100_Q9ZU96 Expressed protein [Arabidopsis thaliana]
Length = 532
Score = 105 bits (262), Expect = 2e-21
Identities = 89/396 (22%), Positives = 171/396 (42%), Gaps = 44/396 (11%)
Query: 27 ETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINKTK 86
+T H A RYG + L++ ++ +D+ G + LH+AV G ++ + ++
Sbjct: 163 KTSLHTAGRYGLLRIVKALIE--KDAAIVGVKDKKGQTALHMAVKGRSLEVVEEILQADY 220
Query: 87 LDINTRNSEGMTALDILDQ-AMDSVESRQLQAIFIRAGGKRSIQSSSFSLELDKNNSPSP 145
+N R+ +G TAL I + A + S L I NN
Sbjct: 221 TILNERDRKGNTALHIATRKARPQITSLLLTFTAIEVNAI--------------NNQKET 266
Query: 146 AYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASERFENGTYNPYYVS 205
A L+ +Y + +E+ +V + + ++R+ + S+
Sbjct: 267 AMDLADKLQYSESALEINEALVEAGAKHGRFIGREDEARALKRAVSDI------------ 314
Query: 206 PTNLVKQKHHHNKGKIENVNHTKR-----KHYHEMHQEALLNARNTIVLVAVLIATVTFA 260
KH ++N +R K ++H+EA+ N N+I +VAVL A++ F
Sbjct: 315 -------KHEVQSQLLQNEKTNRRVSGIAKELRKLHREAVQNTTNSITVVAVLFASIAFL 367
Query: 261 AGISPPGGVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQT 320
A + PG + EG G++ + + F+VF + N +LF SL+VV+V +++V + + Q
Sbjct: 368 AIFNLPGQYFTEGSHVGQANIAGRTGFRVFCLLNATSLFISLAVVVVQITLVAWDTRAQK 427
Query: 321 LLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVM 380
++ + +K+MW A A ++A + ++ +G W+++ + LG L +
Sbjct: 428 KVVSVVNKLMWAACACTFGAFLAIAFAVV--GKGNSWMAITITLLGAPILVGTLASMCYF 485
Query: 381 LVEHSLRKSKWRKKRKEIGDGAAESDKESEN-SDFQ 415
+ R ++R G + S S + SDF+
Sbjct: 486 VFRQRFRSGNDSQRRIRRGSSKSFSWSYSHHVSDFE 521
Score = 55.8 bits (133), Expect = 2e-06
Identities = 31/123 (25%), Positives = 63/123 (51%), Gaps = 2/123 (1%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G + I+ + AA ++ +T H+AV+ + + ++Q +L+ +DR G
Sbjct: 173 GLLRIVKALIEKDAAIVGVKDKKGQTALHMAVKGRSLEVVEEILQADY--TILNERDRKG 230
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRA 122
N+ LH+A R ++T L+ T +++N N++ TA+D+ D+ S + ++ + A
Sbjct: 231 NTALHIATRKARPQITSLLLTFTAIEVNAINNQKETAMDLADKLQYSESALEINEALVEA 290
Query: 123 GGK 125
G K
Sbjct: 291 GAK 293
>UniRef100_Q8H8W7 Putative ankyrin repeat containing protein [Oryza sativa]
Length = 565
Score = 105 bits (261), Expect = 3e-21
Identities = 102/415 (24%), Positives = 183/415 (43%), Gaps = 64/415 (15%)
Query: 27 ETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINKTK 86
+T H A R G + + L++ ++ D+ G + LH+A G R + D L+
Sbjct: 190 KTALHSAARNGHVEVVRALMEAE--PSIAARVDKKGQTALHMAAKGTRLDIVDALLAGEP 247
Query: 87 LDINTRNSEGMTALDILDQ-AMDSVESRQLQAIFIRAGGKRSIQSSSFSLELDKNNSPSP 145
+N +S+G TAL I + A + R L+ + ++F N+ S
Sbjct: 248 TLLNLADSKGNTALHIAARKARTPIVKRLLELPDTDLKAINRSRETAFDTAEKMGNTESV 307
Query: 146 AYRLSPSRRYIPNEMEVLTE--MVSYDCISPPPVSKSSDSRSPQPQASERFENGTYNPYY 203
A VL E + S +SP + R + Q S+
Sbjct: 308 A---------------VLAEHGVPSARAMSPTGGGGGNPGRELKQQVSD----------- 341
Query: 204 VSPTNLVKQKHHHNKGKIENVNHTK------RKHYHEMHQEALLNARNTIVLVAVLIATV 257
+K + H ++E T+ K +++H E L NA N+ +VAVLIATV
Sbjct: 342 ------IKHEVH---SQLEQTRQTRVRMQGIAKQINKLHDEGLNNAINSTTVVAVLIATV 392
Query: 258 TFAAGISPPG------GVYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSI 311
FAA + PG G G G++ + +AF +F + +++ALF SL+VV+V S+
Sbjct: 393 AFAAIFTVPGEYVDDAGSLTPGQALGEANISHQTAFLIFFVFDSVALFISLAVVVVQTSV 452
Query: 312 VPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSLG 371
V RK + ++ + +K+MWVA ++ ++A ++V++ + +WL+V + +G L
Sbjct: 453 VVIERKAKKQMMAVINKLMWVACVLVSVAFLALSFVVV--GKAERWLAVGVTIMGATILV 510
Query: 372 TIFIGLSVMLVEH--------SLRKSKWRKKRKEIGDGAAESDKESENSDFQSSY 418
T + ++ H S+++S + R G +E++ E +F+ Y
Sbjct: 511 TTIGTMLYWVIAHRIEAKRMRSIKRSSLSRSRSFSASGMSEAEWVEE--EFKRMY 563
>UniRef100_Q5QMH5 Ankyrin-like protein [Oryza sativa]
Length = 556
Score = 103 bits (256), Expect = 1e-20
Identities = 92/368 (25%), Positives = 169/368 (45%), Gaps = 42/368 (11%)
Query: 31 HLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINKTKLDIN 90
H A R G + + L++ L D+ G + LH+AV G + L++ +
Sbjct: 202 HFAARQGHVEIVKALLE--KDPQLARRNDKKGQTALHMAVKGTNCDVLRALVDADPAIVM 259
Query: 91 TRNSEGMTALDILDQAMDSVESRQLQAIFIRAGGKRSIQSSSFSLELDKNNSPSPAYRLS 150
+ G TAL + + + ++ A+ +R + +L D AY ++
Sbjct: 260 LPDKNGNTALHVATRK----KRAEIVAVLLRLP-----DTHVNALTRDHKT----AYDIA 306
Query: 151 PSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASERFENGTYNPYYVSPTNLV 210
+ E+ D +S +S + P+ + + + T +
Sbjct: 307 EALPLCEESSEIK------DILSQHGALRSRELNQPRDELRK------------TVTEIK 348
Query: 211 KQKHHH-NKGKIENVN-HTKRKHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGG 268
K H + + N N H K ++H+E + NA N++ +VAVL ATV FAA + PGG
Sbjct: 349 KDVHTQLEQTRKTNKNVHGIAKELRKLHREGINNATNSVTVVAVLFATVAFAAIFTVPGG 408
Query: 269 VYQEGPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHK 328
G ++V + ++F++F I N IALFTSL+VV+V +++V K + ++ + +K
Sbjct: 409 ----NANNGVAVVVQAASFRIFFIFNAIALFTSLAVVVVQITVVRGETKSERKVVEVINK 464
Query: 329 VMWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRK 388
+MW+A ++A+ +++L + QW ++L+ +GG ++ + +G V S R
Sbjct: 465 LMWLASVCTTISFIASCYIVL--GRHFQWAALLVSLIGGITMAGV-LGTMTYYVVKSKRM 521
Query: 389 SKWRKKRK 396
K RKK K
Sbjct: 522 RKIRKKEK 529
Score = 38.9 bits (89), Expect = 0.28
Identities = 31/116 (26%), Positives = 52/116 (44%), Gaps = 2/116 (1%)
Query: 24 REEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLIN 83
++ +T H+AV+ D L LV +L D+ GN+ LH+A R ++ L+
Sbjct: 229 KKGQTALHMAVKGTNCDVLRALVDADPAIVML--PDKNGNTALHVATRKKRAEIVAVLLR 286
Query: 84 KTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRAGGKRSIQSSSFSLELDK 139
+N + TA DI + ES +++ I + G RS + + EL K
Sbjct: 287 LPDTHVNALTRDHKTAYDIAEALPLCEESSEIKDILSQHGALRSRELNQPRDELRK 342
>UniRef100_Q6Z750 Ankyrin-like protein [Oryza sativa]
Length = 526
Score = 100 bits (248), Expect = 1e-19
Identities = 91/393 (23%), Positives = 173/393 (43%), Gaps = 62/393 (15%)
Query: 27 ETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYGNSVLHLAVSGGRHKMTDFLINKTK 86
+T H A R G + + L++ G ++ +DR G + LH+AV G + + L+
Sbjct: 152 KTSLHTAARIGYHRIVKALIERDPG--IVPIRDRKGQTALHMAVKGKNTDVVEELLMADV 209
Query: 87 LDINTRNSEGMTALDI--------LDQAMDSVESRQLQAIFIRAGGKRSIQSSSFSLELD 138
++ R+ + TAL I + Q + S E+ ++ AI
Sbjct: 210 SILDVRDKKANTALHIATRKWRPQMVQLLLSYEALEVNAI-------------------- 249
Query: 139 KNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSDSRSPQPQASERFENGT 198
NN A L+ Y ++ME++ + + V K ++ + S+
Sbjct: 250 -NNQNETAMDLAEKVPYGESKMEIIEWLTEAGAKNARNVGKIDEASELRRTVSDI----- 303
Query: 199 YNPYYVSPTNLVKQKHHHNKGKIENVNHTKR-----KHYHEMHQEALLNARNTIVLVAVL 253
KH+ EN KR K ++H+EA+ N N++ +VA L
Sbjct: 304 --------------KHNVQAQLNENAKTNKRVTGIAKELRKLHREAVQNTINSVTMVATL 349
Query: 254 IATVTFAAGISPPGGVYQE---GPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVS 310
IA++ F A + PG Y + G G++ + + F+VF + N ALF SL+VV+V ++
Sbjct: 350 IASIAFVAIFNLPGQYYVDRDSGGDIGEAHIANLTGFRVFCLLNATALFISLAVVVVQIT 409
Query: 311 IVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVIL-PHNQGMQWLSVLLLALGGGS 369
+V + Q ++ I +K+MW A +++ +V++ P N W++ + A+GG
Sbjct: 410 LVAWETGAQKRVIKIVNKLMWSACLSTCAAFISLAYVVVGPQN---AWMAFTISAIGGPI 466
Query: 370 LGTIFIGLSVMLVEHSLRKSKWRKKRKEIGDGA 402
+ + L+ +L+ + + R++R + G+
Sbjct: 467 MIGTLLFLAYLLLRPRFKFGEDRQRRIKRASGS 499
>UniRef100_Q6ZJG2 Ankyrin-like protein [Oryza sativa]
Length = 528
Score = 98.2 bits (243), Expect = 4e-19
Identities = 94/431 (21%), Positives = 185/431 (42%), Gaps = 55/431 (12%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G I++ + A +TV H A R G + + L+ G L D+ G
Sbjct: 136 GHTEIVNLLLESDANLARIARNNGKTVLHSAARLGHVEIVRSLLSRDPGIGLR--TDKKG 193
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIRA 122
+ LH+A G ++ L+ I+ +++G L + + + V + L ++
Sbjct: 194 QTALHMASKGQNAEIVIELLKPDISVIHLEDNKGNRPLHVATRKANIVIVQTLLSV---- 249
Query: 123 GGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSSD 182
I+ ++ ++++ + A + + N + + + + + PP
Sbjct: 250 ---EGIEVNA----VNRSGHTALAIAEQLNNEELVNILREAGGVTAKEQVHPP------- 295
Query: 183 SRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTK------RKHYHEMH 236
+P Q + V H + +I+ TK +K ++H
Sbjct: 296 --NPAKQLKQT----------------VSDIRHDVQSQIKQTKQTKMQVQKIKKRLEKLH 337
Query: 237 QEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE------GPMRGKSMVGRTSAFKVF 290
L NA N+ +VAVLIATV FAA + PG ++ G G++ V AF VF
Sbjct: 338 IGGLNNAINSNTVVAVLIATVAFAAIFTVPGNFVEDITQAPPGMSLGQAYVASNPAFLVF 397
Query: 291 AISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATWVILP 350
+ + +ALF SL+VV+V S++ +K + ++ + +K+MW+A F++ ++A T+V++
Sbjct: 398 LVFDALALFISLAVVVVQTSLIVVEQKAKRRMVFVMNKLMWLACLFISVAFIALTYVVVG 457
Query: 351 HNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRK---KRKEIGDGAAESDK 407
+ WL+ +A+G + T + ++ H + + K RK + +SD
Sbjct: 458 RDD--WWLAWCTMAIGAVIMLTTLGSMCYCIIAHRMDERKIRKASTSQSRSWSQTVDSDP 515
Query: 408 ESENSDFQSSY 418
+ NS+++ Y
Sbjct: 516 DLLNSEYKKMY 526
>UniRef100_Q8H8W8 Hypothetical protein [Oryza sativa]
Length = 284
Score = 97.4 bits (241), Expect = 7e-19
Identities = 68/255 (26%), Positives = 127/255 (49%), Gaps = 33/255 (12%)
Query: 170 DCISPPPVSKSSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTKR 229
DC+S +++D P+ + + + T + K H ++E T +
Sbjct: 48 DCLSRAGAVRANDLNQPRDELRK------------TVTEIKKDVHT----QLEQARKTNK 91
Query: 230 ------KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQEGPMRGKSMVGR 283
K ++H+E + NA N++ +VAVL ATV FAA + PGG G ++
Sbjct: 92 NVSGIAKELRKLHREGINNATNSVTVVAVLFATVAFAAIFTVPGG----NDNNGVAIAVH 147
Query: 284 TSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVA 343
+FK+F I N IALFTSL+VV+V +++V K + ++ I +K+MW+A +++
Sbjct: 148 AVSFKIFFIFNAIALFTSLAVVVVQITLVRGETKAERRVVEIINKLMWLASVCTTVAFIS 207
Query: 344 ATWVILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRKEI-GDGA 402
+ ++++ + QW ++L+ +G G I G+ + + +R + R RK++
Sbjct: 208 SAYIVV--GKHFQWAALLVTLIG----GVIMAGVLGTMTYYVVRSKRTRSIRKKVKSTRR 261
Query: 403 AESDKESENSDFQSS 417
+ S+ +NS+F S
Sbjct: 262 SGSNSWQQNSEFSDS 276
>UniRef100_Q6AWW5 At5g02620 [Arabidopsis thaliana]
Length = 524
Score = 97.1 bits (240), Expect = 9e-19
Identities = 100/408 (24%), Positives = 177/408 (42%), Gaps = 57/408 (13%)
Query: 2 NGKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRY 61
NG + +LD + + + T H A G + + FL+ G +L
Sbjct: 101 NGNLQVLDVLIEANPELSFTFDSSKTTALHTAASQGHGEIVCFLLD--KGVDLAAIARSN 158
Query: 62 GNSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQLQAIFIR 121
G + LH A G + LI K + + +G TAL + + ++ ++ + +
Sbjct: 159 GKTALHSAARNGHTVIVKKLIEKKAGMVTRVDKKGQTALHMAVKGQNT----EIVDVLME 214
Query: 122 AGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSKSS 181
A G I S+ +K N+P L + R N E++ ++ Y +S V+KS
Sbjct: 215 ADGSL-INSAD-----NKGNTP-----LHIAVR--KNRAEIVQTVLKYCEVSRVAVNKSG 261
Query: 182 DSRSPQPQASERFEN-------GTYNPYYVSPTNLVKQKHHHNKGK-------------I 221
++ + + E G N + P V+ K K +
Sbjct: 262 ETALDIAEKTGLHEIVPLLQKIGMQNARSIKPAEKVEPSGSSRKLKETVSEIGHEVHTQL 321
Query: 222 ENVNHTKR------KHYHEMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE--- 272
E T+R K ++MH E L NA N+ LVA+LIATV FAA + PG +
Sbjct: 322 EQTGRTRREIQGIAKRVNKMHTEGLNNAINSTTLVAILIATVAFAAIFNVPGQYTDDPKD 381
Query: 273 ---GPMRGKSMVGRTSAFKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKV 329
G G++ F +F + ++ ALF SL+VV+V S+V R+ + ++ I +K+
Sbjct: 382 VPPGYSLGEARAAPRPEFLIFVVFDSFALFISLAVVVVQTSVVVIERRAKKQMMAIINKL 441
Query: 330 MWVAVAFMATGYVAATWVILPHNQGMQWLSVLLLALGG----GSLGTI 373
MW+A ++ +V+ ++V++ + + L+V + A+G +LGT+
Sbjct: 442 MWMACIMISVAFVSLSFVVVGEKE--KPLAVGVTAIGALIMVSTLGTM 487
>UniRef100_Q69IU6 Ankyrin-like protein [Oryza sativa]
Length = 562
Score = 93.6 bits (231), Expect = 1e-17
Identities = 94/436 (21%), Positives = 185/436 (41%), Gaps = 63/436 (14%)
Query: 3 GKVSILDDFVSGSAASFHYLTREEETVFHLAVRYGCYDALVFLVQVSNGTNLLHCQDRYG 62
G + I++ + A+ +TV H A R G + + L+ G D+ G
Sbjct: 168 GHIDIVNLLLETDASLARIARNNGKTVLHSAARMGHVEVVTALLNKDPGIGFR--TDKKG 225
Query: 63 NSVLHLAVSGGRHKMTDFLINKTKLDINTRNSEGMTALDILDQAMDSVESRQL---QAIF 119
+ LH+A G ++ L+ I+ +++G AL + + ++V + L + I
Sbjct: 226 QTALHMASKGQNAEILLELLKPDLSVIHVEDNKGNRALHVATRKGNTVIVQTLISVKEIV 285
Query: 120 IRAGGKRSIQSSSFSLELDKNNSPSPAYRLSPSRRYIPNEMEVLTEMVSYDCISPPPVSK 179
I A + ++F++ N LS R + E + + ++PP +K
Sbjct: 286 INAVNRAG--ETAFAIAEKLGNE-----ELSNILREVGGE-------TAKEQVNPPNSAK 331
Query: 180 SSDSRSPQPQASERFENGTYNPYYVSPTNLVKQKHHHNKGKIENVNHTK------RKHYH 233
V H + I+ TK +K
Sbjct: 332 QLKKT-------------------------VSDIRHDVQSGIKQTRQTKMQFQKIKKRIQ 366
Query: 234 EMHQEALLNARNTIVLVAVLIATVTFAAGISPPGGVYQE-------GPMRGKSMVGRTSA 286
++H L NA N+ +VAVLIATV FAA + PG ++ G++ V A
Sbjct: 367 KLHIGGLNNAINSNTVVAVLIATVAFAAIFTIPGNFLEDMKDPHDPNMTLGQAFVASNPA 426
Query: 287 FKVFAISNNIALFTSLSVVIVLVSIVPFRRKPQTLLLIIAHKVMWVAVAFMATGYVAATW 346
F +F + + +ALF SL+VV+V S++ +K + ++ + +K+MW+A ++ ++A T+
Sbjct: 427 FIIFLVFDALALFISLAVVVVQTSLIVVEQKAKKKMVFVMNKLMWMACLCISAAFIALTY 486
Query: 347 VILPHNQGMQWLSVLLLALGGGSLGTIFIGLSVMLVEHSLRKSKWRKKRK----EIGDGA 402
V++ + +WL+ +A+G + + ++ H + + +K R+ + +
Sbjct: 487 VVVGRDD--RWLAWCTMAIGTAIMLATLGSMCYCIIAHRMEEKNMKKIRRSSTSQSWSIS 544
Query: 403 AESDKESENSDFQSSY 418
+SD E N++++ Y
Sbjct: 545 VDSDTELLNNEYKKIY 560
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.133 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 700,076,839
Number of Sequences: 2790947
Number of extensions: 28505214
Number of successful extensions: 94633
Number of sequences better than 10.0: 803
Number of HSP's better than 10.0 without gapping: 178
Number of HSP's successfully gapped in prelim test: 640
Number of HSP's that attempted gapping in prelim test: 92965
Number of HSP's gapped (non-prelim): 2079
length of query: 425
length of database: 848,049,833
effective HSP length: 130
effective length of query: 295
effective length of database: 485,226,723
effective search space: 143141883285
effective search space used: 143141883285
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC138130.16


