

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138087.2 + phase: 0 /pseudo
(296 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
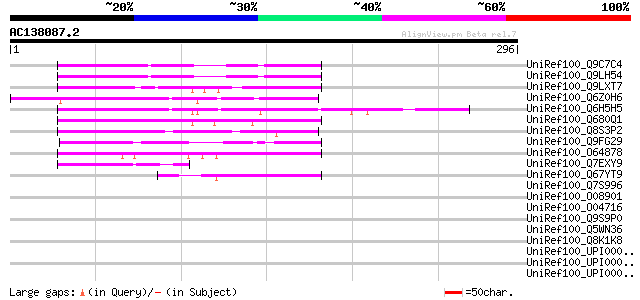
UniRef100_Q9C7C4 Hypothetical protein T21B14.4 [Arabidopsis thal... 136 8e-31
UniRef100_Q9LH54 Emb|CAB88266.1 [Arabidopsis thaliana] 136 8e-31
UniRef100_Q9LXT7 Hypothetical protein T24H18_190 [Arabidopsis th... 135 2e-30
UniRef100_Q6Z0H6 Hypothetical protein OSJNBa0049I01.19 [Oryza sa... 131 2e-29
UniRef100_Q6H5H5 Emsy N terminus domain-containing protein-like ... 129 9e-29
UniRef100_Q680Q1 MRNA, complete cds, clone: RAFL22-41-F10 [Arabi... 110 6e-23
UniRef100_Q8S3P2 Hypothetical protein 24K23.15 [Oryza sativa] 102 2e-20
UniRef100_Q9FG29 Emb|CAB88266.1 [Arabidopsis thaliana] 99 1e-19
UniRef100_O64878 Hypothetical protein At2g44440 [Arabidopsis tha... 78 2e-13
UniRef100_Q7EXY9 Hypothetical protein OSJNBa0016N23.123 [Oryza s... 49 2e-04
UniRef100_Q67YT9 MRNA, partial cds, clone: RAFL24-08-N13 [Arabid... 47 6e-04
UniRef100_Q7S996 Predicted protein [Neurospora crassa] 41 0.044
UniRef100_O08901 Mitotic checkpoint serine/threonine-protein kin... 37 0.48
UniRef100_O04716 DNA mismatch repair protein MSH6-1 [Arabidopsis... 37 0.63
UniRef100_Q9S9P0 T24D18.4 [Arabidopsis thaliana] 37 0.82
UniRef100_Q5WN36 Hypothetical protein CBG08165 [Caenorhabditis b... 37 0.82
UniRef100_Q8K1K8 Budding uninhibited by benzimidazoles 1 homolog... 36 1.1
UniRef100_UPI000021E7B7 UPI000021E7B7 UniRef100 entry 36 1.4
UniRef100_UPI000021DC77 UPI000021DC77 UniRef100 entry 36 1.4
UniRef100_UPI00001D16A1 UPI00001D16A1 UniRef100 entry 36 1.4
>UniRef100_Q9C7C4 Hypothetical protein T21B14.4 [Arabidopsis thaliana]
Length = 327
Score = 136 bits (342), Expect = 8e-31
Identities = 79/154 (51%), Positives = 92/154 (59%), Gaps = 22/154 (14%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL ELRVSD+EHRELL RVN D I R RDWR+ G Q T QP D+LPS
Sbjct: 32 ESLITELRKELRVSDDEHRELLSRVNKDDTIQRIRDWRQGGASQITRHATIQP-FDVLPS 90
Query: 89 PTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAE 148
PT SA+ KK KT S PS+ G TG R F+N SS ++
Sbjct: 91 PTFSAARKKQKTFPSYNPSI------------------GATGNRSFNNRLVSSGISG--- 129
Query: 149 EAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
+ LIG+KVWT+WPED+HFYEA+IT YN DE
Sbjct: 130 NESAEALIGRKVWTKWPEDNHFYEAIITQYNADE 163
>UniRef100_Q9LH54 Emb|CAB88266.1 [Arabidopsis thaliana]
Length = 383
Score = 136 bits (342), Expect = 8e-31
Identities = 79/154 (51%), Positives = 92/154 (59%), Gaps = 22/154 (14%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL ELRVSD+EHRELL RVN D I R RDWR+ G Q T QP D+LPS
Sbjct: 32 ESLITELRKELRVSDDEHRELLSRVNKDDTIQRIRDWRQGGASQITRHATIQP-FDVLPS 90
Query: 89 PTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAE 148
PT SA+ KK KT S PS+ G TG R F+N SS ++
Sbjct: 91 PTFSAARKKQKTFPSYNPSI------------------GATGNRSFNNRLVSSGISG--- 129
Query: 149 EAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
+ LIG+KVWT+WPED+HFYEA+IT YN DE
Sbjct: 130 NESAEALIGRKVWTKWPEDNHFYEAIITQYNADE 163
>UniRef100_Q9LXT7 Hypothetical protein T24H18_190 [Arabidopsis thaliana]
Length = 414
Score = 135 bits (339), Expect = 2e-30
Identities = 84/178 (47%), Positives = 102/178 (57%), Gaps = 33/178 (18%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL ELRVSDEEHRELL RVN+D +I R R+WR+ Q + Q VHD PS
Sbjct: 81 ESLITELRKELRVSDEEHRELLSRVNADEMIRRIREWRKANSLQS---SVPQLVHDA-PS 136
Query: 89 PTVSASNKKPKTSHSVP--------PSLGQSL-------------PGLSSVKP---VQCA 124
P VS S KK KTS S+ PSL S+ PG + KP +Q
Sbjct: 137 PAVSGSRKKQKTSQSIASLAMGPPSPSLHPSMQPSSSALRRGGPPPGPKTKKPKTSMQYP 196
Query: 125 STGPTGARHFSNHNSSSNLNAPAEEAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
STG G + + N P E +DPL+G+KVWT+WP+D+ +YEAVITDYNP E
Sbjct: 197 STGIAG-----RPQAGALTNEPGESGSYDPLVGRKVWTKWPDDNQYYEAVITDYNPVE 249
>UniRef100_Q6Z0H6 Hypothetical protein OSJNBa0049I01.19 [Oryza sativa]
Length = 458
Score = 131 bits (329), Expect = 2e-29
Identities = 91/223 (40%), Positives = 114/223 (50%), Gaps = 48/223 (21%)
Query: 1 MEVQSHQLGRCILCCLVCF*SSVRCSYL---ESLITELGIELRVSDEEHRELLGRVNSD* 57
ME Q HQL + C ++ + + E LITEL ELRVSD+EHRELL RVN D
Sbjct: 52 METQIHQLEQDAYCSVLRAFKAQSDAISWEKEGLITELRKELRVSDKEHRELLNRVNGDD 111
Query: 58 IIHRTRDWRET-GIYQPPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPPSL-------- 108
II R R+WRET G Q +Q HD +PSPT SA K+ KTS S+P +
Sbjct: 112 IIQRIREWRETKGGLQADMVNNAQRSHDRVPSPTTSA-RKRQKTSQSIPSASVPVPSPAV 170
Query: 109 -------------------------------GQSLPGLSSVKPVQCASTGPTGARHFSNH 137
GQ +PG S++K + +S GP+G N
Sbjct: 171 HSQTLTAPMQPLSSATKKVAPPGTKGKKTKPGQKIPGGSAIKTM--SSAGPSGRGPIMNK 228
Query: 138 NSSSNLNAPAEEAQFDPLIGKKVWTRWPEDSHFYEAVITDYNP 180
N S L P E +PLIG+KV TRWP+D+ FYEAVITDY+P
Sbjct: 229 NLSGGL--PTEPISVNPLIGRKVMTRWPDDNSFYEAVITDYDP 269
>UniRef100_Q6H5H5 Emsy N terminus domain-containing protein-like [Oryza sativa]
Length = 409
Score = 129 bits (324), Expect = 9e-29
Identities = 107/293 (36%), Positives = 136/293 (45%), Gaps = 62/293 (21%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
E LITEL ELRVSD +HRELL RVNSD II R+WR G Q +QP+HD+ PS
Sbjct: 32 EGLITELRKELRVSDVDHRELLNRVNSDDIIRSIREWRSAGGPQAMLPNNAQPMHDLAPS 91
Query: 89 PTVSASNKKPKTSHSVP-------------------PSL------------------GQS 111
PT S K+ KTS S P PS GQ
Sbjct: 92 PTTSG-RKRQKTSQSFPALPAPPPVMHSQQLALQGPPSSSTAKKGASSGAKGKKTKPGQK 150
Query: 112 LPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNA--PAEEAQFDPLIGKKVWTRWPEDSH 169
+PG SVK + +S GP+G N N L + P+E +PLI +KV +RWPED+
Sbjct: 151 VPGGPSVKAMT-SSAGPSGRGPHMNRNFPVGLVSFEPSEALHINPLINRKVMSRWPEDNS 209
Query: 170 FYEAVITDYNPDE*HVHILQG*YSLLEVS--------LG*YNIT*G------YTMGR*RS 215
FYEA ITDYNP E ++ L + S +G +I Y R
Sbjct: 210 FYEATITDYNP-ETDLYALAYDINTANESWEWVDLKQMGPEDIRWQGDDPGIYQGVRGAP 268
Query: 216 GHTT*RLTQRARPWS*EILWLGAGRGKGHPRFHSRKELMPPQDGIGNRVPNGI 268
G + + R P G GRG+G P+ SRK+ P Q+G+G R + I
Sbjct: 269 GSGGKKSSSRGGPTP------GTGRGRGLPKHVSRKDFPPSQNGVGKRSSDDI 315
>UniRef100_Q680Q1 MRNA, complete cds, clone: RAFL22-41-F10 [Arabidopsis thaliana]
Length = 429
Score = 110 bits (274), Expect = 6e-23
Identities = 71/192 (36%), Positives = 96/192 (49%), Gaps = 38/192 (19%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ES+ITEL EL +S+EEHRELLGRVNSD I R R+WR++G QP +Q VHD LPS
Sbjct: 88 ESVITELRKELSLSNEEHRELLGRVNSDDTIRRIREWRQSGGMQPSMRNAAQVVHDTLPS 147
Query: 89 PTVSASNKKPKTSHSVP------------PSLGQSLPGLSSV-----------KPVQCAS 125
P+VSAS K K + +P P + P SS K +
Sbjct: 148 PSVSASMKTHKPNQPIPSQPFASSSPSFHPQADPTHPFASSTAKRGPVPIVKGKKHKPVF 207
Query: 126 TGPTGARHFSNHNSS---------------SNLNAPAEEAQFDPLIGKKVWTRWPEDSHF 170
G + +H H S ++ + P + +G++V T+WPED+ F
Sbjct: 208 PGSSSTKHAPYHPSDQPPRGQVMNRLPSVPASSSEPTNGIDPESFLGRRVRTKWPEDNTF 267
Query: 171 YEAVITDYNPDE 182
Y+A+IT YNP E
Sbjct: 268 YDAIITQYNPVE 279
>UniRef100_Q8S3P2 Hypothetical protein 24K23.15 [Oryza sativa]
Length = 397
Score = 102 bits (253), Expect = 2e-20
Identities = 64/155 (41%), Positives = 81/155 (51%), Gaps = 11/155 (7%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL ELRVSD+EHRELL +VN D I R R++R+ G S+ HD P
Sbjct: 80 ESLITELRKELRVSDDEHRELLNKVNEDVAIRRMREFRQGGGSLSAQHRGSRIFHDTEPG 139
Query: 89 PTVSASNKKPKTSHSVPP-SLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPA 147
P K+ +T S+P S G P + S GP + N P
Sbjct: 140 PAA----KRQRTPLSIPSHSAGLQSPAMPSPSVPSSTKWGPFS----GTKGKKTRTNTPL 191
Query: 148 EEAQFDP--LIGKKVWTRWPEDSHFYEAVITDYNP 180
DP LI +K++TRWP+D++FYEA ITDYNP
Sbjct: 192 AVPSADPTSLINRKIYTRWPDDNNFYEATITDYNP 226
>UniRef100_Q9FG29 Emb|CAB88266.1 [Arabidopsis thaliana]
Length = 320
Score = 99.4 bits (246), Expect = 1e-19
Identities = 64/153 (41%), Positives = 84/153 (54%), Gaps = 30/153 (19%)
Query: 30 SLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPSP 89
+++T L ELR+SD+E+R+LL V++D +I R RD R G Q D+ PSP
Sbjct: 40 TVMTNLRKELRISDDENRQLLNNVHNDDLIKRIRDSRPRGGNQ----VVRHQSLDVHPSP 95
Query: 90 TVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAEE 149
T SAS KK KT S P S G T ++ F+N S+N PAE
Sbjct: 96 TFSASRKKQKTFQSYP-------------------SIGSTRSKSFNNRVVSAN--EPAEA 134
Query: 150 AQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
LIG+KVWT+WPED+ FYEAV+T YN +E
Sbjct: 135 -----LIGRKVWTKWPEDNSFYEAVVTQYNANE 162
>UniRef100_O64878 Hypothetical protein At2g44440 [Arabidopsis thaliana]
Length = 343
Score = 78.2 bits (191), Expect = 2e-13
Identities = 65/192 (33%), Positives = 84/192 (42%), Gaps = 38/192 (19%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRD----------------WRETGIY- 71
ES+ITEL EL +S+EEHRELLGRVNSD I R R WR Y
Sbjct: 62 ESVITELRKELSLSNEEHRELLGRVNSDDTIRRIRQDLKSEHLSDEQGMETIWRNAAKYA 121
Query: 72 ----------QPPGCTTSQPVHDILPSPTVSASNKKPKTSHS-----VPPSLGQS----L 112
+P SQP PS A P S + VP G+
Sbjct: 122 QCCSTSMKTHKPNQPIPSQPFASSSPSFHPQADPTHPFASSTAKRGPVPIVKGKKHKPVF 181
Query: 113 PGLSSVK--PVQCASTGPTGARHFSNHNSSSNLNAPAEEAQFDPLIGKKVWTRWPEDSHF 170
PG SS K P + P G + ++ + P + +G++V T+WPED+ F
Sbjct: 182 PGSSSTKHAPYHPSDQPPRGQVMNRLPSVPASSSEPTNGIDPESFLGRRVRTKWPEDNTF 241
Query: 171 YEAVITDYNPDE 182
Y+A+IT YNP E
Sbjct: 242 YDAIITQYNPVE 253
>UniRef100_Q7EXY9 Hypothetical protein OSJNBa0016N23.123 [Oryza sativa]
Length = 272
Score = 48.5 bits (114), Expect = 2e-04
Identities = 34/77 (44%), Positives = 43/77 (55%), Gaps = 8/77 (10%)
Query: 29 ESLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPS 88
ESLITEL EL++SD+EHR LL V + + R R R+TG Q S VH +P+
Sbjct: 74 ESLITELRKELQISDKEHRVLLKGVTEEEAVCRIRQSRQTGGTQ-SSSHHSSVVHTPVPA 132
Query: 89 PTVSASNKKPKTSHSVP 105
K+ K SHSVP
Sbjct: 133 -------KRQKKSHSVP 142
>UniRef100_Q67YT9 MRNA, partial cds, clone: RAFL24-08-N13 [Arabidopsis thaliana]
Length = 251
Score = 47.0 bits (110), Expect = 6e-04
Identities = 30/98 (30%), Positives = 43/98 (43%), Gaps = 14/98 (14%)
Query: 87 PSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVK--PVQCASTGPTGARHFSNHNSSSNLN 144
P P V KP PG SS K P + P G + ++ +
Sbjct: 16 PVPIVKGKKHKPV------------FPGSSSTKHAPYHPSDQPPRGQVMNRLPSVPASSS 63
Query: 145 APAEEAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
P + +G++V T+WPED+ FY+A+IT YNP E
Sbjct: 64 EPTNGIDPESFLGRRVRTKWPEDNTFYDAIITQYNPVE 101
>UniRef100_Q7S996 Predicted protein [Neurospora crassa]
Length = 152
Score = 40.8 bits (94), Expect = 0.044
Identities = 23/76 (30%), Positives = 38/76 (49%), Gaps = 1/76 (1%)
Query: 72 QPPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGA 131
QP + S PV L +S + T SVP ++ LPG++S+ +T T
Sbjct: 9 QPAAASPSLPVPTTLEPEPISRPSST-STQASVPQNMSHMLPGIASIATTTTTTTTTTVT 67
Query: 132 RHFSNHNSSSNLNAPA 147
+ +N+N+++N N PA
Sbjct: 68 TNNNNNNNNNNNNTPA 83
>UniRef100_O08901 Mitotic checkpoint serine/threonine-protein kinase BUB1 [Mus
musculus]
Length = 1058
Score = 37.4 bits (85), Expect = 0.48
Identities = 44/156 (28%), Positives = 65/156 (41%), Gaps = 11/156 (7%)
Query: 30 SLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPSP 89
S I + GI ++ E +ELL + +R R TGI+ P TTS+P+H
Sbjct: 119 SAIFQTGIH---NEAEPKELLQQQ------YRLFQARLTGIHLPAQATTSEPLHSAQILN 169
Query: 90 TVSASNKKP-KTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNAPAE 148
V +N P K S VP S G G++S + ++ + S S+ AP
Sbjct: 170 QVMMTNSSPEKNSACVPKSQGSECSGVASSTCDEKSNMEQRVIMISKSECSVSSSVAPKP 229
Query: 149 EAQFDPLIGKKVWTRWPEDSHFYEAVITDYNPDE*H 184
EAQ + K+ R + F E YN + H
Sbjct: 230 EAQ-QVMYCKEKLIRGDSEFSFEELRAQKYNQRKKH 264
>UniRef100_O04716 DNA mismatch repair protein MSH6-1 [Arabidopsis thaliana]
Length = 1324
Score = 37.0 bits (84), Expect = 0.63
Identities = 32/115 (27%), Positives = 45/115 (38%), Gaps = 19/115 (16%)
Query: 87 PSPTVSASNKK-----------PKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFS 135
PSP+ S SNKK P S S P + P SS P + S GP
Sbjct: 37 PSPSPSLSNKKTPKSNNPNPKSPSPSPSPPKKTPKLNPNPSSNLPARSPSPGPDTPSPVQ 96
Query: 136 NHNSSSNL-----NAPAEEAQF---DPLIGKKVWTRWPEDSHFYEAVITDYNPDE 182
+ L +P + D ++GK+V WP D +Y+ +T Y+ E
Sbjct: 97 SKFKKPLLVIGQTPSPPQSVVITYGDEVVGKQVRVYWPLDKKWYDGSVTFYDKGE 151
>UniRef100_Q9S9P0 T24D18.4 [Arabidopsis thaliana]
Length = 990
Score = 36.6 bits (83), Expect = 0.82
Identities = 28/93 (30%), Positives = 38/93 (40%), Gaps = 15/93 (16%)
Query: 86 LPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLNA 145
+ +P +S K K S + P+ + K Q + P R + SN N
Sbjct: 517 MDTPIPQSSKSKKKDSRATTPA---------TKKSEQAPKSHPKMKR-IAGEEVESNTNE 566
Query: 146 PAEEAQFDPLIGKKVWTRWPEDSHFYEAVITDY 178
EE L+GK+V WP D FYE VI Y
Sbjct: 567 LGEE-----LVGKRVNVWWPLDKKFYEGVIKSY 594
>UniRef100_Q5WN36 Hypothetical protein CBG08165 [Caenorhabditis briggsae]
Length = 672
Score = 36.6 bits (83), Expect = 0.82
Identities = 27/86 (31%), Positives = 42/86 (48%), Gaps = 8/86 (9%)
Query: 85 ILPSPTVSASNKKPKTSHSVPPSLGQSLPGLSSVKPVQCASTGPTGARHFSNHNSSSNLN 144
I P + +S P+ ++S+ SL + P L S + ST H S+ NS SN+N
Sbjct: 337 IKTEPEMESSASSPEMTNSMRESLSSTSPKLISGLDLGSFST------HSSSTNSMSNVN 390
Query: 145 APAEEA--QFDPLIGKKVWTRWPEDS 168
+ A + FDPL V +W E++
Sbjct: 391 SSAARSRLMFDPLTELPVLEKWFEEN 416
>UniRef100_Q8K1K8 Budding uninhibited by benzimidazoles 1 homolog [Mus musculus]
Length = 1059
Score = 36.2 bits (82), Expect = 1.1
Identities = 30/89 (33%), Positives = 42/89 (46%), Gaps = 10/89 (11%)
Query: 30 SLITELGIELRVSDEEHRELLGRVNSD*IIHRTRDWRETGIYQPPGCTTSQPVHDILPSP 89
S I + GI ++ E +ELL + +R R TGI+ P TTS+P+H
Sbjct: 119 SAIFQTGIH---NEAEPKELLQQQ------YRLFQARLTGIHLPAQATTSEPLHSAQILN 169
Query: 90 TVSASNKKP-KTSHSVPPSLGQSLPGLSS 117
V +N P K S VP S G G++S
Sbjct: 170 QVMMTNSSPEKNSACVPKSQGSECSGVAS 198
>UniRef100_UPI000021E7B7 UPI000021E7B7 UniRef100 entry
Length = 946
Score = 35.8 bits (81), Expect = 1.4
Identities = 26/87 (29%), Positives = 41/87 (46%), Gaps = 6/87 (6%)
Query: 49 LLGRVNSD*IIHRTRDWRETGIYQ--PPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPP 106
+L ++ S ++ + R+TG+ Q P G LP T SAS ++P+ + P
Sbjct: 493 MLEQLESQFVLMQQHQMRQTGVAQVRPTGIPNGPTADSSLP--TNSASGQQPQLPLTRMP 550
Query: 107 SLGQSLPGLSSVKPVQCASTGPTGARH 133
S+ Q PG+ P Q + GP A H
Sbjct: 551 SVSQ--PGVQPACPRQALANGPFSAGH 575
>UniRef100_UPI000021DC77 UPI000021DC77 UniRef100 entry
Length = 1279
Score = 35.8 bits (81), Expect = 1.4
Identities = 26/87 (29%), Positives = 41/87 (46%), Gaps = 6/87 (6%)
Query: 49 LLGRVNSD*IIHRTRDWRETGIYQ--PPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPP 106
+L ++ S ++ + R+TG+ Q P G LP T SAS ++P+ + P
Sbjct: 419 MLEQLESQFVLMQQHQMRQTGVAQVRPTGIPNGPTADSSLP--TNSASGQQPQLPLTRMP 476
Query: 107 SLGQSLPGLSSVKPVQCASTGPTGARH 133
S+ Q PG+ P Q + GP A H
Sbjct: 477 SVSQ--PGVQPACPRQALANGPFSAGH 501
>UniRef100_UPI00001D16A1 UPI00001D16A1 UniRef100 entry
Length = 1392
Score = 35.8 bits (81), Expect = 1.4
Identities = 26/87 (29%), Positives = 41/87 (46%), Gaps = 6/87 (6%)
Query: 49 LLGRVNSD*IIHRTRDWRETGIYQ--PPGCTTSQPVHDILPSPTVSASNKKPKTSHSVPP 106
+L ++ S ++ + R+TG+ Q P G LP T SAS ++P+ + P
Sbjct: 338 MLEQLESQFVLMQQHQMRQTGVAQVRPTGIPNGPTADSSLP--TNSASGQQPQLPLTRMP 395
Query: 107 SLGQSLPGLSSVKPVQCASTGPTGARH 133
S+ Q PG+ P Q + GP A H
Sbjct: 396 SVSQ--PGVQPACPRQALANGPFSAGH 420
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.335 0.147 0.499
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 492,813,069
Number of Sequences: 2790947
Number of extensions: 20589546
Number of successful extensions: 82103
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 52
Number of HSP's that attempted gapping in prelim test: 81899
Number of HSP's gapped (non-prelim): 178
length of query: 296
length of database: 848,049,833
effective HSP length: 126
effective length of query: 170
effective length of database: 496,390,511
effective search space: 84386386870
effective search space used: 84386386870
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.6 bits)
S2: 74 (33.1 bits)
Medicago: description of AC138087.2


