

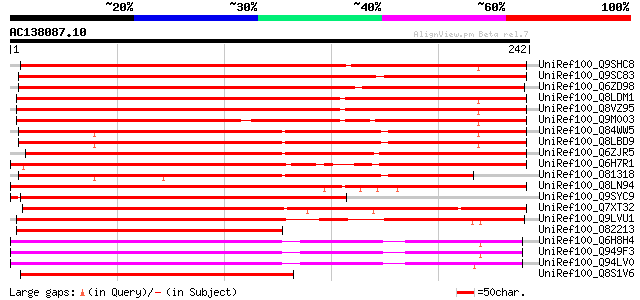
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138087.10 + phase: 0
(242 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SHC8 Putative VAMP-associated protein (Putative VAMP... 329 3e-89
UniRef100_Q9SC83 VAP27 [Nicotiana plumbaginifolia] 329 4e-89
UniRef100_Q6ZD98 Putative vesicle-associated membrane protein-as... 313 2e-84
UniRef100_Q8LDM1 Putative VAMP-associated protein [Arabidopsis t... 311 9e-84
UniRef100_Q8VZ95 Hypothetical protein At3g60600; T4C21_10 [Arabi... 310 3e-83
UniRef100_Q9M003 Hypothetical protein T4C21_10 [Arabidopsis thal... 297 1e-79
UniRef100_Q84WW5 Putative proline-rich protein [Arabidopsis thal... 280 2e-74
UniRef100_Q8LBD9 Putative proline-rich protein [Arabidopsis thal... 277 2e-73
UniRef100_Q6ZJR5 Putative 27k vesicle-associated membrane protei... 268 1e-70
UniRef100_Q6H7R1 Putative vesicle-associated membrane protein-as... 245 8e-64
UniRef100_O81318 F6N15.21 protein [Arabidopsis thaliana] 241 1e-62
UniRef100_Q8LN94 Putative vesicle-associated membrane protein [O... 218 1e-55
UniRef100_Q9SYC9 F11M15.13 protein [Arabidopsis thaliana] 209 5e-53
UniRef100_Q7XT32 OSJNBa0010H02.20 protein [Oryza sativa] 186 3e-46
UniRef100_Q9LVU1 VAMP (Vesicle-associated membrane protein)-asso... 184 2e-45
UniRef100_O82213 Hypothetical protein At2g23830 [Arabidopsis tha... 174 2e-42
UniRef100_Q6H8H4 Putative vesicle-associated membrane protein-as... 170 3e-41
UniRef100_Q949F3 Putative vesicle-associated membrane protein [O... 168 1e-40
UniRef100_Q94LV0 Putative membrane protein [Oryza sativa] 165 8e-40
UniRef100_Q8S1V6 P0504E02.2 protein [Oryza sativa] 159 8e-38
>UniRef100_Q9SHC8 Putative VAMP-associated protein (Putative VAMP (Vesicle-associated
membrane protein)-associated protein) [Arabidopsis
thaliana]
Length = 239
Score = 329 bits (844), Expect = 3e-89
Identities = 160/237 (67%), Positives = 205/237 (85%), Gaps = 3/237 (1%)
Query: 6 DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LL+I+P++L+FPFELKKQISCSL L NKTD+YVAFKVKTTNP+KYCVRPNTG+V PRS+
Sbjct: 4 ELLTIDPVDLQFPFELKKQISCSLYLGNKTDNYVAFKVKTTNPKKYCVRPNTGVVHPRSS 63
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVV 125
+V+VTMQAQKEAPAD+QCKDKFLLQ V + G +PKD++ EMF+KEAGH VEE KLRVV
Sbjct: 64 SEVLVTMQAQKEAPADLQCKDKFLLQCVVASPGATPKDVTHEMFSKEAGHRVEETKLRVV 123
Query: 126 YVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARALI 185
YV+PP+PPSPV EGSEEGSSPR S S+NGNA+ +F R SA+R +AQD S+EARAL+
Sbjct: 124 YVAPPRPPSPVREGSEEGSSPRASVSDNGNAS--DFTAAPRFSADRVDAQDNSSEARALV 181
Query: 186 SRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
++LTEEKN+A+Q +RL+QEL+ L+RE R++ GG+ F+ V+L+GL+GLI+GY+MK+
Sbjct: 182 TKLTEEKNSAVQLNNRLQQELDQLRRESKRSKSGGIPFMYVLLVGLIGLILGYIMKR 238
>UniRef100_Q9SC83 VAP27 [Nicotiana plumbaginifolia]
Length = 240
Score = 329 bits (843), Expect = 4e-89
Identities = 163/237 (68%), Positives = 197/237 (82%), Gaps = 3/237 (1%)
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRS 64
G+LL IEPLEL+FPFELKKQISCS+QL+NK+D+YVAFKVKTTNP+KYCVRPNTG+V+P S
Sbjct: 6 GELLQIEPLELQFPFELKKQISCSIQLTNKSDNYVAFKVKTTNPKKYCVRPNTGVVMPHS 65
Query: 65 TCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRV 124
DV+VTMQAQKEAPADMQCKDKFLLQSV + G + KDI+ EMFNKE G+ VE+CKLRV
Sbjct: 66 ASDVIVTMQAQKEAPADMQCKDKFLLQSVVASPGATAKDITPEMFNKEEGNHVEDCKLRV 125
Query: 125 VYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARAL 184
+YV P QPPSPV EGSEEGSSPR S SENG N EF ++R E+ QD S+E +AL
Sbjct: 126 IYVPPQQPPSPVQEGSEEGSSPRASVSENGAVNTSEFNNISRAYNEQ---QDSSSETKAL 182
Query: 185 ISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMKK 241
IS+LTEEK + IQQ ++L+QEL LL+RE N +RGG+ I V++IGLLG+I+GYL+KK
Sbjct: 183 ISKLTEEKISVIQQNNKLQQELALLRRERNSSRGGIPTIYVLIIGLLGIILGYLLKK 239
>UniRef100_Q6ZD98 Putative vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 243
Score = 313 bits (802), Expect = 2e-84
Identities = 155/236 (65%), Positives = 194/236 (81%), Gaps = 3/236 (1%)
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRS 64
G+LL I+P+EL+FPFELKKQISCS+QLSN +D Y+AFKVKTT+P+KY VRPNTG+VLPRS
Sbjct: 10 GELLRIDPVELRFPFELKKQISCSMQLSNLSDDYIAFKVKTTSPKKYSVRPNTGVVLPRS 69
Query: 65 TCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRV 124
TCDV+VTMQAQ+EAP DMQCKDKFL+QSV GV+ KDI+ EMF KE+G+ VEE KLRV
Sbjct: 70 TCDVVVTMQAQREAPPDMQCKDKFLVQSVIAPSGVTVKDITGEMFTKESGNKVEEVKLRV 129
Query: 125 VYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARAL 184
Y++PPQPPSPVPE SEEGS R S SENG++ G F TR ER E Q+ S EA AL
Sbjct: 130 TYIAPPQPPSPVPEESEEGSPSRVSESENGDSLGGGF---TRALRERIEPQENSLEAGAL 186
Query: 185 ISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMK 240
I++L EEKN+AIQQ ++RQEL++++RE ++ RGG SFIIVI++ L+G+ +GY+MK
Sbjct: 187 INKLNEEKNSAIQQNHKIRQELDMMRREISKKRGGFSFIIVIIVALIGIFLGYMMK 242
>UniRef100_Q8LDM1 Putative VAMP-associated protein [Arabidopsis thaliana]
Length = 240
Score = 311 bits (797), Expect = 9e-84
Identities = 154/239 (64%), Positives = 200/239 (83%), Gaps = 3/239 (1%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ +LL++EPL+L+FPFELKKQISCSL L+NKTD+ VAFKVKTTNP+KYCVRPNTG+VLPR
Sbjct: 3 NSELLTVEPLDLQFPFELKKQISCSLYLTNKTDNNVAFKVKTTNPKKYCVRPNTGVVLPR 62
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
STC+V+VTMQAQKEAP+DMQCKDKFLLQ V + GV+ K+++ EMF+KEAGH VEE KLR
Sbjct: 63 STCEVLVTMQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEMFSKEAGHRVEETKLR 122
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V YV+PPQPPSPV EGSEEGSSPR S S+NG +G EF+ + Q+ ++EARA
Sbjct: 123 VTYVAPPQPPSPVHEGSEEGSSPRASVSDNG--HGSEFSFERFIVDNKAGHQENTSEARA 180
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
LI++LTEEK +AIQ +RL++EL+ L+RE +++ GG+ F+ V+L+GL+GLI+GY+MK+
Sbjct: 181 LITKLTEEKQSAIQLNNRLQRELDQLRRESKKSQSGGIPFMYVLLVGLIGLILGYIMKR 239
>UniRef100_Q8VZ95 Hypothetical protein At3g60600; T4C21_10 [Arabidopsis thaliana]
Length = 256
Score = 310 bits (793), Expect = 3e-83
Identities = 153/239 (64%), Positives = 200/239 (83%), Gaps = 3/239 (1%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ +LL++EPL+L+FPFELKKQISCSL L+NKTD+ VAFKVKTTNP+KYCVRPNTG+VLPR
Sbjct: 19 NSELLTVEPLDLQFPFELKKQISCSLYLTNKTDNNVAFKVKTTNPKKYCVRPNTGVVLPR 78
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
STC+V+VTMQAQKEAP+DMQCKDKFLLQ V + GV+ K+++ EMF+KEAGH VEE KLR
Sbjct: 79 STCEVLVTMQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEMFSKEAGHRVEETKLR 138
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V YV+PP+PPSPV EGSEEGSSPR S S+NG +G EF+ + Q+ ++EARA
Sbjct: 139 VTYVAPPRPPSPVHEGSEEGSSPRASVSDNG--HGSEFSFERFIVDNKAGHQENTSEARA 196
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
LI++LTEEK +AIQ +RL++EL+ L+RE +++ GG+ F+ V+L+GL+GLI+GY+MK+
Sbjct: 197 LITKLTEEKQSAIQLNNRLQRELDQLRRESKKSQSGGIPFMYVLLVGLIGLILGYIMKR 255
>UniRef100_Q9M003 Hypothetical protein T4C21_10 [Arabidopsis thaliana]
Length = 250
Score = 297 bits (761), Expect = 1e-79
Identities = 150/239 (62%), Positives = 198/239 (82%), Gaps = 9/239 (3%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ +LL++EPL+L+FPFELKKQISCSL L+NKTD+ VAFKVKTTNP+KYCVRPNTG+VLPR
Sbjct: 19 NSELLTVEPLDLQFPFELKKQISCSLYLTNKTDNNVAFKVKTTNPKKYCVRPNTGVVLPR 78
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
STC+V+VTMQAQKEAP+DMQCKDKFLLQ V + GV+ K+++ EM AGH VEE KLR
Sbjct: 79 STCEVLVTMQAQKEAPSDMQCKDKFLLQGVIASPGVTAKEVTPEM----AGHRVEETKLR 134
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V YV+PP+PPSPV EGSEEGSSPR S S+NG +G EF++ + + Q+ ++EARA
Sbjct: 135 VTYVAPPRPPSPVHEGSEEGSSPRASVSDNG--HGSEFSRFIVDN--KAGHQENTSEARA 190
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
LI++LTEEK +AIQ +RL++EL+ L+RE +++ GG+ F+ V+L+GL+GLI+GY+MK+
Sbjct: 191 LITKLTEEKQSAIQLNNRLQRELDQLRRESKKSQSGGIPFMYVLLVGLIGLILGYIMKR 249
>UniRef100_Q84WW5 Putative proline-rich protein [Arabidopsis thaliana]
Length = 239
Score = 280 bits (717), Expect = 2e-74
Identities = 143/239 (59%), Positives = 188/239 (77%), Gaps = 6/239 (2%)
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSY-VAFKVKTTNPRKYCVRPNTGIVLPR 63
GDL++I P ELKFPFELKKQ SCS+QL+NKT + VAFKVKTTNPRKYCVRPNTG+VLP
Sbjct: 4 GDLVNIHPTELKFPFELKKQSSCSMQLTNKTTTQCVAFKVKTTNPRKYCVRPNTGVVLPG 63
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
+C+V VTMQAQKEAP DMQCKDKFL+Q+V +DG + K++ AEMFNKEAG V+E+ KLR
Sbjct: 64 DSCNVTVTMQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLR 123
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
VVY+ P PPSPVPEGSEEG+SP S ++ + + F V+R E E KS+EA +
Sbjct: 124 VVYI-PANPPSPVPEGSEEGNSPMASLNDIASQSASLFDDVSRTFEETSE---KSSEAWS 179
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
+IS+LTEEK +A QQ+ +LR ELE+L++E ++ + GG S ++++L+GLLG ++GYL+ +
Sbjct: 180 MISKLTEEKTSATQQSQKLRLELEMLRKETSKKQSGGHSLLLMLLVGLLGCVIGYLLNR 238
>UniRef100_Q8LBD9 Putative proline-rich protein [Arabidopsis thaliana]
Length = 239
Score = 277 bits (708), Expect = 2e-73
Identities = 141/239 (58%), Positives = 187/239 (77%), Gaps = 6/239 (2%)
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSY-VAFKVKTTNPRKYCVRPNTGIVLPR 63
GDL++I P ELKFPFELKKQ SCS+QL+NKT + VAFKVKTTNPRKYCVRPNTG+VLP
Sbjct: 4 GDLVNIHPTELKFPFELKKQSSCSMQLTNKTTTQCVAFKVKTTNPRKYCVRPNTGVVLPG 63
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
+C+V VTMQAQKEAP DMQCKDKFL+Q+V +DG + K++ AEMFNKEAG V+E+ KLR
Sbjct: 64 DSCNVTVTMQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNKEAGRVIEDFKLR 123
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
VVY+ P PPSPVPEGSEEG+SP S ++ + + F V++ E E KS+ A +
Sbjct: 124 VVYI-PANPPSPVPEGSEEGNSPMASLNDIASQSASLFDDVSKTFEETSE---KSSXAWS 179
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGNRNR-GGVSFIIVILIGLLGLIMGYLMKK 241
+IS+LTEEK +A QQ+ +LR ELE+L++E ++ + GG S ++++L+GLLG ++GYL+ +
Sbjct: 180 MISKLTEEKTSATQQSQKLRLELEMLRKETSKKQSGGHSLLLMLLVGLLGCVIGYLLNR 238
>UniRef100_Q6ZJR5 Putative 27k vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 237
Score = 268 bits (684), Expect = 1e-70
Identities = 140/234 (59%), Positives = 172/234 (72%), Gaps = 3/234 (1%)
Query: 8 LSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCD 67
L I+P EL FPFEL KQ SCS+QL+NKTD YVAFKVKTTNP++YCVRPN G+VLP STCD
Sbjct: 6 LEIQPSELSFPFELLKQSSCSMQLTNKTDHYVAFKVKTTNPKQYCVRPNIGVVLPGSTCD 65
Query: 68 VMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYV 127
V VTMQAQ+EAP DMQCKDKFL+QSV +G + +DISAEMFNK AG VVEE KLRVVYV
Sbjct: 66 VTVTMQAQREAPPDMQCKDKFLVQSVAAENGATTQDISAEMFNKVAGKVVEEFKLRVVYV 125
Query: 128 SPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARALISR 187
P S +PE SE+GSS R ENG N V R SAE +++ E ++IS+
Sbjct: 126 -PTTTSSAMPEDSEQGSSARPFAQENGIHNSTMPQPVFRSSAE--PTKERPTEPSSMISK 182
Query: 188 LTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMKK 241
L EE AIQQ +LR ELELL++E +++ GG S + ++GLLG+I+GY++KK
Sbjct: 183 LNEENRVAIQQNQKLRHELELLRKESSKSSGGFSLTFLAIVGLLGIIVGYILKK 236
>UniRef100_Q6H7R1 Putative vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 259
Score = 245 bits (625), Expect = 8e-64
Identities = 131/242 (54%), Positives = 176/242 (72%), Gaps = 18/242 (7%)
Query: 1 MSPDG-DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGI 59
MS D +LL I+P EL FPFELKKQISCSL L+NKTD YV FKVKTT+P+KYCVRPN GI
Sbjct: 35 MSSDSRELLGIDPPELIFPFELKKQISCSLHLTNKTDEYVTFKVKTTSPKKYCVRPNNGI 94
Query: 60 VLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEE 119
V P+ST +V+VTMQAQ+EAP DMQCKDKFL+QS ++PKDI+ +MF KE+G+VV+E
Sbjct: 95 VAPQSTSNVLVTMQAQREAPPDMQCKDKFLVQSAIVTQELTPKDITGDMFTKESGNVVDE 154
Query: 120 CKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSA 179
KL+VVY P P+ + GSEEG GS S + + T+GS E++ ++
Sbjct: 155 VKLKVVYTQP--HPTSLNGGSEEG---LGSLS---------YQEATKGSR---ESETVTS 197
Query: 180 EARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLM 239
E ALIS+L EEK++AIQQ +LR+EL+LL+R+ GG S + V++I +LG+++G+L+
Sbjct: 198 EPLALISKLKEEKSSAIQQNMKLREELDLLRRQMGSQHGGFSLVFVLVIAILGILLGFLI 257
Query: 240 KK 241
K+
Sbjct: 258 KR 259
>UniRef100_O81318 F6N15.21 protein [Arabidopsis thaliana]
Length = 399
Score = 241 bits (615), Expect = 1e-62
Identities = 131/225 (58%), Positives = 162/225 (71%), Gaps = 17/225 (7%)
Query: 5 GDLLSIEPLELKFPFELKKQISCSLQLSNKTDSY-VAFKVKTTNPRKYCVRPNTGIVLPR 63
GDL++I P ELKFPFELKKQ SCS+QL+NKT + VAFKVKTTNPRKYCVRPNTG+VLP
Sbjct: 4 GDLVNIHPTELKFPFELKKQSSCSMQLTNKTTTQCVAFKVKTTNPRKYCVRPNTGVVLPG 63
Query: 64 STCDVMV------------TMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNK 111
+C+V V TMQAQKEAP DMQCKDKFL+Q+V +DG + K++ AEMFNK
Sbjct: 64 DSCNVTVNNTKDDCCFVSVTMQAQKEAPLDMQCKDKFLVQTVVVSDGTTSKEVLAEMFNK 123
Query: 112 EAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAER 171
EAG V+E+ KLRVVY+ P PPSPVPEGSEEG+SP S ++ + + F V+R E
Sbjct: 124 EAGRVIEDFKLRVVYI-PANPPSPVPEGSEEGNSPMASLNDIASQSASLFDDVSRTFEET 182
Query: 172 PEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRN 216
E KS+EA ++IS+LTEEK +A QQ+ +LR EL + RN
Sbjct: 183 SE---KSSEAWSMISKLTEEKTSATQQSQKLRLELVRVDPSALRN 224
>UniRef100_Q8LN94 Putative vesicle-associated membrane protein [Oryza sativa]
Length = 275
Score = 218 bits (555), Expect = 1e-55
Identities = 125/276 (45%), Positives = 177/276 (63%), Gaps = 36/276 (13%)
Query: 1 MSPDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIV 60
M+ DL+ ++P EL+FPFEL KQISC L+++NKT+ VAFKVKTT+P+KYCVRPN G+V
Sbjct: 1 MAASCDLVDVDPPELQFPFELDKQISCPLRIANKTERTVAFKVKTTSPKKYCVRPNNGVV 60
Query: 61 LPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEEC 120
PRS V+VTMQAQ AP D+QCKDKFL+QSV +DG+S KDI+++MF ++ ++VEE
Sbjct: 61 RPRSASVVVVTMQAQIVAPPDLQCKDKFLVQSVVVDDGLSAKDITSQMFLRDENNMVEEV 120
Query: 121 KLRVVYVSPPQPPSPVPEGSEEGSS---PRGSFSENGNANGPEFA--QVTRGSAE----- 170
KL+V YV PP+P + E S+ P +NG +G E + V+ SAE
Sbjct: 121 KLKVSYVMPPEPAMEIAEESDIPKRILVPMQRILDNGR-SGSELSSGNVSLRSAEMGTEL 179
Query: 171 --------RPEAQDKSA-----------------EARALISRLTEEKNNAIQQTSRLRQE 205
R E K+A E AL+++LTEEK +A++Q +LR+E
Sbjct: 180 GSPLGRFVRNEDMLKTASPVVETRVHAGPDEQYLELSALVAKLTEEKKSALEQNRKLREE 239
Query: 206 LELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMKK 241
LEL +R+ ++++GG S V++IGLL +I+G L+KK
Sbjct: 240 LELARRQASQHQGGFSLAFVLVIGLLSIILGCLVKK 275
>UniRef100_Q9SYC9 F11M15.13 protein [Arabidopsis thaliana]
Length = 571
Score = 209 bits (532), Expect = 5e-53
Identities = 96/152 (63%), Positives = 125/152 (82%)
Query: 6 DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LL I+P++++FP EL K++SCSL L+NKT++YVAFK KTTN +KY VRPN G+VLPRS+
Sbjct: 175 ELLIIDPVDVQFPIELNKKVSCSLNLTNKTENYVAFKAKTTNAKKYYVRPNVGVVLPRSS 234
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVV 125
C+V+V MQA KEAPADMQC+DK L Q + KD+++EMF+KEAGH EE +L+V+
Sbjct: 235 CEVLVIMQALKEAPADMQCRDKLLFQCKVVEPETTAKDVTSEMFSKEAGHPAEETRLKVM 294
Query: 126 YVSPPQPPSPVPEGSEEGSSPRGSFSENGNAN 157
YV+PPQPPSPV EG+EEGSSPR S S+NGNA+
Sbjct: 295 YVTPPQPPSPVQEGTEEGSSPRASVSDNGNAS 326
Score = 193 bits (491), Expect = 3e-48
Identities = 91/157 (57%), Positives = 120/157 (75%), Gaps = 1/157 (0%)
Query: 1 MSPDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIV 60
MS D +LL+ + ++++FP EL KQ SCSL L+NKTD+YVAFK +TT P+ YCV+P+ G+V
Sbjct: 1 MSTD-ELLTFDHVDIRFPIELNKQGSCSLNLTNKTDNYVAFKAQTTKPKMYCVKPSVGVV 59
Query: 61 LPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEEC 120
LPRS+C+V+V MQA KEAPAD QCKDK L Q G K++++EMF+KEAGH VEE
Sbjct: 60 LPRSSCEVLVVMQALKEAPADRQCKDKLLFQCKVVEPGTMDKEVTSEMFSKEAGHRVEET 119
Query: 121 KLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNAN 157
+++YV+PPQP SPV EG E+GSSP S S+ GNA+
Sbjct: 120 IFKIIYVAPPQPQSPVQEGLEDGSSPSASVSDKGNAS 156
>UniRef100_Q7XT32 OSJNBa0010H02.20 protein [Oryza sativa]
Length = 244
Score = 186 bits (473), Expect = 3e-46
Identities = 106/239 (44%), Positives = 149/239 (61%), Gaps = 6/239 (2%)
Query: 7 LLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTC 66
LL + P ELK P+E K++ SC +QL+NKT+ YVAFKVKTTNPRKY VR GI+ PRS+C
Sbjct: 5 LLRVHPSELKIPYEYKRKRSCCMQLTNKTNQYVAFKVKTTNPRKYSVRHACGILPPRSSC 64
Query: 67 DVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVY 126
D+ VTMQA E +D CKDKFL+QSV G + +D E+F K G V+EE KLRVVY
Sbjct: 65 DITVTMQAPVEMLSDYHCKDKFLVQSVAVGYGATMRDFVPELFTKAPGRVIEEFKLRVVY 124
Query: 127 VSPPQPPSPVP---EGSEEGSSPRGSFSENGNANGPEFAQVTRGS-AERPEAQDKSAEAR 182
V+ PPSPVP E EE +SP+ +G F VT + +R + SAE
Sbjct: 125 VA-ANPPSPVPEEEEEEEEDASPQSEVMSHGVKMTSVFDAVTVSTLTDRSADKVSSAEGV 183
Query: 183 ALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLGLIMGYLMKK 241
++ S L E+ +++ +L+Q++ELL R ++ G S + V+L+ + + +G+ MK+
Sbjct: 184 SVESMLVAEREYPVEENQKLQQQMELL-RAARSSQQGFSAMFVLLVFMSSVCIGHFMKQ 241
>UniRef100_Q9LVU1 VAMP (Vesicle-associated membrane protein)-associated protein-like
[Arabidopsis thaliana]
Length = 220
Score = 184 bits (466), Expect = 2e-45
Identities = 101/239 (42%), Positives = 148/239 (61%), Gaps = 33/239 (13%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ L+SI+P ELKF FEL+KQ C L+++NKT++YVAFKVKTT+P+KY VRPNTG++ P
Sbjct: 6 ENQLISIQPDELKFLFELEKQSYCDLKVANKTENYVAFKVKTTSPKKYFVRPNTGVIQPW 65
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
+C + VT+QAQ+E P DMQCKDKFLLQS ++ + F K++G + ECKL+
Sbjct: 66 DSCIIRVTLQAQREYPPDMQCKDKFLLQSTIVPPHTDVDELPQDTFTKDSGKTLTECKLK 125
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V Y++P S SE+G NG +S+E +
Sbjct: 126 VSYITP---------------STTQRSSESGATNG----------------DGQSSETIS 154
Query: 184 LISRLTEEKNNAIQQTSRLRQELELLKREGN-RNRG-GVSFIIVILIGLLGLIMGYLMK 240
I RL EE++ A++QT +L+ ELE ++R N RN G G+S + ++GL+GLI+G+++K
Sbjct: 155 TIQRLKEERDAAVKQTQQLQHELETVRRRRNQRNSGNGLSLKLAAMVGLIGLIIGFILK 213
>UniRef100_O82213 Hypothetical protein At2g23830 [Arabidopsis thaliana]
Length = 149
Score = 174 bits (441), Expect = 2e-42
Identities = 82/124 (66%), Positives = 104/124 (83%)
Query: 4 DGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPR 63
+ +LL IEP+ L+FPFELKKQ+SCSL L+NKTD+ VAFKVKTTN YCVRPN G++LP+
Sbjct: 3 NNELLEIEPMYLQFPFELKKQMSCSLYLTNKTDNNVAFKVKTTNRNNYCVRPNYGLILPK 62
Query: 64 STCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLR 123
STC V+VTMQAQKE P+DMQ +KF++QSV + GV+ K+++ EMF+KE+GHVVEE KLR
Sbjct: 63 STCKVLVTMQAQKEVPSDMQSFEKFMIQSVLASPGVTAKEVTREMFSKESGHVVEETKLR 122
Query: 124 VVYV 127
V YV
Sbjct: 123 VTYV 126
>UniRef100_Q6H8H4 Putative vesicle-associated membrane protein-associated protein
[Oryza sativa]
Length = 233
Score = 170 bits (431), Expect = 3e-41
Identities = 95/240 (39%), Positives = 137/240 (56%), Gaps = 19/240 (7%)
Query: 1 MSPDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIV 60
M G L+SI P +L F FEL K C+L++ N ++ +VAFKVKTT+PRKY VRPN IV
Sbjct: 1 MGGGGTLISIYPEDLTFLFELDKPCYCNLKVVNNSEHHVAFKVKTTSPRKYFVRPNANIV 60
Query: 61 LPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEEC 120
P +C + +T+QAQKE P DMQCKDKFL+QS + +I + FNKE V+EE
Sbjct: 61 QPWDSCTITITLQAQKEYPQDMQCKDKFLIQSTRVAASTDMDEIPPDTFNKEVDKVIEEI 120
Query: 121 KLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAE 180
KL+VVY VP GS + S S + + +F + S E +
Sbjct: 121 KLKVVYT--------VPSGSSDDSGITSLGSRSFKSLSDDFTMLKNASIEEIQT------ 166
Query: 181 ARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRG-GVSFIIVILIGLLGLIMGYLM 239
I RL +E++N +QQ ++++EL++++R +R G S +GL+GL++G LM
Sbjct: 167 ----IQRLKDERDNMLQQNQQMQRELDVIRRRRSRKSDTGFSLTFAAFVGLIGLLVGLLM 222
>UniRef100_Q949F3 Putative vesicle-associated membrane protein [Oryza sativa]
Length = 233
Score = 168 bits (425), Expect = 1e-40
Identities = 94/240 (39%), Positives = 136/240 (56%), Gaps = 19/240 (7%)
Query: 1 MSPDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIV 60
M G L+SI P +L F FEL K C+L++ N ++ +VAFKVKTT+PRKY VRPN IV
Sbjct: 1 MGGGGTLISIYPEDLTFLFELDKPCYCNLKVVNNSEHHVAFKVKTTSPRKYFVRPNANIV 60
Query: 61 LPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEEC 120
P +C + +T+QAQK+ P DMQCKDKFL+QS + +I + FNKE V+EE
Sbjct: 61 QPWDSCTITITLQAQKDYPQDMQCKDKFLIQSTRVAASTDMDEIPPDTFNKEVDKVIEEI 120
Query: 121 KLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAE 180
KL+VVY VP GS + S S + + +F + S E +
Sbjct: 121 KLKVVYT--------VPSGSSDDSGITSLGSRSFKSLSDDFTMLKNASIEEIQT------ 166
Query: 181 ARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRG-GVSFIIVILIGLLGLIMGYLM 239
I RL +E++N +QQ ++++EL +++R +R G S +GL+GL++G LM
Sbjct: 167 ----IQRLKDERDNMLQQNQQMQRELVMIRRRRSRKSDTGFSLTFAAFVGLIGLLVGLLM 222
>UniRef100_Q94LV0 Putative membrane protein [Oryza sativa]
Length = 233
Score = 165 bits (418), Expect = 8e-40
Identities = 92/240 (38%), Positives = 134/240 (55%), Gaps = 19/240 (7%)
Query: 1 MSPDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIV 60
M G L+SI P +L F FEL K C+L++ N ++ +VAFKVKTT+PRKY VRPN I+
Sbjct: 1 MGASGRLISIYPEDLTFLFELDKPCYCNLKVVNNSEHHVAFKVKTTSPRKYFVRPNASII 60
Query: 61 LPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEEC 120
P +C + +T+QAQKE P DMQCKDKFL+QS K +I FNKE V+EE
Sbjct: 61 QPWDSCTITITLQAQKEYPPDMQCKDKFLIQSTKVAASTDMDEIPPNTFNKEVDKVIEEM 120
Query: 121 KLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAE 180
KL+VVY VP GS + S S + + + S E+ +
Sbjct: 121 KLKVVYT--------VPSGSSDDSGITSLGSRSFKLGSDDLTMLKNASIEKIQT------ 166
Query: 181 ARALISRLTEEKNNAIQQTSRLRQELELLKREGNR-NRGGVSFIIVILIGLLGLIMGYLM 239
I RL +E++ +QQ ++++EL++++R +R + G S GL+G+++G LM
Sbjct: 167 ----IQRLKDERDTTLQQNQQMQRELDVIRRRRSRKSDAGFSLTFAAFAGLIGVLIGLLM 222
>UniRef100_Q8S1V6 P0504E02.2 protein [Oryza sativa]
Length = 394
Score = 159 bits (401), Expect = 8e-38
Identities = 76/127 (59%), Positives = 95/127 (73%)
Query: 6 DLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
DL I P EL+F FE+KKQ SC++ L NK++ YVAFKVKTT+P++YCVRPNTG++LPRST
Sbjct: 4 DLAEIHPRELQFTFEVKKQSSCTVHLVNKSNEYVAFKVKTTSPKRYCVRPNTGVILPRST 63
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVV 125
CD VTMQAQ+ AP DMQ KDKFL+Q+ G S +D+ F+KE+G +EE KLRVV
Sbjct: 64 CDFTVTMQAQRTAPPDMQLKDKFLVQTTVVPYGTSDEDLVPSYFSKESGRYIEESKLRVV 123
Query: 126 YVSPPQP 132
VS P
Sbjct: 124 LVSASHP 130
Score = 45.8 bits (107), Expect = 0.001
Identities = 21/62 (33%), Positives = 38/62 (60%), Gaps = 1/62 (1%)
Query: 180 EARALISRLTEEKNNAIQQTSRLRQELELLKREGN-RNRGGVSFIIVILIGLLGLIMGYL 238
+A +I +L EE Q+ +L+QE+ L+++G RN+ G + V+ + LLG +GYL
Sbjct: 332 QAEKMIIKLREESRTTTQERDKLQQEMMFLRKKGTPRNQVGFPLLFVVYVALLGTSLGYL 391
Query: 239 MK 240
++
Sbjct: 392 LR 393
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 407,620,052
Number of Sequences: 2790947
Number of extensions: 17335392
Number of successful extensions: 67129
Number of sequences better than 10.0: 330
Number of HSP's better than 10.0 without gapping: 224
Number of HSP's successfully gapped in prelim test: 110
Number of HSP's that attempted gapping in prelim test: 66510
Number of HSP's gapped (non-prelim): 597
length of query: 242
length of database: 848,049,833
effective HSP length: 124
effective length of query: 118
effective length of database: 501,972,405
effective search space: 59232743790
effective search space used: 59232743790
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138087.10


