

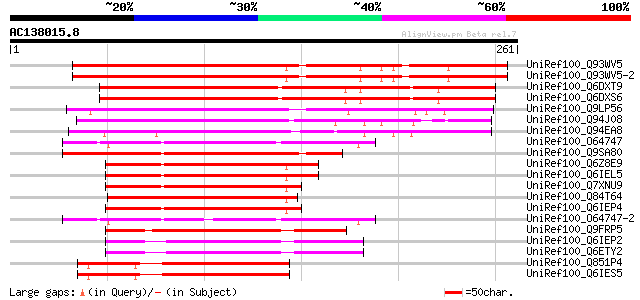
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138015.8 + phase: 0
(261 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q93WV5 Probable WRKY transcription factor 69 [Arabidop... 234 2e-60
UniRef100_Q93WV5-2 Splice isoform 2 of Q93WV5 [Arabidopsis thali... 229 4e-59
UniRef100_Q6DXT9 Putative WRKY transcription factor [Gossypium h... 215 1e-54
UniRef100_Q6DXS6 Putative WRKY transcription factor [Gossypium h... 213 5e-54
UniRef100_Q9LP56 Probable WRKY transcription factor 65 [Arabidop... 211 1e-53
UniRef100_Q94J08 P0481E12.40 protein [Oryza sativa] 177 2e-43
UniRef100_Q94EA8 P0435H01.23 protein [Oryza sativa] 161 1e-38
UniRef100_O64747 Probable WRKY transcription factor 35 [Arabidop... 149 9e-35
UniRef100_Q9SA80 Probable WRKY transcription factor 14 [Arabidop... 138 2e-31
UniRef100_Q6Z8E9 Putative WRKY DNA-binding protein [Oryza sativa] 134 2e-30
UniRef100_Q6IEL5 WRKY transcription factor 66 [Oryza sativa] 134 2e-30
UniRef100_Q7XNU9 OSJNBa0093F12.9 protein [Oryza sativa] 133 4e-30
UniRef100_Q84T64 Putative DNA-binding protein [Oryza sativa] 133 4e-30
UniRef100_Q6IEP4 WRKY transcription factor 37 [Oryza sativa] 133 4e-30
UniRef100_O64747-2 Splice isoform 2 of O64747 [Arabidopsis thali... 133 4e-30
UniRef100_Q9FRP5 Putative DNA-binding protein [Oryza sativa] 129 1e-28
UniRef100_Q6IEP2 WRKY transcription factor 39 [Oryza sativa] 125 8e-28
UniRef100_Q6ETY2 Putative WRKY transcription factor [Oryza sativa] 125 8e-28
UniRef100_Q851P4 Putative WRKY DNA-binding protein [Oryza sativa] 123 5e-27
UniRef100_Q6IES5 WRKY transcription factor 6 [Oryza sativa] 123 5e-27
>UniRef100_Q93WV5 Probable WRKY transcription factor 69 [Arabidopsis thaliana]
Length = 271
Score = 234 bits (597), Expect = 2e-60
Identities = 129/240 (53%), Positives = 158/240 (65%), Gaps = 22/240 (9%)
Query: 33 PASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKP 92
P SPSSCED+KI + +PKKRR ++KRVV++PIADVEGSKSRGE YPPSDSWAWRKYGQKP
Sbjct: 23 PESPSSCEDSKISKPTPKKRRNVEKRVVSVPIADVEGSKSRGEVYPPSDSWAWRKYGQKP 82
Query: 93 IKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPL----PKSHHSS 148
IKGSPYPRGYYRCSSSKGCPARKQVERSRVDP+ L++TYA +HNH P KSHH S
Sbjct: 83 IKGSPYPRGYYRCSSSKGCPARKQVERSRVDPSKLMITYACDHNHPFPSSSANTKSHHRS 142
Query: 149 TNAVTATVTAAVDTPSPVESPAATPQPEDRP---IFVTHPDFDLT--GDHHAV--FGWFD 201
+ TA + E T + P + ++H D L G + + FGWF
Sbjct: 143 S---VVLKTAKKEEEYEEEEEELTVTAAEEPPAGLDLSHVDSPLLLGGCYSEIGEFGWFY 199
Query: 202 DIVSTGVLVSPICGGVEDVTLTM-----REEDESLFADLGELPECSTVFRQRNIPSVFQY 256
D + S DVTL +EEDESLF DLG+LP+C++VFR+ + + Q+
Sbjct: 200 D---ASISSSSGSSNFLDVTLERGFSVGQEEDESLFGDLGDLPDCASVFRRGTVATEEQH 256
>UniRef100_Q93WV5-2 Splice isoform 2 of Q93WV5 [Arabidopsis thaliana]
Length = 272
Score = 229 bits (585), Expect = 4e-59
Identities = 129/241 (53%), Positives = 158/241 (65%), Gaps = 23/241 (9%)
Query: 33 PASPSSCEDTKIEESSPKK-RREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQK 91
P SPSSCED+KI + +PKK RR ++KRVV++PIADVEGSKSRGE YPPSDSWAWRKYGQK
Sbjct: 23 PESPSSCEDSKISKPTPKKSRRNVEKRVVSVPIADVEGSKSRGEVYPPSDSWAWRKYGQK 82
Query: 92 PIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPL----PKSHHS 147
PIKGSPYPRGYYRCSSSKGCPARKQVERSRVDP+ L++TYA +HNH P KSHH
Sbjct: 83 PIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPSKLMITYACDHNHPFPSSSANTKSHHR 142
Query: 148 STNAVTATVTAAVDTPSPVESPAATPQPEDRP---IFVTHPDFDLT--GDHHAV--FGWF 200
S+ TA + E T + P + ++H D L G + + FGWF
Sbjct: 143 SS---VVLKTAKKEEEYEEEEEELTVTAAEEPPAGLDLSHVDSPLLLGGCYSEIGEFGWF 199
Query: 201 DDIVSTGVLVSPICGGVEDVTLTM-----REEDESLFADLGELPECSTVFRQRNIPSVFQ 255
D + S DVTL +EEDESLF DLG+LP+C++VFR+ + + Q
Sbjct: 200 YD---ASISSSSGSSNFLDVTLERGFSVGQEEDESLFGDLGDLPDCASVFRRGTVATEEQ 256
Query: 256 Y 256
+
Sbjct: 257 H 257
>UniRef100_Q6DXT9 Putative WRKY transcription factor [Gossypium hirsutum]
Length = 272
Score = 215 bits (547), Expect = 1e-54
Identities = 123/226 (54%), Positives = 148/226 (65%), Gaps = 25/226 (11%)
Query: 47 SSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCS 106
++ K +R M+KRVV++PI DVEGS+ +GE PPSDSWAWRKYGQKPIKGSPYPRGYYRCS
Sbjct: 43 TTKKGKRSMQKRVVSVPIKDVEGSRLKGEGAPPSDSWAWRKYGQKPIKGSPYPRGYYRCS 102
Query: 107 SSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSPV 166
SSKGCPARKQVERSRV+PT L++TY+ EHNH+ P S H++T+A A A + SP
Sbjct: 103 SSKGCPARKQVERSRVNPTMLVITYSCEHNHA--WPASRHNNTSAKQAAAAAGEASESPT 160
Query: 167 ESPAA------TPQPEDRP--------IFVTHPDFDLTGDHHAVFGWFDDIVSTGVLVSP 212
+S A T QP+ P +T TGD A FG + ST VL S
Sbjct: 161 KSTTAVKHDPSTSQPDTEPDSGMEDGFACLTEDSILTTGDEFAWFGEMETTSST-VLESS 219
Query: 213 ICGGVED-------VTLTMREEDESLFADLGELPECSTVFR-QRNI 250
+ ++ V MREEDESLFADLGELPECS VFR QRN+
Sbjct: 220 LFSERDNSEADDTAVISPMREEDESLFADLGELPECSFVFRHQRNV 265
>UniRef100_Q6DXS6 Putative WRKY transcription factor [Gossypium hirsutum]
Length = 273
Score = 213 bits (541), Expect = 5e-54
Identities = 123/227 (54%), Positives = 150/227 (65%), Gaps = 26/227 (11%)
Query: 47 SSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCS 106
++ K +R M+KRVV++PI DVEGS+ +GE PPSDSWAWRKYGQKPIKGSPYPRGYYRCS
Sbjct: 43 TTKKGKRSMQKRVVSVPIKDVEGSRLKGEGAPPSDSWAWRKYGQKPIKGSPYPRGYYRCS 102
Query: 107 SSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTATVTAAVD-TPSP 165
SSKGCPARKQVERSRV+PT L++TY+ EHNH+ P S H++T+A A AA + + SP
Sbjct: 103 SSKGCPARKQVERSRVNPTMLVITYSCEHNHA--WPASRHNNTSAKQAAAAAAGEASESP 160
Query: 166 VESPAA------TPQPEDRP--------IFVTHPDFDLTGDHHAVFGWFDDIVSTGVLVS 211
+S A T QP+ P +T TGD A FG + ST VL S
Sbjct: 161 TKSSTAVKHEPSTSQPDTEPDSGMEDGFACLTEDSILTTGDEFAWFGEMETTSST-VLES 219
Query: 212 PICGGVED-------VTLTMREEDESLFADLGELPECSTVFR-QRNI 250
P+ ++ + MREEDESLFADL ELPECS VFR QRN+
Sbjct: 220 PLFSERDNSEADDTAMIFPMREEDESLFADLEELPECSFVFRHQRNV 266
>UniRef100_Q9LP56 Probable WRKY transcription factor 65 [Arabidopsis thaliana]
Length = 259
Score = 211 bits (537), Expect = 1e-53
Identities = 120/239 (50%), Positives = 143/239 (59%), Gaps = 27/239 (11%)
Query: 30 EDGPASPSSCE----DTKIEESSPKK-RREMKKRVVTIPIADVEGSKSRGETYPPSDSWA 84
E GP SP+S I SPK+ RR ++KRVV +P+ ++EGS+ +G+T PPSDSWA
Sbjct: 19 ETGPESPNSSTFNGMKALISSHSPKRSRRSVEKRVVNVPMKEMEGSRHKGDTTPPSDSWA 78
Query: 85 WRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKS 144
WRKYGQKPIKGSPYPRGYYRCSS+KGCPARKQVERSR DPT +L+TY EHNH PL S
Sbjct: 79 WRKYGQKPIKGSPYPRGYYRCSSTKGCPARKQVERSRDDPTMILITYTSEHNHPWPLTSS 138
Query: 145 HHSSTNAVTATVTAAVDTPSPVESPAATP--QPEDRPIFVTHPDFDLTGDHHAVFGWFDD 202
T P P P P + ED V + T F WF +
Sbjct: 139 --------TRNGPKPKPEPKPEPEPEVEPEAEEEDNKFMVLGRGIETTPSCVDEFAWFTE 190
Query: 203 IVSTG--VLVSPI--------CGGVEDVTL--TMREEDESLFADLGELPECSTVFRQRN 249
+ +T +L SPI G +DV + M EEDESLFADLGELPECS VFR R+
Sbjct: 191 METTSSTILESPIFSSEKKTAVSGADDVAVFFPMGEEDESLFADLGELPECSVVFRHRS 249
>UniRef100_Q94J08 P0481E12.40 protein [Oryza sativa]
Length = 316
Score = 177 bits (450), Expect = 2e-43
Identities = 112/261 (42%), Positives = 140/261 (52%), Gaps = 55/261 (21%)
Query: 35 SPSSCEDTKIEESSPKKRREMKKRVVTIPIADV-EGSKSRGETYPPSDSWAWRKYGQKPI 93
SPS+ + + ++ + RR ++KRVV++PIA+ + K GE PPSDSWAWRKYGQKPI
Sbjct: 52 SPSASPSSPLPPAAKRSRRSVEKRVVSVPIAECGDRPKGAGEGPPPSDSWAWRKYGQKPI 111
Query: 94 KGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVT 153
KGSPYPRGYYRCSSSKGCPARKQVERSR DPT LLVTY+ EHNH P PKS SS +A
Sbjct: 112 KGSPYPRGYYRCSSSKGCPARKQVERSRADPTVLLVTYSFEHNHPWPQPKS--SSCHASK 169
Query: 154 ATVTAAVDTPSPV-------------------ESPAATPQPEDRPIF------------V 182
++ + P PV E P P+ E P+ V
Sbjct: 170 SSPRSTAPKPEPVADGQHPEPAENESSASAELEVPEPEPEQESEPVVKQEEEQKEEQKAV 229
Query: 183 THPDFDLT----------GDHHAVFGWFDDIVST-----GVLVSPICGGVEDVTLTMREE 227
P T D + FGW D T L+ P E+ + +
Sbjct: 230 VEPAAVTTTVAPAPAVEEEDENFDFGWIDQYHPTWHRSYAPLLPP-----EEWEREL-QG 283
Query: 228 DESLFADLGELPECSTVFRQR 248
D++LFA LGELPEC+ VF +R
Sbjct: 284 DDALFAGLGELPECAVVFGRR 304
>UniRef100_Q94EA8 P0435H01.23 protein [Oryza sativa]
Length = 319
Score = 161 bits (408), Expect = 1e-38
Identities = 107/267 (40%), Positives = 133/267 (49%), Gaps = 54/267 (20%)
Query: 31 DGPASPSSCEDTKIEES--------SPKKRREMKKRVVTIPIADVEGSKSRG--ETYPPS 80
D P SP S + S S + RR +KRVVT+P+ADV G + +G E P+
Sbjct: 35 DCPGSPVSPSPAAAQRSAAGAAASPSGRSRRSAQKRVVTVPLADVTGPRPKGVGEGNTPT 94
Query: 81 DSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLP 140
DSWAWRKYGQKPIKGSP+PR YYRCSSSKGCPARKQVERSR DP ++VTY+ EHNHS
Sbjct: 95 DSWAWRKYGQKPIKGSPFPRAYYRCSSSKGCPARKQVERSRNDPDTVIVTYSFEHNHSAT 154
Query: 141 LPKSHHSSTNAVTATVTAAVDTPSPVESPAATPQPEDRPIF--VTHPDFDLTGDHHAV-- 196
+P++ N A SP E P +PE+ + P G A+
Sbjct: 155 VPRAQ----NRQAAPQKPKAQACSPPE-PVVEVEPEETHQYGVTAGPATGGGGGAAAIEV 209
Query: 197 ---FGWFDDIVS--------------------------------TGVLVSPICGGVEDVT 221
F W D+VS T L+ G V +
Sbjct: 210 RDEFRWLYDVVSVPATSTSPSDIDAADEMQLYDQPMFFGGAVVGTAALLPDEFGDVGGLG 269
Query: 222 LTMREEDESLFADLGELPECSTVFRQR 248
E+E+LF LGELPEC+ VFR+R
Sbjct: 270 GEGLGEEEALFEGLGELPECAMVFRRR 296
>UniRef100_O64747 Probable WRKY transcription factor 35 [Arabidopsis thaliana]
Length = 427
Score = 149 bits (375), Expect = 9e-35
Identities = 84/182 (46%), Positives = 104/182 (56%), Gaps = 25/182 (13%)
Query: 28 ILEDGPASPSSCEDTKIEESSP-----KKRREMKKRVVTIPIADVEGSKSRGETYPPSDS 82
+L D + SSC +I SSP K+R+ K+VV IP S+S GE P SD
Sbjct: 160 LLVDNNNNTSSCSQVQIS-SSPRNLGIKRRKSQAKKVVCIPAPAAMNSRSSGEVVP-SDL 217
Query: 83 WAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLP 142
WAWRKYGQKPIKGSPYPRGYYRCSSSKGC ARKQVERSR DP L++TY EHNH P P
Sbjct: 218 WAWRKYGQKPIKGSPYPRGYYRCSSSKGCSARKQVERSRTDPNMLVITYTSEHNH--PWP 275
Query: 143 KSHHSSTNAVTATVTAAVDTPSPVESPAATPQPEDR----------------PIFVTHPD 186
++ + ++ +++++ S + AAT P R P THP
Sbjct: 276 TQRNALAGSTRSSSSSSLNPSSKSSTAAATTSPSSRVFQNNSSKDEPNNSNLPSSSTHPP 335
Query: 187 FD 188
FD
Sbjct: 336 FD 337
>UniRef100_Q9SA80 Probable WRKY transcription factor 14 [Arabidopsis thaliana]
Length = 430
Score = 138 bits (347), Expect = 2e-31
Identities = 70/144 (48%), Positives = 90/144 (61%), Gaps = 4/144 (2%)
Query: 28 ILEDGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRK 87
+L DG S + + K+R+ K+VV IP S+S GE P SD WAWRK
Sbjct: 166 LLVDGTTFSSQIQISSPRNLGLKRRKSQAKKVVCIPAPAAMNSRSSGEVVP-SDLWAWRK 224
Query: 88 YGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHS 147
YGQKPIKGSP+PRGYYRCSSSKGC ARKQVERSR DP L++TY EHNH P+ ++ +
Sbjct: 225 YGQKPIKGSPFPRGYYRCSSSKGCSARKQVERSRTDPNMLVITYTSEHNHPWPIQRNALA 284
Query: 148 STNAVTATVTAAVDTPSPVESPAA 171
+ T + T++ P+P + A
Sbjct: 285 GS---TRSSTSSSSNPNPSKPSTA 305
>UniRef100_Q6Z8E9 Putative WRKY DNA-binding protein [Oryza sativa]
Length = 506
Score = 134 bits (337), Expect = 2e-30
Identities = 64/118 (54%), Positives = 79/118 (66%), Gaps = 9/118 (7%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ ++VV IP G ++ GE P SD WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 210 KRRKNQARKVVCIPAPAAAGGRTSGEVVP-SDLWAWRKYGQKPIKGSPYPRGYYRCSSSK 268
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPL--------PKSHHSSTNAVTATVTAA 159
GC ARKQVERSR DP L++TY EHNH P +SHH+ ++ ++ + A
Sbjct: 269 GCSARKQVERSRTDPNMLVITYTSEHNHPWPTQRNALAGSTRSHHAKNSSSNSSSSGA 326
>UniRef100_Q6IEL5 WRKY transcription factor 66 [Oryza sativa]
Length = 503
Score = 134 bits (337), Expect = 2e-30
Identities = 64/118 (54%), Positives = 79/118 (66%), Gaps = 9/118 (7%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ ++VV IP G ++ GE P SD WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 203 KRRKNQARKVVCIPAPAAAGGRTSGEVVP-SDLWAWRKYGQKPIKGSPYPRGYYRCSSSK 261
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPL--------PKSHHSSTNAVTATVTAA 159
GC ARKQVERSR DP L++TY EHNH P +SHH+ ++ ++ + A
Sbjct: 262 GCSARKQVERSRTDPNMLVITYTSEHNHPWPTQRNALAGSTRSHHAKNSSSNSSSSGA 319
>UniRef100_Q7XNU9 OSJNBa0093F12.9 protein [Oryza sativa]
Length = 514
Score = 133 bits (335), Expect = 4e-30
Identities = 64/109 (58%), Positives = 73/109 (66%), Gaps = 9/109 (8%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ ++VV IP G + GE P SD WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 234 KRRKNQARKVVCIPAPTAAGGRPSGEVVP-SDLWAWRKYGQKPIKGSPYPRGYYRCSSSK 292
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPL--------PKSHHSSTN 150
GC ARKQVERSR DP L++TY EHNH P +SHHS +
Sbjct: 293 GCSARKQVERSRTDPNMLVITYTSEHNHPWPTQRNALAGSTRSHHSKNS 341
>UniRef100_Q84T64 Putative DNA-binding protein [Oryza sativa]
Length = 387
Score = 133 bits (335), Expect = 4e-30
Identities = 65/106 (61%), Positives = 73/106 (68%), Gaps = 8/106 (7%)
Query: 51 KRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKG 110
KR+ K+VV IP S+ G PSD WAWRKYGQKPIKGSPYPRGYYRCSSSKG
Sbjct: 170 KRKNEVKKVVCIPAPPATSSRGGGGEVIPSDLWAWRKYGQKPIKGSPYPRGYYRCSSSKG 229
Query: 111 CPARKQVERSRVDPTNLLVTYAHEHNHSLPL--------PKSHHSS 148
C ARKQVERSR DP L++TYA EHNH P+ +SHHS+
Sbjct: 230 CMARKQVERSRSDPNMLVITYAAEHNHPWPMQRNVLAGYARSHHST 275
>UniRef100_Q6IEP4 WRKY transcription factor 37 [Oryza sativa]
Length = 489
Score = 133 bits (335), Expect = 4e-30
Identities = 64/109 (58%), Positives = 73/109 (66%), Gaps = 9/109 (8%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ ++VV IP G + GE P SD WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 209 KRRKNQARKVVCIPAPTAAGGRPSGEVVP-SDLWAWRKYGQKPIKGSPYPRGYYRCSSSK 267
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPL--------PKSHHSSTN 150
GC ARKQVERSR DP L++TY EHNH P +SHHS +
Sbjct: 268 GCSARKQVERSRTDPNMLVITYTSEHNHPWPTQRNALAGSTRSHHSKNS 316
>UniRef100_O64747-2 Splice isoform 2 of O64747 [Arabidopsis thaliana]
Length = 357
Score = 133 bits (335), Expect = 4e-30
Identities = 80/182 (43%), Positives = 100/182 (53%), Gaps = 29/182 (15%)
Query: 28 ILEDGPASPSSCEDTKIEESSP-----KKRREMKKRVVTIPIADVEGSKSRGETYPPSDS 82
+L D + SSC +I SSP K+R+ K+VV IP S+S GE P SD
Sbjct: 94 LLVDNNNNTSSCSQVQIS-SSPRNLGIKRRKSQAKKVVCIPAPAAMNSRSSGEVVP-SDL 151
Query: 83 WAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLP 142
WAWRKYGQKPIKGSPYPR CSSSKGC ARKQVERSR DP L++TY EHNH P P
Sbjct: 152 WAWRKYGQKPIKGSPYPR----CSSSKGCSARKQVERSRTDPNMLVITYTSEHNH--PWP 205
Query: 143 KSHHSSTNAVTATVTAAVDTPSPVESPAATPQPEDR----------------PIFVTHPD 186
++ + ++ +++++ S + AAT P R P THP
Sbjct: 206 TQRNALAGSTRSSSSSSLNPSSKSSTAAATTSPSSRVFQNNSSKDEPNNSNLPSSSTHPP 265
Query: 187 FD 188
FD
Sbjct: 266 FD 267
>UniRef100_Q9FRP5 Putative DNA-binding protein [Oryza sativa]
Length = 299
Score = 129 bits (323), Expect = 1e-28
Identities = 66/124 (53%), Positives = 77/124 (61%), Gaps = 9/124 (7%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ K+VV IP G G PSD WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 125 KRRKSQTKKVVCIPAGASGGG---GGEVVPSDLWAWRKYGQKPIKGSPYPRGYYRCSSSK 181
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSPVESP 169
GC ARKQVERSR DPT L+VTY +HNH P + NA+ + + S +
Sbjct: 182 GCSARKQVERSRADPTMLVVTYTSDHNHPWP------THRNALAGSTRPSSSNSSNIRLQ 235
Query: 170 AATP 173
+TP
Sbjct: 236 DSTP 239
>UniRef100_Q6IEP2 WRKY transcription factor 39 [Oryza sativa]
Length = 361
Score = 125 bits (315), Expect = 8e-28
Identities = 65/133 (48%), Positives = 80/133 (59%), Gaps = 16/133 (12%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ +K+VV AD + D WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 155 KRRKNQQKKVVRHVPADGVSA----------DVWAWRKYGQKPIKGSPYPRGYYRCSSSK 204
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSPVESP 169
GCPARKQVERSR DP ++TY EHNHS P + N++ T + + S +
Sbjct: 205 GCPARKQVERSRSDPNTFILTYTGEHNHSAP------THRNSLAGTTRNKLPSSSSASAA 258
Query: 170 AATPQPEDRPIFV 182
+A PQP + V
Sbjct: 259 SAQPQPPPPSVVV 271
>UniRef100_Q6ETY2 Putative WRKY transcription factor [Oryza sativa]
Length = 361
Score = 125 bits (315), Expect = 8e-28
Identities = 65/133 (48%), Positives = 80/133 (59%), Gaps = 16/133 (12%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
K+R+ +K+VV AD + D WAWRKYGQKPIKGSPYPRGYYRCSSSK
Sbjct: 155 KRRKNQQKKVVRHVPADGVSA----------DVWAWRKYGQKPIKGSPYPRGYYRCSSSK 204
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSPVESP 169
GCPARKQVERSR DP ++TY EHNHS P + N++ T + + S +
Sbjct: 205 GCPARKQVERSRSDPNTFILTYTGEHNHSAP------THRNSLAGTTRNKLPSSSAASAA 258
Query: 170 AATPQPEDRPIFV 182
+A PQP + V
Sbjct: 259 SAQPQPPPPSVVV 271
>UniRef100_Q851P4 Putative WRKY DNA-binding protein [Oryza sativa]
Length = 215
Score = 123 bits (308), Expect = 5e-27
Identities = 64/121 (52%), Positives = 79/121 (64%), Gaps = 23/121 (19%)
Query: 36 PSSC-------EDTKIEESSPKKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSW 83
PSSC +D + PKK++ +K+VVT+P +AD+ PSD++
Sbjct: 95 PSSCARKATADDDAGGKCHCPKKKKPREKKVVTVPAISDKVADI-----------PSDNY 143
Query: 84 AWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPK 143
+WRKYGQKPIKGSP+PRGYYRCSS K CPARK VER R DP LLVTY +EHNH+ PL
Sbjct: 144 SWRKYGQKPIKGSPHPRGYYRCSSKKDCPARKHVERCRSDPAMLLVTYENEHNHAQPLDL 203
Query: 144 S 144
S
Sbjct: 204 S 204
>UniRef100_Q6IES5 WRKY transcription factor 6 [Oryza sativa]
Length = 380
Score = 123 bits (308), Expect = 5e-27
Identities = 64/121 (52%), Positives = 79/121 (64%), Gaps = 23/121 (19%)
Query: 36 PSSC-------EDTKIEESSPKKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSW 83
PSSC +D + PKK++ +K+VVT+P +AD+ PSD++
Sbjct: 260 PSSCARKATADDDAGGKCHCPKKKKPREKKVVTVPAISDKVADI-----------PSDNY 308
Query: 84 AWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPK 143
+WRKYGQKPIKGSP+PRGYYRCSS K CPARK VER R DP LLVTY +EHNH+ PL
Sbjct: 309 SWRKYGQKPIKGSPHPRGYYRCSSKKDCPARKHVERCRSDPAMLLVTYENEHNHAQPLDL 368
Query: 144 S 144
S
Sbjct: 369 S 369
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.313 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 493,003,007
Number of Sequences: 2790947
Number of extensions: 22729297
Number of successful extensions: 77652
Number of sequences better than 10.0: 539
Number of HSP's better than 10.0 without gapping: 384
Number of HSP's successfully gapped in prelim test: 163
Number of HSP's that attempted gapping in prelim test: 76536
Number of HSP's gapped (non-prelim): 946
length of query: 261
length of database: 848,049,833
effective HSP length: 125
effective length of query: 136
effective length of database: 499,181,458
effective search space: 67888678288
effective search space used: 67888678288
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 73 (32.7 bits)
Medicago: description of AC138015.8


