

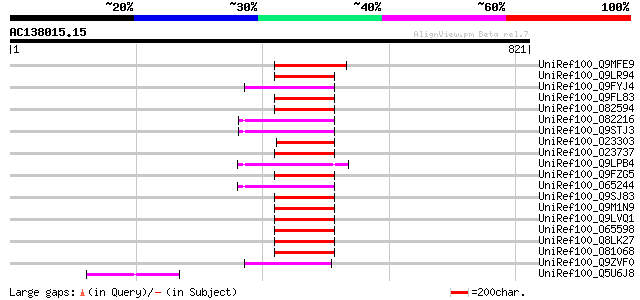
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138015.15 + phase: 0 /pseudo
(821 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9MFE9 Orf129a protein [Beta vulgaris subsp. vulgaris] 109 4e-22
UniRef100_Q9LR94 T23E23.16 [Arabidopsis thaliana] 97 2e-18
UniRef100_Q9FYJ4 F17F8.5 [Arabidopsis thaliana] 92 8e-17
UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like... 90 2e-16
UniRef100_O82594 F11O4.2 protein [Arabidopsis thaliana] 89 7e-16
UniRef100_O82216 Putative non-LTR retroelement reverse transcrip... 89 7e-16
UniRef100_Q9STJ3 Putative reverse transcriptase [Arabidopsis tha... 89 7e-16
UniRef100_O23303 Reverse transcriptase like protein [Arabidopsis... 85 8e-15
UniRef100_O23737 Reverse transcriptase [Beta lomatogona] 85 1e-14
UniRef100_Q9LPB4 T32E20.6 [Arabidopsis thaliana] 84 2e-14
UniRef100_Q9FZG5 T2E6.4 [Arabidopsis thaliana] 84 2e-14
UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana] 83 3e-14
UniRef100_Q9SJ83 Putative non-LTR retroelement reverse transcrip... 83 4e-14
UniRef100_Q9M1N9 Hypothetical protein T18B22.50 [Arabidopsis tha... 82 5e-14
UniRef100_Q9LVQ1 Non-LTR retroelement reverse transcriptase-like... 82 5e-14
UniRef100_O65598 Hypothetical protein M3E9.210 [Arabidopsis thal... 82 8e-14
UniRef100_Q8LK27 Putative AP endonuclease/reverse transcriptase ... 82 8e-14
UniRef100_O81068 Putative non-LTR retroelement reverse transcrip... 81 1e-13
UniRef100_Q9ZVF0 Putative non-LTR retroelement reverse transcrip... 80 2e-13
UniRef100_Q5U6J8 Orf177 protein [Beta vulgaris subsp. vulgaris] 80 2e-13
>UniRef100_Q9MFE9 Orf129a protein [Beta vulgaris subsp. vulgaris]
Length = 129
Score = 109 bits (272), Expect = 4e-22
Identities = 52/114 (45%), Positives = 77/114 (66%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ G+ I DNI+L+ EL+K YS+K +SPRCM+K+DL+KAY S+E F+ ++ E+GFP +
Sbjct: 16 FIPGKHIGDNILLATELIKGYSQKAVSPRCMIKVDLRKAYDSIEWSFLQCMLSELGFPSQ 75
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGDLSLHISM*YVWNILTDAF 532
F+ M C+TT SY+ +NG T PF +KKG+RQ + W+I DA+
Sbjct: 76 FITWIMECVTTVSYSILLNGKPTTPFPSKKGVRQVTPCHPSCLLLGWSICLDAY 129
>UniRef100_Q9LR94 T23E23.16 [Arabidopsis thaliana]
Length = 653
Score = 97.1 bits (240), Expect = 2e-18
Identities = 44/95 (46%), Positives = 68/95 (71%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y ++ +S RC +KID+ KA+ SV+ FI ++L M FP +
Sbjct: 92 FVKDRLLIENLLLATELVKDYHKESVSSRCAIKIDISKAFNSVQWSFIRNILLSMDFPME 151
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
FV+ M C++T S++ VNG+L F +K+GLRQG
Sbjct: 152 FVHWIMLCISTASFSVQVNGELVGFFQSKRGLRQG 186
>UniRef100_Q9FYJ4 F17F8.5 [Arabidopsis thaliana]
Length = 872
Score = 91.7 bits (226), Expect = 8e-17
Identities = 50/142 (35%), Positives = 78/142 (54%)
Query: 372 PKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKGRVIFDNIIL 431
PKKL ++R P+ ++ K K + + FVK R++ +N++L
Sbjct: 165 PKKLAAKEMRDYRPISCCNVLYKVISKIIANRLKLLLPRFIAENQSAFVKDRLLIENLLL 224
Query: 432 SHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN*AMACLTTTS 491
+ ELVK Y + IS RC +KID+ KA+ SV+ F+ ++ M F F++ C+TT S
Sbjct: 225 ATELVKDYHKDSISARCAIKIDISKAFDSVQWSFLTNTLVAMNFSPTFIHWINLCITTAS 284
Query: 492 YTYNVNGDLTRPFAAKKGLRQG 513
++ VNGDL F +K+GLRQG
Sbjct: 285 FSVQVNGDLVGYFQSKRGLRQG 306
>UniRef100_Q9FL83 Non-LTR retroelement reverse transcriptase-like protein
[Arabidopsis thaliana]
Length = 1223
Score = 90.1 bits (222), Expect = 2e-16
Identities = 42/95 (44%), Positives = 65/95 (68%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y + IS RC +KID+ KA+ SV+ PF+ + +GFP +
Sbjct: 565 FVKDRLLIENLLLATELVKDYHKDTISTRCAIKIDISKAFDSVQWPFLINVFTILGFPRE 624
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F++ C+TT S++ VNG+L F + +GLRQG
Sbjct: 625 FIHWINICITTASFSVQVNGELAGYFQSSRGLRQG 659
>UniRef100_O82594 F11O4.2 protein [Arabidopsis thaliana]
Length = 382
Score = 88.6 bits (218), Expect = 7e-16
Identities = 43/95 (45%), Positives = 65/95 (68%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y ++ IS R +KID+ KA+ SV+ F+ ++L M P +
Sbjct: 15 FVKDRLLIENLLLATELVKDYHKESISSRYAIKIDISKAFDSVQWSFLRNVLLAMDIPSE 74
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
FV+ M C+TT S++ VNG+L F +K GLRQG
Sbjct: 75 FVHWIMLCVTTASFSVQVNGELAGYFQSKCGLRQG 109
>UniRef100_O82216 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1216
Score = 88.6 bits (218), Expect = 7e-16
Identities = 50/151 (33%), Positives = 83/151 (54%), Gaps = 2/151 (1%)
Query: 363 STILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKG 422
STIL + PKK +++ P+ + K K +V FVK
Sbjct: 232 STILALI--PKKKEAREIKDYRPISCCNVLYKAISKILANRLKRILPKFIVGNQSAFVKD 289
Query: 423 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
R++ +N++L+ ELVK Y + IS RC +KID+ KA+ S++ F+ +++ M FP +F++
Sbjct: 290 RLLIENVLLATELVKDYHKDSISTRCAMKIDISKAFDSLQWSFLTHVLAAMNFPGEFIHW 349
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
C++T S++ VNG+L F + +GLRQG
Sbjct: 350 ISLCMSTASFSIQVNGELAGYFRSARGLRQG 380
>UniRef100_Q9STJ3 Putative reverse transcriptase [Arabidopsis thaliana]
Length = 662
Score = 88.6 bits (218), Expect = 7e-16
Identities = 50/151 (33%), Positives = 84/151 (55%), Gaps = 2/151 (1%)
Query: 363 STILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKG 422
STIL + PKK+ +++ P+ ++ K K + F+K
Sbjct: 29 STILALI--PKKMEAKEIKDYRPISCCNVLYKVISKIIANRLKRVLPQFIAGNQSAFIKD 86
Query: 423 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
R++ +N++L+ ELVK Y + +S RC +KID+ KA+ SV+ F+ ++L + FP +FV+
Sbjct: 87 RLLIENLLLATELVKDYHKDSVSERCAIKIDISKAFDSVQWSFLRNVLLTLDFPQEFVHW 146
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
M C+TT S++ VN +L F + +GLRQG
Sbjct: 147 IMLCVTTASFSVQVNRELAGYFNSLRGLRQG 177
>UniRef100_O23303 Reverse transcriptase like protein [Arabidopsis thaliana]
Length = 318
Score = 85.1 bits (209), Expect = 8e-15
Identities = 39/91 (42%), Positives = 62/91 (67%)
Query: 423 RVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN* 482
R++ +N++L+ ELVK Y + +S RC +KID+ KA+ SV+ F+ ++L + FP FV+
Sbjct: 5 RLLIENLLLATELVKDYHKDSVSERCAIKIDISKAFDSVQWTFLKNVLLTLDFPQVFVHW 64
Query: 483 AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
M C+TT S++ VNG+L F + +GLRQG
Sbjct: 65 IMLCVTTASFSVQVNGELAGYFNSSRGLRQG 95
>UniRef100_O23737 Reverse transcriptase [Beta lomatogona]
Length = 143
Score = 84.7 bits (208), Expect = 1e-14
Identities = 40/96 (41%), Positives = 59/96 (60%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FV R I N+++ L+K Y++ P C++K+DL+KAY ++E F+ +M E+GFP
Sbjct: 45 FVSRRSILHNVLICQGLIKMYNQGKAPPSCLMKLDLRKAYDTIEWAFVEEIMTELGFPSN 104
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
F M CL+TT Y+ +NG T K+GLRQGD
Sbjct: 105 FTQLKMICLSTTRYSILLNGSPTDLLQPKRGLRQGD 140
>UniRef100_Q9LPB4 T32E20.6 [Arabidopsis thaliana]
Length = 458
Score = 84.0 bits (206), Expect = 2e-14
Identities = 56/175 (32%), Positives = 95/175 (54%), Gaps = 4/175 (2%)
Query: 361 ILSTILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFV 420
I STIL + PKK ++++ P+ ++ K K +V FV
Sbjct: 250 INSTILALI--PKKATTNEMKDYRPISCCNVIFKVILKIIANPLKRVLPNFVVGNQSAFV 307
Query: 421 KGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFV 480
+ R++ +N++L+ ELVK Y ++ +S C +KID+ KA+ SV+ + ++ M FP F+
Sbjct: 308 QDRLLIENVLLATELVKDYHKESVSTGCAIKIDISKAFDSVQWSVLRKVLEAMKFPLFFI 367
Query: 481 N*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGDLSLHISM*YVWNILTDAFYSL 535
+ M C+TTTS++ VNG+L F + +GLRQ SL +S+ Y++ + D L
Sbjct: 368 HWIMLCVTTTSFSVQVNGELVGYFNSVRGLRQ-RCSLSLSL-YLFVVSMDVLSKL 420
>UniRef100_Q9FZG5 T2E6.4 [Arabidopsis thaliana]
Length = 740
Score = 84.0 bits (206), Expect = 2e-14
Identities = 39/95 (41%), Positives = 63/95 (66%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FV+ R++ +N++L+ ELVK Y + ISPRC +KID+ KA+ SV+ F+ + + FP
Sbjct: 138 FVRERLLIENVLLATELVKDYHKDSISPRCAMKIDISKAFDSVQWQFLLNTLEALNFPEN 197
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F + C++T +++ VNG+L F +K+GLRQG
Sbjct: 198 FCHWIKLCISTATFSVQVNGELAGFFGSKRGLRQG 232
>UniRef100_O65244 F21E10.5 protein [Arabidopsis thaliana]
Length = 928
Score = 83.2 bits (204), Expect = 3e-14
Identities = 50/153 (32%), Positives = 83/153 (53%), Gaps = 2/153 (1%)
Query: 361 ILSTILM*LCYPKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFV 420
I STIL + PKK +++ P+ ++ K K +V FV
Sbjct: 397 INSTILALI--PKKKEAKEMKDYRPISCCNVLYKVISKIIANRLKLVLPKFIVGNQSAFV 454
Query: 421 KGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFV 480
K R++ +N++L+ E+VK Y + +S RC +KID+ KA+ SV+ F+ ++ M FP +F
Sbjct: 455 KDRLLIENVLLATEIVKDYHKDSVSSRCALKIDISKAFDSVQWKFLINVLEAMNFPPEFT 514
Query: 481 N*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
+ C+TT S++ VNG+L F++ + LRQG
Sbjct: 515 HWITLCITTASFSVQVNGELAGVFSSARELRQG 547
>UniRef100_Q9SJ83 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 631
Score = 82.8 bits (203), Expect = 4e-14
Identities = 40/95 (42%), Positives = 62/95 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y + ISPRC +KID+ KA+ SV+ F+ + + FP K
Sbjct: 135 FVKERLLMENVLLATELVKDYHKDSISPRCAMKIDISKAFDSVQWQFLLNTLEALNFPEK 194
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
+ C++T +++ VNG+L F K+GLRQG
Sbjct: 195 IRHWIKLCISTATFSVQVNGELAGFFGNKRGLRQG 229
>UniRef100_Q9M1N9 Hypothetical protein T18B22.50 [Arabidopsis thaliana]
Length = 762
Score = 82.4 bits (202), Expect = 5e-14
Identities = 40/95 (42%), Positives = 61/95 (64%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ +LVK Y + IS RC +KID+ KA SV+ F+ + M FP
Sbjct: 118 FVKDRLLIENVLLATDLVKDYHKDSISERCAIKIDISKASDSVQWSFLINTLTAMHFPEM 177
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F++ C+TT S++ VNG+L F + +GLRQG
Sbjct: 178 FIHWIRLCITTPSFSVQVNGELAGFFQSSRGLRQG 212
>UniRef100_Q9LVQ1 Non-LTR retroelement reverse transcriptase-like [Arabidopsis
thaliana]
Length = 1072
Score = 82.4 bits (202), Expect = 5e-14
Identities = 40/96 (41%), Positives = 63/96 (64%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ GR + +N++L+ E+V Y+R ISPR M+K+DL+KA+ SV+ F+ + + P +
Sbjct: 419 FLPGRSLAENVLLATEMVHGYNRLNISPRGMLKVDLKKAFDSVKWEFVTAALRALAIPER 478
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
++N C+TT S+T +VNG F + KGLRQGD
Sbjct: 479 YINWIHQCITTPSFTISVNGATGGFFRSTKGLRQGD 514
Score = 41.2 bits (95), Expect = 0.13
Identities = 24/90 (26%), Positives = 45/90 (49%), Gaps = 7/90 (7%)
Query: 145 KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIEAEKVCLSSLKK 204
KLKA+KK +K+ + ++ G + ++A L+ Q ++P + + L + +K
Sbjct: 145 KLKAMKKPIKDFSRLNYSGIELRTKEAHELLITCQNLTLANPS----VSNAALELEAQRK 200
Query: 205 W---STIEEQIWI*KSRANWIQLGDSTQNF 231
W S EE + +SR +W GDS ++
Sbjct: 201 WVLLSCAEESFFHQRSRVSWFAEGDSNTHY 230
>UniRef100_O65598 Hypothetical protein M3E9.210 [Arabidopsis thaliana]
Length = 1141
Score = 81.6 bits (200), Expect = 8e-14
Identities = 41/96 (42%), Positives = 64/96 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
F+ GR++ +N++L+ E+V Y+ + IS R M+K+DL+KA+ SV FI +L +G P K
Sbjct: 537 FLPGRLLAENVLLATEMVHGYNWRNISLRGMLKVDLRKAFDSVRWEFIIAALLALGVPTK 596
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
F+N C++T ++T +VNG F + KGLRQGD
Sbjct: 597 FINWIHQCISTPTFTVSVNGCCGGFFKSAKGLRQGD 632
Score = 44.3 bits (103), Expect = 0.015
Identities = 35/128 (27%), Positives = 59/128 (45%), Gaps = 24/128 (18%)
Query: 137 NLLTNIWF--------------KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQL 182
NL+ ++W+ KLKALKK +K+ + ++ K+ E+A L+ Q L
Sbjct: 255 NLVWDVWYSTNVVGSSMFRVSKKLKALKKPIKDFSRLNYSNLEKRTEEAHETLLSFQ-NL 313
Query: 183 SSDPMNLDLIEAEKVCLSSLKKW---STIEEQIWI*KSRANWIQLGDSTQNFSMPLQRKE 239
+ D +L+ E L + +KW +T EE + +SR W GD + R
Sbjct: 314 TLDNPSLENAAHE---LEAQRKWQILATAEESFFRQRSRVTWFAEGDGNTRY---FHRMA 367
Query: 240 DVRISLSS 247
D R S+++
Sbjct: 368 DSRKSVNT 375
>UniRef100_Q8LK27 Putative AP endonuclease/reverse transcriptase [Brassica napus]
Length = 1214
Score = 81.6 bits (200), Expect = 8e-14
Identities = 42/96 (43%), Positives = 63/96 (64%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVKGR++ +N++L+ ELV+ + + IS R ++K+DL+KA+ SV FI + P +
Sbjct: 558 FVKGRLLTENVLLATELVQGFGQANISSRGVLKVDLRKAFDSVGWGFIIETLKAANAPPR 617
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQGD 514
FVN C+T+TS++ NV+G L F KGLRQGD
Sbjct: 618 FVNWIKQCITSTSFSINVSGSLCGYFKGSKGLRQGD 653
Score = 41.2 bits (95), Expect = 0.13
Identities = 28/87 (32%), Positives = 39/87 (44%), Gaps = 1/87 (1%)
Query: 145 KLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIEAEKVCLSSLKK 204
K K LK ++ N H+ G K+V A L Q L + P + L EK S +
Sbjct: 283 KSKFLKGTIRTFNREHYSGLEKRVVQAAQNLKTCQNNLLAAPSSY-LAGLEKEAHRSWAE 341
Query: 205 WSTIEEQIWI*KSRANWIQLGDSTQNF 231
+ EE+ KSR W++ GDS F
Sbjct: 342 LALAEERFLCQKSRVLWLKCGDSNTTF 368
>UniRef100_O81068 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1529
Score = 81.3 bits (199), Expect = 1e-13
Identities = 37/95 (38%), Positives = 63/95 (65%)
Query: 419 FVKGRVIFDNIILSHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCK 478
FVK R++ +N++L+ ELVK Y ++ ++PRC +KID+ KA+ SV+ F+ + + FP
Sbjct: 862 FVKERLLMENVLLATELVKDYHKESVTPRCAMKIDISKAFDSVQWQFLLNTLEALNFPET 921
Query: 479 FVN*AMACLTTTSYTYNVNGDLTRPFAAKKGLRQG 513
F + C++T +++ VNG+L F + +GLRQG
Sbjct: 922 FRHWIKLCISTATFSVQVNGELAGFFGSSRGLRQG 956
Score = 36.2 bits (82), Expect = 4.1
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 1/96 (1%)
Query: 136 TNLLTNIWFKLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLIEAE 195
T+ L KLK LK L+EL K+ +A L + Q ++P + + I E
Sbjct: 578 TSALYRFSKKLKTLKPHLRELGKEKLGDLPKRTREAHILLCEKQATTLANP-SQETIAEE 636
Query: 196 KVCLSSLKKWSTIEEQIWI*KSRANWIQLGDSTQNF 231
+ S +EE KS+ +W+ +GD ++
Sbjct: 637 LKAYTDWTHLSELEEGFLKQKSKLHWMNVGDGNNSY 672
>UniRef100_Q9ZVF0 Putative non-LTR retroelement reverse transcriptase [Arabidopsis
thaliana]
Length = 1449
Score = 80.5 bits (197), Expect = 2e-13
Identities = 44/137 (32%), Positives = 74/137 (53%)
Query: 372 PKKLMIHQLRILGPLPVVQ*YIRLFPKF*QVECKES*IVLLVKINMLFVKGRVIFDNIIL 431
PKKL +++ P+ ++ K K + FVK R++ +N++L
Sbjct: 933 PKKLEAKEMKDYRPISCCNVIYKVISKIIANRLKHVLPNFIAGNQSAFVKDRLLIENLLL 992
Query: 432 SHELVKSYSRKGISPRCMVKIDLQKAYYSVE*PFINYLMLEMGFPCKFVN*AMACLTTTS 491
+ ELVK Y + IS RC +KID+ KA+ SV+ F+ ++ + FP +FV+ M C+TT S
Sbjct: 993 ATELVKDYHKDTISGRCAIKIDISKAFDSVQWSFLKNVLSALDFPPEFVHWVMLCVTTAS 1052
Query: 492 YTYNVNGDLTRPFAAKK 508
++ VNG+L F + +
Sbjct: 1053 FSVQVNGELAGYFQSSR 1069
Score = 35.4 bits (80), Expect = 7.0
Identities = 25/94 (26%), Positives = 45/94 (47%), Gaps = 1/94 (1%)
Query: 133 NLHTNLLTNIWFKLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQLSSDPMNLDLI 192
++ T+ L KLKALK L+ L K+ +A +L Q+ S +P + +
Sbjct: 693 HMSTSSLFRFTKKLKALKPKLRGLAKEKMGNLVKRTREAYLSLCQAQQSNSQNP-SQRAM 751
Query: 193 EAEKVCLSSLKKWSTIEEQIWI*KSRANWIQLGD 226
E E + ++IEE+ S+ +W+++GD
Sbjct: 752 EIESEAYVRWDRIASIEEKYLKQVSKLHWLKVGD 785
>UniRef100_Q5U6J8 Orf177 protein [Beta vulgaris subsp. vulgaris]
Length = 177
Score = 80.1 bits (196), Expect = 2e-13
Identities = 47/147 (31%), Positives = 76/147 (50%), Gaps = 2/147 (1%)
Query: 122 FPSLIQEK*GQNLHTNLLTNIWFKLKALKKDLKELNSTHFQGTTKKVEDARTALVDVQRQ 181
F ++++ G +H + +W KL LK LK+L+S+ F G T ++E AR L VQ
Sbjct: 7 FEDVVRDSWGAVVHGTSMYRVWRKLSLLKVGLKQLHSSQFAGITSRIEKAREDLEVVQSS 66
Query: 182 LSSDPMNLDLIEAEKVCLSSLKKWSTIEEQIWI*KSRANWIQLGDSTQNFSMPLQRKEDV 241
L + P+N L EK L+KW++IEE +SR W+ LGDS +F + ++ +
Sbjct: 67 LQAAPLNPSLHSQEKT--EQLRKWNSIEESALRQQSRVQWLTLGDSNNHFFVSAIKERNF 124
Query: 242 RISLSSS*QKMAPKLTSITSSRRRLEG 268
R S+ KLT+ ++ + G
Sbjct: 125 RNSIDVLFDDQDVKLTTHQDIQKEVVG 151
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.348 0.153 0.523
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,164,235,288
Number of Sequences: 2790947
Number of extensions: 43141847
Number of successful extensions: 185742
Number of sequences better than 10.0: 258
Number of HSP's better than 10.0 without gapping: 158
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 185324
Number of HSP's gapped (non-prelim): 389
length of query: 821
length of database: 848,049,833
effective HSP length: 136
effective length of query: 685
effective length of database: 468,481,041
effective search space: 320909513085
effective search space used: 320909513085
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 79 (35.0 bits)
Medicago: description of AC138015.15


