

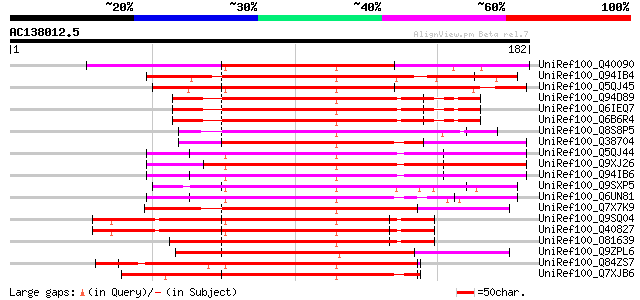
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138012.5 + phase: 0
(182 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q40090 SPF1 protein [Ipomoea batatas] 120 1e-26
UniRef100_Q94IB4 WRKY DNA-binding protein [Nicotiana tabacum] 116 2e-25
UniRef100_Q5QJ45 WRKY3 [Nicotiana attenuata] 115 7e-25
UniRef100_Q94D89 WRKY8 [Oryza sativa] 111 1e-23
UniRef100_Q6IEQ7 WRKY transcription factor 24 [Oryza sativa] 111 1e-23
UniRef100_Q6B6R4 Transcription factor WRKY07 [Oryza sativa] 111 1e-23
UniRef100_Q8S8P5 Probable WRKY transcription factor 33 [Arabidop... 110 2e-23
UniRef100_Q38704 DNA-binding protein [Avena fatua] 108 9e-23
UniRef100_Q5QJ44 WRKY6 [Nicotiana attenuata] 107 1e-22
UniRef100_Q9XJ26 NtWRKY1 [Nicotiana tabacum] 107 1e-22
UniRef100_Q94IB6 WRKY DNA-binding protein [Nicotiana tabacum] 107 1e-22
UniRef100_Q9SXP5 Transcription factor NtWRKY2 [Nicotiana tabacum] 107 1e-22
UniRef100_Q6UN81 WRKY-type transcription factor [Solanum chacoense] 107 1e-22
UniRef100_Q7X7K9 WRKY transcription factor 33 [Capsella rubella] 106 2e-22
UniRef100_Q9SQ04 Zinc-finger type transcription factor WRKY1 [Pe... 106 3e-22
UniRef100_Q40827 WRKY1 [Petroselinum crispum] 106 3e-22
UniRef100_O81639 Zinc finger protein [Pimpinella brachycarpa] 104 9e-22
UniRef100_Q9ZPL6 DNA-binding protein 2 [Nicotiana tabacum] 101 1e-20
UniRef100_Q84ZS7 SUSIBA2-like protein [Oryza sativa] 98 1e-19
UniRef100_Q7XJB6 Putative WRKY-type DNA binding protein [Glycine... 97 2e-19
>UniRef100_Q40090 SPF1 protein [Ipomoea batatas]
Length = 549
Score = 120 bits (302), Expect = 1e-26
Identities = 66/162 (40%), Positives = 94/162 (57%), Gaps = 7/162 (4%)
Query: 28 RHEIGTLRVEDDVAIEIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNH-DDGYNWKKYEEK 86
+ E+ +L+ V N S+Y+N ++R DDGYNW+KY +K
Sbjct: 162 KDELNSLQSLPPVTTSTQMSSQNNGGSYSEYNNQCCPPSQTLREQRRSDDGYNWRKYGQK 221
Query: 87 VAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYL 146
KGSEN RSYYKCT PNC KKKVER +DG++ E +YKG HNH KP S+ + +SS+
Sbjct: 222 QVKGSENPRSYYKCTHPNCPTKKKVERALDGQITEIVYKGAHNHPKPQSTRRSSSSTASS 281
Query: 147 YSLLPSET---GSIDLQDQSF---GSEQLDSDEEPTKSSVSI 182
S L +++ + D+ DQS+ G+ Q+DS P SS+S+
Sbjct: 282 ASTLAAQSYNAPASDVPDQSYWSNGNGQMDSVATPENSSISV 323
Score = 69.7 bits (169), Expect = 3e-11
Identities = 33/62 (53%), Positives = 41/62 (65%), Gaps = 1/62 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 386 DDGYRWRKYGQKVVKGNPNPRSYYKCTSQGCPVRKHVERASHDIRSVITTYEGKHNHDVP 445
Query: 134 TS 135
+
Sbjct: 446 AA 447
>UniRef100_Q94IB4 WRKY DNA-binding protein [Nicotiana tabacum]
Length = 378
Score = 116 bits (291), Expect = 2e-25
Identities = 64/137 (46%), Positives = 84/137 (60%), Gaps = 14/137 (10%)
Query: 49 NNRNEFQSDYSNGR--QQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCF 106
N +E QS +NG+ Q NK S R +DGYNW+KY +K KGSEN RSYYKCT+PNC
Sbjct: 44 NGTSELQSLKNNGQSNQYNKQSSRS---EDGYNWRKYGQKQVKGSENPRSYYKCTFPNCP 100
Query: 107 VKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGS 166
KKKVER +DG++ E +YKG HNH KPT S +R+SS ++ P T + ++ D
Sbjct: 101 TKKKVERCLDGQITEIVYKGNHNHPKPTQSTRRSSS----LAIQPYNTQTNEIPDHQSTP 156
Query: 167 EQL-----DSDEEPTKS 178
E D D E ++S
Sbjct: 157 ENSSISFGDDDHEKSRS 173
Score = 77.0 bits (188), Expect = 2e-13
Identities = 44/97 (45%), Positives = 56/97 (57%), Gaps = 8/97 (8%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT P C V+K VER + D + T Y+G HNH P
Sbjct: 228 DDGYRWRKYGQKVVKGNPNPRSYYKCTSPGCPVRKHVERASQDIRSVITTYEGKHNHDVP 287
Query: 134 T---SSMKRNSSSEYLYS----LLPSETGSIDLQDQS 163
S++ R + Y+ + PS T I L QS
Sbjct: 288 AARGSAINRPVAPTITYNNAIPIRPSVTSQIPLPQQS 324
>UniRef100_Q5QJ45 WRKY3 [Nicotiana attenuata]
Length = 354
Score = 115 bits (287), Expect = 7e-25
Identities = 60/134 (44%), Positives = 86/134 (63%), Gaps = 8/134 (5%)
Query: 51 RNEFQSDYSNG-RQQNKSSVRRMNH-DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVK 108
+N QS+ N Q+ S+R +DGYNW+KY +K KGSEN RSYYKCT+PNC K
Sbjct: 5 KNNAQSNGGNQYNNQSSQSIREQKRSEDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTK 64
Query: 109 KKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQD-QSFGSE 167
KKVER++DG++ E +YKG HNH KP S+ + +S++ ++ P T + ++ D QS+G
Sbjct: 65 KKVERSLDGQITEIVYKGNHNHPKPQSTRRSSSTASSSSAVQPYNTQTNEIPDHQSYG-- 122
Query: 168 QLDSDEEPTKSSVS 181
S+ P SS+S
Sbjct: 123 ---SNATPENSSIS 133
Score = 72.8 bits (177), Expect = 4e-12
Identities = 34/62 (54%), Positives = 42/62 (66%), Gaps = 1/62 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT P C V+K VER + D + T Y+G HNH P
Sbjct: 199 DDGYRWRKYGQKVVKGNPNPRSYYKCTSPGCPVRKHVERASQDIRSVITTYEGKHNHDVP 258
Query: 134 TS 135
+
Sbjct: 259 AA 260
>UniRef100_Q94D89 WRKY8 [Oryza sativa]
Length = 357
Score = 111 bits (277), Expect = 1e-23
Identities = 57/108 (52%), Positives = 74/108 (67%), Gaps = 11/108 (10%)
Query: 58 YSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDG 117
YS + Q +SS DDGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER++DG
Sbjct: 11 YSQPQSQRRSS------DDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSLDG 64
Query: 118 EVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFG 165
++ E +YKGTHNH KP ++ +RNS S L ++G D+ + SFG
Sbjct: 65 QITEIVYKGTHNHAKPQNT-RRNSGSSAAQVL---QSGG-DMSEHSFG 107
Score = 71.2 bits (173), Expect = 1e-11
Identities = 36/72 (50%), Positives = 46/72 (63%), Gaps = 4/72 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 187 DDGYRWRKYGQKVVKGNPNPRSYYKCTTAGCPVRKHVERASHDLRAVITTYEGKHNHDVP 246
Query: 134 TSSMKRNSSSEY 145
+ R S++ Y
Sbjct: 247 AA---RGSAALY 255
>UniRef100_Q6IEQ7 WRKY transcription factor 24 [Oryza sativa]
Length = 555
Score = 111 bits (277), Expect = 1e-23
Identities = 57/108 (52%), Positives = 74/108 (67%), Gaps = 11/108 (10%)
Query: 58 YSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDG 117
YS + Q +SS DDGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER++DG
Sbjct: 209 YSQPQSQRRSS------DDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSLDG 262
Query: 118 EVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFG 165
++ E +YKGTHNH KP ++ +RNS S L ++G D+ + SFG
Sbjct: 263 QITEIVYKGTHNHAKPQNT-RRNSGSSAAQVL---QSGG-DMSEHSFG 305
Score = 71.2 bits (173), Expect = 1e-11
Identities = 36/72 (50%), Positives = 46/72 (63%), Gaps = 4/72 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 385 DDGYRWRKYGQKVVKGNPNPRSYYKCTTAGCPVRKHVERASHDLRAVITTYEGKHNHDVP 444
Query: 134 TSSMKRNSSSEY 145
+ R S++ Y
Sbjct: 445 AA---RGSAALY 453
>UniRef100_Q6B6R4 Transcription factor WRKY07 [Oryza sativa]
Length = 555
Score = 111 bits (277), Expect = 1e-23
Identities = 57/108 (52%), Positives = 74/108 (67%), Gaps = 11/108 (10%)
Query: 58 YSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDG 117
YS + Q +SS DDGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER++DG
Sbjct: 209 YSQPQSQRRSS------DDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSLDG 262
Query: 118 EVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFG 165
++ E +YKGTHNH KP ++ +RNS S L ++G D+ + SFG
Sbjct: 263 QITEIVYKGTHNHAKPQNT-RRNSGSSAAQVL---QSGG-DMSEHSFG 305
Score = 71.2 bits (173), Expect = 1e-11
Identities = 36/72 (50%), Positives = 46/72 (63%), Gaps = 4/72 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 385 DDGYRWRKYGQKVVKGNPNPRSYYKCTTAGCPVRKHVERASHDLRAVITTYEGKHNHDVP 444
Query: 134 TSSMKRNSSSEY 145
+ R S++ Y
Sbjct: 445 AA---RGSAALY 453
>UniRef100_Q8S8P5 Probable WRKY transcription factor 33 [Arabidopsis thaliana]
Length = 512
Score = 110 bits (275), Expect = 2e-23
Identities = 57/127 (44%), Positives = 77/127 (59%), Gaps = 23/127 (18%)
Query: 60 NGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEV 119
NGR+Q K +DGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER+++G++
Sbjct: 169 NGREQRKG-------EDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERSLEGQI 221
Query: 120 IETLYKGTHNHWKPTSSMKRNSSSEYLYSLL---------------PSETGSIDLQDQSF 164
E +YKG+HNH KP S+ + +SSS +S + P+ S Q SF
Sbjct: 222 TEIVYKGSHNHPKPQSTRRSSSSSSTFHSAVYNASLDHNRQASSDQPNSNNSFH-QSDSF 280
Query: 165 GSEQLDS 171
G +Q D+
Sbjct: 281 GMQQEDN 287
Score = 72.8 bits (177), Expect = 4e-12
Identities = 37/87 (42%), Positives = 51/87 (58%), Gaps = 6/87 (6%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 355 DDGYRWRKYGQKVVKGNPNPRSYYKCTTIGCPVRKHVERASHDMRAVITTYEGKHNHDVP 414
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQ 160
+ S Y + P ++ S+ ++
Sbjct: 415 AA-----RGSGYATNRAPQDSSSVPIR 436
>UniRef100_Q38704 DNA-binding protein [Avena fatua]
Length = 402
Score = 108 bits (269), Expect = 9e-23
Identities = 53/122 (43%), Positives = 73/122 (59%)
Query: 60 NGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEV 119
+G ++ +R + DDGYNW+KY +K KGSEN RSYYKCT+PNC KKKVE +I+G++
Sbjct: 38 SGGVYRQTHSQRRSSDDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVETSIEGQI 97
Query: 120 IETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSS 179
E +YKGTHNH KP S+ + + + + G D + SFG+ P SS
Sbjct: 98 TEIVYKGTHNHAKPLSTRRGSGGGGGGAAQVLQSGGGGDASEHSFGAMSGAPVSTPENSS 157
Query: 180 VS 181
S
Sbjct: 158 AS 159
Score = 70.9 bits (172), Expect = 2e-11
Identities = 36/72 (50%), Positives = 46/72 (63%), Gaps = 4/72 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 225 DDGYRWRKYGQKVVKGNPNPRSYYKCTTVGCPVRKHVERASHDLRAVITTYEGKHNHDVP 284
Query: 134 TSSMKRNSSSEY 145
+ R S++ Y
Sbjct: 285 AA---RGSAALY 293
>UniRef100_Q5QJ44 WRKY6 [Nicotiana attenuata]
Length = 563
Score = 107 bits (268), Expect = 1e-22
Identities = 56/118 (47%), Positives = 70/118 (58%), Gaps = 2/118 (1%)
Query: 64 QNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETL 123
Q VR +DGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER +DG + E +
Sbjct: 217 QPSQYVREQKAEDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERNLDGHITEIV 276
Query: 124 YKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVS 181
YKG HNH KP S+ R SSS+ + +L S + + + Q DS SS S
Sbjct: 277 YKGNHNHPKPQST--RRSSSQSIQNLAYSNLDITNQPNAFLDNAQRDSFAGTDNSSAS 332
Score = 73.2 bits (178), Expect = 3e-12
Identities = 43/109 (39%), Positives = 59/109 (53%), Gaps = 9/109 (8%)
Query: 49 NNRNEFQSDYSNGRQQNKSSVRRMNH----DDGYNWKKYEEKVAKGSENQRSYYKCTWPN 104
+N NE S S ++ + V+ + DDGY W+KY +KV KG+ N RSYYKCT+
Sbjct: 363 DNENEVISSASRTVREPRIVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTFTG 422
Query: 105 CFVKKKVER-TIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPS 152
C V+K VER + D + T Y+G HNH P + S Y + PS
Sbjct: 423 CPVRKHVERASHDLRAVITTYEGKHNHDVPAA----RGSGSYAMNKPPS 467
>UniRef100_Q9XJ26 NtWRKY1 [Nicotiana tabacum]
Length = 477
Score = 107 bits (268), Expect = 1e-22
Identities = 55/113 (48%), Positives = 69/113 (60%), Gaps = 2/113 (1%)
Query: 69 VRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTH 128
VR +DGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER +DG + E +YKG H
Sbjct: 137 VREQKAEDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERNLDGHITEIVYKGNH 196
Query: 129 NHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVS 181
NH KP S+ R SSS+ + +L S + + + Q DS SS S
Sbjct: 197 NHPKPQST--RRSSSQSIQNLAYSNLDITNQSNAFLDNAQRDSFAGTDNSSAS 247
Score = 74.7 bits (182), Expect = 1e-12
Identities = 44/109 (40%), Positives = 60/109 (54%), Gaps = 9/109 (8%)
Query: 49 NNRNEFQSDYSNGRQQNKSSVRRMNH----DDGYNWKKYEEKVAKGSENQRSYYKCTWPN 104
+N NE S S ++ + V+ + DDGY W+KY +KVAKG+ N RSYYKCT+
Sbjct: 278 DNENEVISSASRTVREPRIVVQTTSDIDILDDGYRWRKYGQKVAKGNPNPRSYYKCTFTG 337
Query: 105 CFVKKKVER-TIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPS 152
C V+K VER + D + T Y+G HNH P + S Y + PS
Sbjct: 338 CPVRKHVERASHDLRAVITTYEGKHNHDVPAA----RGSGSYAMNKPPS 382
>UniRef100_Q94IB6 WRKY DNA-binding protein [Nicotiana tabacum]
Length = 559
Score = 107 bits (268), Expect = 1e-22
Identities = 56/118 (47%), Positives = 70/118 (58%), Gaps = 2/118 (1%)
Query: 64 QNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETL 123
Q VR +DGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER +DG + E +
Sbjct: 214 QPSQYVREQKAEDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERNLDGHITEIV 273
Query: 124 YKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEPTKSSVS 181
YKG HNH KP S+ R SSS+ + +L S + + + Q DS SS S
Sbjct: 274 YKGNHNHPKPQST--RRSSSQSIQNLAYSNLDITNQPNAFLDNAQRDSFAGTDNSSAS 329
Score = 73.2 bits (178), Expect = 3e-12
Identities = 43/109 (39%), Positives = 59/109 (53%), Gaps = 9/109 (8%)
Query: 49 NNRNEFQSDYSNGRQQNKSSVRRMNH----DDGYNWKKYEEKVAKGSENQRSYYKCTWPN 104
+N NE S S ++ + V+ + DDGY W+KY +KV KG+ N RSYYKCT+
Sbjct: 360 DNENEVISSASRTVREPRIVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTFTG 419
Query: 105 CFVKKKVER-TIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPS 152
C V+K VER + D + T Y+G HNH P + S Y + PS
Sbjct: 420 CPVRKHVERASHDLRAVITTYEGKHNHDVPAA----RGSGSYAMNKPPS 464
>UniRef100_Q9SXP5 Transcription factor NtWRKY2 [Nicotiana tabacum]
Length = 353
Score = 107 bits (267), Expect = 1e-22
Identities = 59/143 (41%), Positives = 84/143 (58%), Gaps = 17/143 (11%)
Query: 51 RNEFQSDYSNGRQQNKSSVRRMNH-DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKK 109
+N QS+ N Q+ S+R +DGYNW+KY +K KGSEN RSYYKCT+PNC KK
Sbjct: 5 KNNGQSNQYNN--QSSQSIREQKRSEDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKK 62
Query: 110 KVERTIDGEVIETLYKGTHNHWKPTSSMKRNSS-----------SEYLYSLLPSETGSID 158
KVER++DG++ E +YKG HNH KP S+ + +S+ ++ + S + S GS
Sbjct: 63 KVERSLDGQITEIVYKGNHNHPKPQSTRRSSSTASSLTRPTLQYTKLMKSQIISSYGSNA 122
Query: 159 LQDQ---SFGSEQLDSDEEPTKS 178
+ SFG + D ++ KS
Sbjct: 123 TPENSSISFGDDDHDHEQSSQKS 145
Score = 73.2 bits (178), Expect = 3e-12
Identities = 42/97 (43%), Positives = 53/97 (54%), Gaps = 11/97 (11%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N R YYKCT P C V+K VER + D + T Y+G HNH P
Sbjct: 196 DDGYRWRKYGQKVVKGNPNPRGYYKCTSPGCPVRKHVERASQDIRSVITTYEGKHNHDVP 255
Query: 134 T---SSMKRNSSSEYLY-------SLLPSETGSIDLQ 160
S + R + Y ++ PS T I LQ
Sbjct: 256 AARGSGINRPVAPNITYNNGANAMAIRPSVTSQIPLQ 292
>UniRef100_Q6UN81 WRKY-type transcription factor [Solanum chacoense]
Length = 525
Score = 107 bits (267), Expect = 1e-22
Identities = 60/133 (45%), Positives = 74/133 (55%), Gaps = 20/133 (15%)
Query: 64 QNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETL 123
Q VR +DGYNW+KY +K KGSEN RSYYKCT+PNC KKKVER +DG + E +
Sbjct: 184 QPSQYVREQKAEDGYNWRKYGQKQVKGSENPRSYYKCTFPNCPTKKKVERNLDGHITEIV 243
Query: 124 YKGTHNHWKPTSSMKRNSSSEYLYSLLPS------------ETGS------IDLQDQSFG 165
YKG+HNH KP S+ R SSS+ + +L S E G D SFG
Sbjct: 244 YKGSHNHPKPQST--RRSSSQSIQNLAYSNLDVTNQPNAFHENGQRDSFAVTDNSSASFG 301
Query: 166 SEQLDSDEEPTKS 178
E +D +KS
Sbjct: 302 DEDVDQGSPISKS 314
Score = 73.9 bits (180), Expect = 2e-12
Identities = 46/113 (40%), Positives = 63/113 (55%), Gaps = 11/113 (9%)
Query: 49 NNRNEFQSDYSNGRQQNKSSVRRMNH----DDGYNWKKYEEKVAKGSENQRSYYKCTWPN 104
+N NE S S ++ + V+ + DDGY W+KY +KV KG+ N RSYYKCT+
Sbjct: 330 DNENEVISSASRTVREPRIVVQTTSDIDILDDGYRWRKYGQKVVKGNPNPRSYYKCTFTG 389
Query: 105 CFVKKKVER-TIDGEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSLLPSETGS 156
C V+K VER + D + T Y+G HNH P + R S S Y++ TGS
Sbjct: 390 CPVRKHVERASHDLRAVITTYEGKHNHDVPAA---RGSGS---YAMNRPPTGS 436
>UniRef100_Q7X7K9 WRKY transcription factor 33 [Capsella rubella]
Length = 514
Score = 106 bits (265), Expect = 2e-22
Identities = 49/96 (51%), Positives = 68/96 (70%), Gaps = 7/96 (7%)
Query: 48 DNNRNEFQSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFV 107
+ +R Q+ NGR+Q K +DGYNW+KY +K KGSEN RSYYKCT+P+C
Sbjct: 154 NESRQNNQAVSYNGREQRKG-------EDGYNWRKYGQKQVKGSENPRSYYKCTFPSCPT 206
Query: 108 KKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSS 143
KKKVER+++G++ E +YKG+HNH KP S+ + +SSS
Sbjct: 207 KKKVERSLEGQITEIVYKGSHNHPKPQSTRRSSSSS 242
Score = 73.6 bits (179), Expect = 2e-12
Identities = 38/102 (37%), Positives = 53/102 (51%), Gaps = 6/102 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 353 DDGYRWRKYGQKVVKGNPNPRSYYKCTTIGCPVRKHVERASHDMRAVITTYEGKHNHDVP 412
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEP 175
+ S Y + P + S+ ++ + + P
Sbjct: 413 AA-----RGSGYATNRAPQDASSVPIRPAAIAGHSNSTTSSP 449
>UniRef100_Q9SQ04 Zinc-finger type transcription factor WRKY1 [Petroselinum crispum]
Length = 514
Score = 106 bits (264), Expect = 3e-22
Identities = 58/124 (46%), Positives = 78/124 (62%), Gaps = 6/124 (4%)
Query: 30 EIGTL-RVEDDVAIEIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNH---DDGYNWKKYEE 85
+I TL R+ ++ + + +N QS+ +N Q ++SS + DDGYNW+KY +
Sbjct: 143 DIATLQRISPEMTMNHANMQSNA-ALQSNLNNYAQSSQSSQTNRDQSKLDDGYNWRKYGQ 201
Query: 86 KVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEY 145
K KGSEN RSYYKCT+ NC KKKVE T DG + E +YKG HNH KP S+ KR+SS Y
Sbjct: 202 KQVKGSENPRSYYKCTYLNCPTKKKVETTFDGHITEIVYKGNHNHPKPQST-KRSSSQSY 260
Query: 146 LYSL 149
S+
Sbjct: 261 QNSI 264
Score = 70.1 bits (170), Expect = 3e-11
Identities = 33/60 (55%), Positives = 40/60 (66%), Gaps = 1/60 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 353 DDGYRWRKYGQKVVKGNPNPRSYYKCTQVGCPVRKHVERASHDLRAVITTYEGKHNHDVP 412
>UniRef100_Q40827 WRKY1 [Petroselinum crispum]
Length = 514
Score = 106 bits (264), Expect = 3e-22
Identities = 58/124 (46%), Positives = 78/124 (62%), Gaps = 6/124 (4%)
Query: 30 EIGTL-RVEDDVAIEIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNH---DDGYNWKKYEE 85
+I TL R+ ++ + + +N QS+ +N Q ++SS + DDGYNW+KY +
Sbjct: 143 DIATLQRISPEMTMNQANMQSNA-ALQSNLNNYAQSSQSSQTNRDQSKLDDGYNWRKYGQ 201
Query: 86 KVAKGSENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSSEY 145
K KGSEN RSYYKCT+ NC KKKVE T DG + E +YKG HNH KP S+ KR+SS Y
Sbjct: 202 KQVKGSENPRSYYKCTYLNCPTKKKVETTFDGHITEIVYKGNHNHPKPQST-KRSSSQSY 260
Query: 146 LYSL 149
S+
Sbjct: 261 QNSI 264
Score = 70.1 bits (170), Expect = 3e-11
Identities = 33/60 (55%), Positives = 40/60 (66%), Gaps = 1/60 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 353 DDGYRWRKYGQKVVKGNPNPRSYYKCTQVGCPVRKHVERASHDLRAVITTYEGKHNHDVP 412
>UniRef100_O81639 Zinc finger protein [Pimpinella brachycarpa]
Length = 515
Score = 104 bits (260), Expect = 9e-22
Identities = 49/93 (52%), Positives = 64/93 (68%), Gaps = 1/93 (1%)
Query: 57 DYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTID 116
+Y+ Q ++++ + DDGYNW+KY +K KGSEN RSYYKCT+ NC KKKVE T D
Sbjct: 173 NYAQSSQSSQTNRDQSKLDDGYNWRKYGQKQVKGSENPRSYYKCTYLNCPTKKKVETTFD 232
Query: 117 GEVIETLYKGTHNHWKPTSSMKRNSSSEYLYSL 149
G + E +YKG HNH KP S+ KR+SS Y S+
Sbjct: 233 GHITEIVYKGNHNHPKPQST-KRSSSQSYQNSI 264
Score = 70.1 bits (170), Expect = 3e-11
Identities = 33/60 (55%), Positives = 40/60 (66%), Gaps = 1/60 (1%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ N RSYYKCT C V+K VER + D + T Y+G HNH P
Sbjct: 353 DDGYRWRKYGQKVVKGNPNPRSYYKCTQVGCPVRKHVERASHDLRAVITTYEGKHNHDVP 412
>UniRef100_Q9ZPL6 DNA-binding protein 2 [Nicotiana tabacum]
Length = 528
Score = 101 bits (251), Expect = 1e-20
Identities = 48/84 (57%), Positives = 57/84 (67%)
Query: 59 SNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERTIDGE 118
S+ R + SS DDGYNW+KY +K KGSE RSYYKCT PNC VKKKVER++DG+
Sbjct: 219 SDQRSEPASSAVDKPADDGYNWRKYGQKHVKGSEYPRSYYKCTHPNCPVKKKVERSLDGQ 278
Query: 119 VIETLYKGTHNHWKPTSSMKRNSS 142
V E +YKG HNH P SS + S
Sbjct: 279 VTEIIYKGQHNHQPPQSSKRSKES 302
Score = 73.2 bits (178), Expect = 3e-12
Identities = 37/102 (36%), Positives = 57/102 (55%), Gaps = 1/102 (0%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVERT-IDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV KG+ RSYYKCT C V+K VER D + + T Y+G HNH P
Sbjct: 415 DDGYRWRKYGQKVVKGNPYPRSYYKCTSQGCNVRKHVERAPSDPKAVITTYEGEHNHDVP 474
Query: 134 TSSMKRNSSSEYLYSLLPSETGSIDLQDQSFGSEQLDSDEEP 175
+ ++++ S + +D QD + +++++P
Sbjct: 475 AARNSSHNTTNNSVSQMRPHNPVVDKQDATRRIGFSNNEQQP 516
>UniRef100_Q84ZS7 SUSIBA2-like protein [Oryza sativa]
Length = 618
Score = 97.8 bits (242), Expect = 1e-19
Identities = 45/113 (39%), Positives = 70/113 (61%), Gaps = 2/113 (1%)
Query: 31 IGTLRVEDDVAIEIDGVDNNRNEFQSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKG 90
+G + + D E+ + ++ S + ++ SV + +DGYNW+KY +K KG
Sbjct: 194 VGMVGLTDSSPAEVG--TSELHQMNSSGNAMQESQPESVAEKSAEDGYNWRKYGQKHVKG 251
Query: 91 SENQRSYYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSS 143
SEN RSYYKCT PNC VKK +ER++DG++ E +YKG HNH KP + + ++ +
Sbjct: 252 SENPRSYYKCTHPNCDVKKLLERSLDGQITEVVYKGRHNHPKPQPNRRLSAGA 304
Score = 75.9 bits (185), Expect = 5e-13
Identities = 46/117 (39%), Positives = 66/117 (56%), Gaps = 14/117 (11%)
Query: 39 DVAIEIDGVDNNRNEFQSDYSNGRQQNKSS------VRRMNH----DDGYNWKKYEEKVA 88
D A+E D +++ R + +S + K + V+ ++ DDGY W+KY +KV
Sbjct: 364 DDAVEDDDLESKRRKMESAAIDAALMGKPNREPRVVVQTVSEVDILDDGYRWRKYGQKVV 423
Query: 89 KGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKPTSSMKRNSSSE 144
KG+ N RSYYKCT C V+K VER + D + + T Y+G HNH P S RN+S E
Sbjct: 424 KGNPNPRSYYKCTNTGCPVRKHVERASHDPKSVITTYEGKHNHEVPAS---RNASHE 477
>UniRef100_Q7XJB6 Putative WRKY-type DNA binding protein [Glycine max]
Length = 493
Score = 97.1 bits (240), Expect = 2e-19
Identities = 46/107 (42%), Positives = 66/107 (60%), Gaps = 3/107 (2%)
Query: 40 VAIEIDGVDNNRNE---FQSDYSNGRQQNKSSVRRMNHDDGYNWKKYEEKVAKGSENQRS 96
V +++D +++ N Q+ + R S DDGYNW+KY +K+ KGSE RS
Sbjct: 99 VDVDLDDINHKGNTATGLQASHVEVRGSGLSVAAEKTSDDGYNWRKYGQKLVKGSEFPRS 158
Query: 97 YYKCTWPNCFVKKKVERTIDGEVIETLYKGTHNHWKPTSSMKRNSSS 143
YYKCT PNC VKK ER+ DG++ E +YKGTH+H KP S + ++ +
Sbjct: 159 YYKCTHPNCEVKKLFERSHDGQITEIVYKGTHDHPKPQPSCRYSTGT 205
Score = 72.4 bits (176), Expect = 5e-12
Identities = 36/71 (50%), Positives = 47/71 (65%), Gaps = 4/71 (5%)
Query: 75 DDGYNWKKYEEKVAKGSENQRSYYKCTWPNCFVKKKVER-TIDGEVIETLYKGTHNHWKP 133
DDGY W+KY +KV +G+ N RSYYKCT C V+K VER + D + + T Y+G HNH P
Sbjct: 313 DDGYRWRKYGQKVVRGNPNPRSYYKCTNAGCPVRKHVERASHDPKAVITTYEGKHNHDVP 372
Query: 134 TSSMKRNSSSE 144
+ RNSS +
Sbjct: 373 AA---RNSSHD 380
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.311 0.129 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 300,730,929
Number of Sequences: 2790947
Number of extensions: 12259094
Number of successful extensions: 32880
Number of sequences better than 10.0: 378
Number of HSP's better than 10.0 without gapping: 311
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 32271
Number of HSP's gapped (non-prelim): 486
length of query: 182
length of database: 848,049,833
effective HSP length: 119
effective length of query: 63
effective length of database: 515,927,140
effective search space: 32503409820
effective search space used: 32503409820
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 71 (32.0 bits)
Medicago: description of AC138012.5


