

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137995.11 + phase: 0
(1094 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
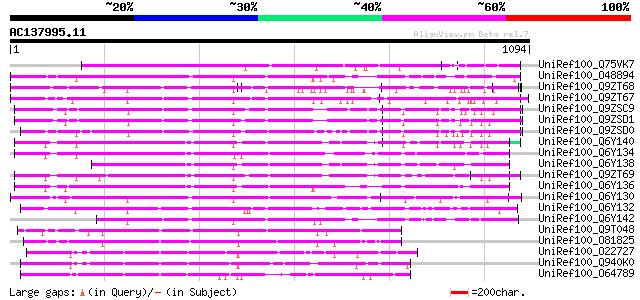
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q75VK7 CC-NB-LRR protein [Solanum tuberosum] 295 5e-78
UniRef100_O48894 Resistance protein candidate [Lactuca sativa] 272 5e-71
UniRef100_Q9ZT68 Resistance protein candidate RGC2K [Lactuca sat... 260 2e-67
UniRef100_Q9ZT67 Resistance protein candidate RGC20 [Lactuca sat... 251 7e-65
UniRef100_Q9ZSC9 Resistance protein candidate RGC2S [Lactuca sat... 247 1e-63
UniRef100_Q9ZSD1 Resistance protein candidate RGC2B [Lactuca sat... 243 2e-62
UniRef100_Q9ZSD0 Resistance protein candidate RGC2C [Lactuca sat... 243 3e-62
UniRef100_Q6Y140 Resistance protein RGC2 [Lactuca sativa] 242 4e-62
UniRef100_Q6Y134 Resistance protein RGC2 [Lactuca sativa] 242 4e-62
UniRef100_Q6Y138 Resistance protein RGC2 [Lactuca sativa] 237 1e-60
UniRef100_Q9ZT69 Resistance protein candidate RGC2J [Lactuca sat... 237 2e-60
UniRef100_Q6Y136 Resistance protein RGC2 [Lactuca sativa] 236 4e-60
UniRef100_Q6Y130 Resistance protein candidate RGC2 [Lactuca sativa] 232 5e-59
UniRef100_Q6Y132 Resistance protein RGC2 [Lactuca sativa] 226 2e-57
UniRef100_Q6Y142 Resistance protein RGC2 [Lactuca sativa] 221 7e-56
UniRef100_Q9T048 Putative disease resistance protein At4g27190 [... 219 5e-55
UniRef100_O81825 Putative disease resistance protein At4g27220 [... 208 8e-52
UniRef100_O22727 Putative disease resistance protein At1g61190 [... 206 3e-51
UniRef100_Q940K0 Probable disease resistance protein At1g61180 [... 204 9e-51
UniRef100_O64789 Putative disease resistance protein At1g61310 [... 202 3e-50
>UniRef100_Q75VK7 CC-NB-LRR protein [Solanum tuberosum]
Length = 1036
Score = 295 bits (755), Expect = 5e-78
Identities = 249/873 (28%), Positives = 413/873 (46%), Gaps = 88/873 (10%)
Query: 152 FDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKRC--GNLFDQVLFVPISS 209
F SRKP +E+M ALKD+ +++ +YGMGG GKT + + R FD+V+ +S
Sbjct: 151 FTSRKPTMDEIMNALKDEGRSIVRVYGMGGVGKTYMVKALASRALKEKKFDRVVESVVSQ 210
Query: 210 TVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIP 269
TV++ +IQ IA L E + DR+ L +L+ILD +W+ ++ IGIP
Sbjct: 211 TVDLRKIQGDIAHGLGVELTSTEVQDRADDLRNLFNDHGNILLILDGLWETINLSTIGIP 270
Query: 270 SIEHHKGCKILITSRSEAVCTLMDCQ-KKIQLSTLTNDETWDLFQKQALISEGTWISIKN 328
CKILIT+R VC +D Q IQ++ L+ D+ W LF ++A + +
Sbjct: 271 QYSERCKCKILITTRQMNVCDDLDRQYSAIQINVLSGDDPWTLFTQKAGDNLKVPPGFEE 330
Query: 329 MAREISNECKGLPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPY--KCLQ 386
+ ++I EC+GLP+A + S+L K W+ A RL SSK +I++ N KC++
Sbjct: 331 IGKKIVEECRGLPIALSTIGSALYKKDLTYWETAATRLHSSKTASIKEDDLNSVIRKCIE 390
Query: 387 LSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKN 446
LSY L + K +FL+CS+FPED IP E LTR +GL ++ + + + AR ++
Sbjct: 391 LSYSFLPNDTCKRVFLMCSIFPEDYNIPKETLTRYVMGLALIRGIETVKEARGDIHQIVE 450
Query: 447 KLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENEIKCAS-EKDIMTLEH----------- 494
+L ++ LLLD ++ + VKMHD++R+++ I N+ K S K M LE+
Sbjct: 451 ELKAASLLLDGDKEETVKMHDVIRDISIQIGYNQEKPKSIVKASMKLENWPGEILTNSCG 510
Query: 495 -TSLRYLWCEKFPNSLDCSNLDFLQIH---TYTQVSDEIFKGMRMLRVL-FLYNKGRERR 549
SL +K P+ +DC + L + V DE F+GMR L+VL F K +
Sbjct: 511 AISLISNHLKKLPDRVDCPETEILLLQDNKNLRLVPDEFFQGMRALKVLDFTGVKFKS-- 568
Query: 550 PLLTTSLKSLTNLRCILFSKWD-LVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLR 608
L +S + L+ LR + L D+S +G++ +LE +TL LP+ L LR
Sbjct: 569 --LPSSTRQLSLLRLLSLDNCRFLKDVSMIGELNRLEILTLRMSGITSLPESFANLKELR 626
Query: 609 LLDLS-ECGMERNPFEVIARHTELEELFFADCRSKWEV------EFLKEFSVPQVLQRYQ 661
+LD++ E P VI+ +LEEL+ C + WE+ +E L +
Sbjct: 627 ILDITLSLQCENVPPGVISSMDKLEELYMQGCFADWEITNENRKTNFQEILTLGSLTILK 686
Query: 662 IQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAK-NIIPDV 720
+ + ++ D + + D+ + + A++A G+ G P+
Sbjct: 687 VDIKNVCCLPPDSVAPNWEKFDICVSDSEECRLANAAQQASF--TRGLTTGVNLEAFPEW 744
Query: 721 F-QSMNH----------------LKELLIRDSKGIECL-VDTC-----LIEVGT-----L 752
F Q+++H L+E L + ++ L +D C LI++G
Sbjct: 745 FRQAVSHKAEKLSYQFCGNLSNILQEYLYGNFDEVKSLYIDQCADIAQLIKLGNGLPNQP 804
Query: 753 FFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKL-TRLFTLAVAQNLAQL 811
F KL L I HM+ + ++P G + ++ + +S CPKL L + Q ++ L
Sbjct: 805 VFPKLEKLNIHHMQKTEGICTEELP-PGSLQQVKMVEVSECPKLKDSLLPPNLIQRMSNL 863
Query: 812 EKLQVLSCP------------------ELQHILIDDDRDEISAY--DYRLLLFPKLKKFH 851
E+++V +L+ + + + S + L++F +L+
Sbjct: 864 EEVKVTGTSINAVFGFDGITFQGGQLRKLKRLTLLNLSQLTSLWKGPSELVMFHRLEVVK 923
Query: 852 VRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALE 911
V + L YI P T+ L L+ L + L+ V G T +G ++ + I L L
Sbjct: 924 VSQRENLRYIFPYTVCDYLCHLQVLWLEDCSGLEKVIGGHTDENGVHEVP-ESITLPRLT 982
Query: 912 ELTLVNLPNINSICPEDCYLMWPSLLQFNLQNC 944
LTL LP++ ++ YL P L + + Q+C
Sbjct: 983 TLTLQRLPHLTDFYTQEAYLRCPELQRLHKQDC 1015
Score = 48.9 bits (115), Expect = 8e-04
Identities = 44/168 (26%), Positives = 79/168 (46%), Gaps = 16/168 (9%)
Query: 910 LEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASH 969
LE+L + ++ IC E+ + P LQ + MV ++ C L ++ +
Sbjct: 809 LEKLNIHHMQKTEGICTEE---LPPGSLQ-------QVKMVEVSECPKLKDS-LLPPNLI 857
Query: 970 QTLQNITEVRVNNCELEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVEST 1029
Q + N+ EV+V + +F G+T G + L+ L L NL QL L K E
Sbjct: 858 QRMSNLEEVKVTGTSINAVFGFDGITFQGGQ---LRKLKRLTLLNLSQLTSLWKGPSEL- 913
Query: 1030 NLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIV 1077
++F L+ +++S L+ IF + L L+ L +E C+ L++++
Sbjct: 914 -VMFHRLEVVKVSQRENLRYIFPYTVCDYLCHLQVLWLEDCSGLEKVI 960
>UniRef100_O48894 Resistance protein candidate [Lactuca sativa]
Length = 1139
Score = 272 bits (695), Expect = 5e-71
Identities = 291/1162 (25%), Positives = 499/1162 (42%), Gaps = 189/1162 (16%)
Query: 1 MEYLYGFASAISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQ 60
M+ + + L+ V + Y +++++ + L A R V++ V R
Sbjct: 1 MDVVNAILKPVVETLMVPVKKHIGYLISCRQYMREMGIKMRGLNATRLGVEEHVNRNISN 60
Query: 61 TRKTAEVVEKWLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNL 120
+ V W ++ ++ ++ S+ SCF N R+ VG++ SK ++
Sbjct: 61 QLEVPAQVRGWFEEVG----KINAKVENFPSDVGSCF----NLKVRHGVGKRASKIIEDI 112
Query: 121 KLYIEEGRQYIEIERPASL--------SAGYFSAERCWEFDSRKPAYEELMCALKDDDVT 172
+ E I + L S S + EF SR+ + E + AL + +
Sbjct: 113 DSVMREHSIIIWNDHSIPLGRIDSTKASTSIPSTDHHDEFQSREQTFTEALNALDPNHKS 172
Query: 173 -MIGLYGMGGCGKTMLAMEVGK--RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQ 229
MI L+GMGG GKT + + K + +F+ ++ + + IQ +A L E
Sbjct: 173 HMIALWGMGGVGKTTMMHRLKKVVKEKKMFNFIIEAVVGEKTDPIAIQSAVADYLGIELN 232
Query: 230 EKDEMDRSKRLCMRLTQED---RVLVILDDVWQMLDFDAIGIPSIEHHK-GCKILITSRS 285
EK + R+++L ++LVILDDVWQ +D + IG+ + + K+L+TSR
Sbjct: 233 EKTKPARTEKLRKWFVDNSGGKKILVILDDVWQFVDLNDIGLSPLPNQGVDFKVLLTSRD 292
Query: 286 EAVCTLMDCQ--KKIQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVA 343
+ VCT M + + L E LF + IS+ + N+ I +C GLP+A
Sbjct: 293 KDVCTEMGAEVNSTFNVKMLIETEAQSLFHQFIEISDDVDPELHNIGVNIVRKCGGLPIA 352
Query: 344 TVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLL 403
+A +L+GK++ WK AL RL NI G+ ++SYDNL EE KS FLL
Sbjct: 353 IKTMACTLRGKSKDAWKNALLRLEHYDIENIVNGV------FKMSYDNLQDEETKSTFLL 406
Query: 404 CSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCV 463
C ++PED +I E L R GL + +V++ AR + +LI + LL++V++ +C+
Sbjct: 407 CGMYPEDFDILTEELVRYGWGLKLFKKVYTIGEARTRLNTCIERLIHTNLLMEVDDVRCI 466
Query: 464 KMHDLVR--------NVAHWIA---ENEIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCS 512
KMHDLVR V H N ++ ++ + + SL KFP L
Sbjct: 467 KMHDLVRAFVLDMYSKVEHASIVNHSNTLEWHADNMHDSCKRLSLTCKGMSKFPTDLKFP 526
Query: 513 NLDFLQI---HTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSK 569
NL L++ + ++ M L V+ + + + PLL +S + NLR K
Sbjct: 527 NLSILKLMHEDISLRFPKNFYEEMEKLEVI---SYDKMKYPLLPSSPQCSVNLRVFHLHK 583
Query: 570 WDLV--DISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIAR 627
LV D S +G++ LE ++ D + LP + +L LRLLDL+ C R V+ +
Sbjct: 584 CSLVMFDCSCIGNLSNLEVLSFADSAIDRLPSTIGKLKKLRLLDLTNCYGVRIDNGVLKK 643
Query: 628 HTELEELFFA--------------DC-----RSK----WEVEFLKEFSVP-----QVLQR 659
+LEEL+ +C RSK E+EF + + P + LQR
Sbjct: 644 LVKLEELYMTVVDRGRKAISLTDDNCKEMAERSKDIYALELEFFENDAQPKNMSFEKLQR 703
Query: 660 YQIQLGSMFSGFQDEFLNHHR-----TLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAK 714
+QI +G G D + H L L + A + +L +K EVLC++
Sbjct: 704 FQISVGRYLYG--DSIKSRHSYENTLKLVLEKGELLEARMNELFKKTEVLCLS------- 754
Query: 715 NIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNG 774
MN L+++ ++ S
Sbjct: 755 ------VGDMNDLEDIEVKSS--------------------------------------S 770
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEI 834
Q+ S F NL L +S C +L FT VA L +LE L+V C ++ ++ +E
Sbjct: 771 QLLQSSSFNNLRVLVVSKCAELKHFFTPGVANTLKKLEHLEVYKCDNMEELIRSRGSEE- 829
Query: 835 SAYDYRLLLFPKLKKFHV----RECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQ 890
+ FPKLK + + G+ + + I L Q L++LE +I ++ +
Sbjct: 830 -----ETITFPKLKFLSLCGLPKLSGLCDNVKIIELPQ-LMELELDDIPGFTSIYPMKKF 883
Query: 891 STHNDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMV 950
T + + E+ I +L L ++ NL I WP +FN+ +F +
Sbjct: 884 ETFS--LLKEEVLIPKLEKLHVSSMWNLKEI-----------WP--CEFNMSEEVKFREI 928
Query: 951 SINTCMALHNNPRINEASHQ---TLQNITEVRVNNC-ELEGIFQL----VGLTNDGEKDP 1002
++ C L +N H+ L ++ E++V NC +E +F + VG T D +
Sbjct: 929 KVSNCDKL-----VNLFPHKPISLLHHLEELKVKNCGSIESLFNIHLDCVGATGDEYNNS 983
Query: 1003 LTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCM--AGGLP 1060
++++ + L L + + +L+++E+ C ++ +F+ + AG +
Sbjct: 984 GVRIIKVISCDKLVNL------FPHNPMSILHHLEELEVENCGSIESLFNIDLDCAGAIG 1037
Query: 1061 Q------LKALKIEKCNQLDQI 1076
Q L+ +K+E +L ++
Sbjct: 1038 QEDNSISLRNIKVENLGKLREV 1059
Score = 39.3 bits (90), Expect = 0.66
Identities = 94/439 (21%), Positives = 168/439 (37%), Gaps = 82/439 (18%)
Query: 566 LFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVI 625
LF K +++ +S VGDM LE I + S + + NLR+L +S+C ++ F
Sbjct: 744 LFKKTEVLCLS-VGDMNDLEDIEVKSSSQLLQS---SSFNNLRVLVVSKCAELKHFFTPG 799
Query: 626 ARHT--ELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLF 683
+T +LE L C + E+ + R G ++E + + F
Sbjct: 800 VANTLKKLEHLEVYKCDNMEEL----------IRSR----------GSEEETITFPKLKF 839
Query: 684 LSYLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVD 743
LS + L + +++ + + + IP F S+ +K+ + L +
Sbjct: 840 LSLCGLPK--LSGLCDNVKIIELPQLMELELDDIPG-FTSIYPMKKF-----ETFSLLKE 891
Query: 744 TCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLA 803
LI KL L + M +L ++ + +S + ++ +S+C KL LF
Sbjct: 892 EVLIP-------KLEKLHVSSMWNLKEIWPCEFNMSEEVK-FREIKVSNCDKLVNLFPHK 943
Query: 804 VAQNLAQLEKLQVLSCPELQHIL------IDDDRDEISAYDYRLL----------LFPK- 846
L LE+L+V +C ++ + + DE + R++ LFP
Sbjct: 944 PISLLHHLEELKVKNCGSIESLFNIHLDCVGATGDEYNNSGVRIIKVISCDKLVNLFPHN 1003
Query: 847 -------LKKFHVRECGVLEYIIPITL-AQGLVQLE----CLEIVCNENLKYVFGQSTHN 894
L++ V CG +E + I L G + E L + ENL +
Sbjct: 1004 PMSILHHLEELEVENCGSIESLFNIDLDCAGAIGQEDNSISLRNIKVENLGKLREVWRIK 1063
Query: 895 DGQNQNELKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINT 954
G N L + ++E + + ++ P+ FNL G +SI+
Sbjct: 1064 GGDNSRPL-VHGFQSVESIRVTKCKKFRNV-------FTPTTTNFNL---GALLEISIDD 1112
Query: 955 CMALHNNPRINEASHQTLQ 973
C N E+SH+ Q
Sbjct: 1113 CGENRGNDESEESSHEQEQ 1131
Score = 37.0 bits (84), Expect = 3.3
Identities = 19/77 (24%), Positives = 42/77 (53%), Gaps = 1/77 (1%)
Query: 1006 CLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKAL 1065
CL + + +L + S + ++ F NL+ + +S C LK F+ +A L +L+ L
Sbjct: 752 CLSVGDMNDLEDIEVKSSSQLLQSSS-FNNLRVLVVSKCAELKHFFTPGVANTLKKLEHL 810
Query: 1066 KIEKCNQLDQIVEDIGT 1082
++ KC+ +++++ G+
Sbjct: 811 EVYKCDNMEELIRSRGS 827
>UniRef100_Q9ZT68 Resistance protein candidate RGC2K [Lactuca sativa]
Length = 1715
Score = 260 bits (664), Expect = 2e-67
Identities = 291/1160 (25%), Positives = 513/1160 (44%), Gaps = 190/1160 (16%)
Query: 1 MEYLYG-FASAISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKK 59
ME + G F++ ++ L+ V L + +V D+ + L A +D V++R + +
Sbjct: 1 MECITGIFSNPFAQCLIAPVKEHLCLLIFYTQYVGDMLTAMTELNAAKDIVEERKNQNVE 60
Query: 60 QTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNW---IWRYSVGRKLSKK 116
+ + V +WL+D V+++L ++ + F C + + + +++
Sbjct: 61 KCFEVPNHVNRWLEDVQTINRKVERVL----NDNCNWFNLCNRYMLAVKALEITQEIDHA 116
Query: 117 KRNLKLYIEEGRQYIEIERPASLSAGYFSAERCW-EFDSRKPAYEELMCALKDDDVT-MI 174
+ L IE + + R S A + + +F+SR+ + + + AL + + M+
Sbjct: 117 MKQLSR-IEWTDDSVPLGRNDSTKASTSTPSSDYNDFESREHTFRKALEALGSNHTSHMV 175
Query: 175 GLYGMGGCGKTMLAMEVGKRCGNL------FDQVLFVPISSTVEVERIQEKIAGSLEFEF 228
L+GMGG GKT + KR N+ F ++ V I +++ IQ+ +A L+ +
Sbjct: 176 ALWGMGGVGKTTMM----KRLKNIIKEKRTFHYIVLVVIKENMDLISIQDAVADYLDMKL 231
Query: 229 QEKDEMDRSKRLCMRLTQE-----DRVLVILDDVWQMLDFDAIGI-PSIEHHKGCKILIT 282
E +E +R+ +L + +R L+ILDDVWQ ++ + IG+ P K+L+T
Sbjct: 232 TESNESERADKLREGFQAKSDGGKNRFLIILDDVWQSVNMEDIGLSPFPNQGVDFKVLLT 291
Query: 283 SRSEAVCTLMDCQKKI--QLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGL 340
S ++ VC M + + + LT +E LF + +S+ + + + I C GL
Sbjct: 292 SENKDVCAKMGVEANLIFDVKFLTEEEAQSLFYQFVKVSD---THLDKIGKAIVRNCGGL 348
Query: 341 PVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSL 400
P+A +A++LK + + WK AL R+ I + Q+SYDNL EEA+S+
Sbjct: 349 PIAIKTIANTLKNRNKDVWKDALSRIEHHDIETIA------HVVFQMSYDNLQNEEAQSI 402
Query: 401 FLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEG 460
FLLC +FPED +IP E L R GL + V++ AR+ + L S LL++ ++
Sbjct: 403 FLLCGLFPEDFDIPTEELVRYGWGLRVFNGVYTIGEARHRLNAYIELLKDSNLLIESDDV 462
Query: 461 KCVKMHDLVR---------------------NVAHWIAENEIKCASEKDIMTLEHTSLRY 499
C+KMHDLVR + W EN++ +S K I + +
Sbjct: 463 HCIKMHDLVRAFVLDTFNRFKHSLIVNHGNGGMLGW-PENDMSASSCKRISLICKGMSDF 521
Query: 500 LWCEKFPNSLDCSNLDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSL 559
KFPN L L + + + + M+ L+V+ + + PLL TS +
Sbjct: 522 PRDVKFPNLL---ILKLMHADKSLKFPQDFYGEMKKLQVI---SYDHMKYPLLPTSPQCS 575
Query: 560 TNLRCILFSKWDLV-DISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGME 618
TNLR + + L+ D S +G++ LE ++ + LP + L LR+LDL+ C
Sbjct: 576 TNLRVLHLHQCSLMFDCSSIGNLLNLEVLSFANSGIEWLPSTIGNLKELRVLDLTNCDGL 635
Query: 619 RNPFEVIARHTELEELF------------FAD--CR---------SKWEVEFLKEFSVP- 654
R V+ + +LEEL+ F D C S E EF K + P
Sbjct: 636 RIDNGVLKKLVKLEELYMRVGGRYQKAISFTDENCNEMAERSKNLSALEFEFFKNNAQPK 695
Query: 655 ----QVLQRYQIQLGSMFSG-FQDEFLNHHRTLFL--SYLDTSNAAIKDLAEKAEVLCIA 707
+ L+R++I +G F G F F + TL L + + + + +L EK +VL ++
Sbjct: 696 NMSFENLERFKISVGCYFKGDFGKIFHSFENTLRLVTNRTEVLESRLNELFEKTDVLYLS 755
Query: 708 GIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKH 767
G N + DV + HL
Sbjct: 756 ---VGDMNDLEDVEVKLAHL---------------------------------------- 772
Query: 768 LGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILI 827
P S F NL L IS C +L LFTL VA L++LE LQV C ++ I+
Sbjct: 773 ---------PKSSSFHNLRVLIISECIELRYLFTLDVANTLSKLEHLQVYECDNMEEIIH 823
Query: 828 DDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQ-GLVQLECLEIVCNENLKY 886
+ R E++ + FPKLK + CG +P L G V + L + L
Sbjct: 824 TEGRGEVT------ITFPKLKFLSL--CG-----LPNLLGLCGNVHIINLPQLTELKLNG 870
Query: 887 VFG-QSTHNDGQNQNELKIIELSAL--EELTLVNLPNINSICPEDCYLMWPSLLQFNLQ- 942
+ G S + + K +E S+L +E+ + NL ++ +D +WP L + +
Sbjct: 871 IPGFTSIYPE-------KDVETSSLLNKEVVIPNLEKLDISYMKDLKEIWPCELGMSQEV 923
Query: 943 NCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRVNNC-ELEGIFQL----VGLTND 997
+ ++ +++C L N N + ++ E++V C +E +F + +G +
Sbjct: 924 DVSTLRVIKVSSCDNLVNLFPCNPM--PLIHHLEELQVIFCGSIEVLFNIELDSIGQIGE 981
Query: 998 GEKDPLTSCLEMLYLENLPQLRYLCK-SSVESTNLL---FQNLQQMEISGCRRLKCIFSS 1053
G + S L ++ L+NL +L + + ++++LL FQ ++ + ++ C+ + +F+
Sbjct: 982 GINN---SSLRIIQLQNLGKLSEVWRIKGADNSSLLISGFQGVESIIVNKCKMFRNVFTP 1038
Query: 1054 CMAG-GLPQLKALKIEKCNQ 1072
L L ++I+ C +
Sbjct: 1039 TTTNFDLGALMEIRIQDCGE 1058
Score = 78.2 bits (191), Expect = 1e-12
Identities = 153/732 (20%), Positives = 279/732 (37%), Gaps = 155/732 (21%)
Query: 480 EIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQIHTYTQVSDEIFKGMRMLRVL 539
E+ + E D+ TL ++ C+ N C+ + + QV IF G + VL
Sbjct: 916 ELGMSQEVDVSTLR--VIKVSSCDNLVNLFPCNPMPLIHHLEELQV---IFCGS--IEVL 968
Query: 540 F--------LYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCD 591
F +G L L++L L + K + + +ESI +
Sbjct: 969 FNIELDSIGQIGEGINNSSLRIIQLQNLGKLSEVWRIKGADNSSLLISGFQGVESIIVNK 1028
Query: 592 CSFVELPDVVTQLTN------LRLLDLSECGMERNPFEVIARHTELEE------LFFADC 639
C +V T T L + + +CG +R E++ E E+ +F+ C
Sbjct: 1029 CKMFR--NVFTPTTTNFDLGALMEIRIQDCGEKRRNNELVESSQEQEQFYQAGGVFWTLC 1086
Query: 640 RSKWEVEFLKEFSVPQVLQRY---QIQ---------LGSMFSGFQDEFLNHHRTLFLSYL 687
+ E+ + +++ V+ Y Q+Q SM F+ + +N++ S
Sbjct: 1087 QYSREINIRECYALSSVIPCYAAGQMQNVQVLNIYRCNSMKELFETQGMNNNNGD--SGC 1144
Query: 688 DTSNAAIKDLAEKAEVLCIAGIE-------GGAKNIIP-DVFQSMNHLKELLIRDSKGIE 739
D N I + V+ + ++ G +++ S+ L+EL I K ++
Sbjct: 1145 DEGNGCIPAIPRLNNVIMLPNLKILKIEDCGHLEHVFTFSALGSLRQLEELTIEKCKAMK 1204
Query: 740 CLVD---------TCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYI 790
+V T + F +L + +E+++ L Y G+ + + +L+ + I
Sbjct: 1205 VIVKEEDEYGEQTTKASSKEVVVFPRLKSIELENLQELMGFYLGKNEIQ--WPSLDKVMI 1262
Query: 791 SHCPKLTRLFTLAVAQNLAQLEKL-----------QVLSCPELQHILIDDDRDEISAYDY 839
+CP++ A ++ K +VL + + D+ D+ +
Sbjct: 1263 KNCPEM---MVFAPGESTVPKRKYINTSFGIYGMEEVLETQGMNNNNDDNCCDDGNGGIP 1319
Query: 840 RL---LLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDG 896
RL ++FP +K + CG LE+I + + L+QL+ L I + +K + + D
Sbjct: 1320 RLNNVIMFPNIKILQISNCGSLEHIFTFSALESLMQLKELTIADCKAMKVIVKEEY--DV 1377
Query: 897 QNQNELKIIELSALEELTLVNLPNINSICPEDCYLMWPSL-------------------- 936
+ LK + S L+ +TL +LP + WPSL
Sbjct: 1378 EQTRVLKAVVFSCLKSITLCHLPELVGFFLGKNEFWWPSLDKVTIIDCPQMMGFTPGGST 1437
Query: 937 ----------LQFNLQNCGEFFMVS---------INTCMA--------LHNNPRI----- 964
L + CG F V+ +++C A HN I
Sbjct: 1438 TSHLKYIHSSLGKHTLECGLNFQVTTTAYHQTPFLSSCPATSEGMPWSFHNLIEISLMFN 1497
Query: 965 --------NEASHQTLQNITEVRVNNCE-LEGIFQLV--GLTNDGEKDPLTSCLEMLYLE 1013
NE H LQ + +V V +C +E +F+ + G + D ++ L
Sbjct: 1498 DVEKIIPSNELLH--LQKLEKVHVRHCNGVEEVFEALEAGANSSNGFDESLQTTTLVKLP 1555
Query: 1014 NLPQ--------LRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKAL 1065
NL Q LRY+ K++ + T F NL + I C L+ +F+S M G L QL+ L
Sbjct: 1556 NLTQVELEYLDCLRYIWKTN-QWTTFEFPNLTTVTIRECHGLEHVFTSSMVGSLLQLQEL 1614
Query: 1066 KIEKCNQLDQIV 1077
I C +++++
Sbjct: 1615 HIYNCKYMEEVI 1626
Score = 56.6 bits (135), Expect = 4e-06
Identities = 128/636 (20%), Positives = 232/636 (36%), Gaps = 138/636 (21%)
Query: 490 MTLEHTSLRYLWCEKFPNSLD-CSNLDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRER 548
+T+ L++L PN L C N+ + + T++ G +
Sbjct: 830 VTITFPKLKFLSLCGLPNLLGLCGNVHIINLPQLTELKLNGIPGFTSIY----------- 878
Query: 549 RPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLR 608
P SL N ++ + +DIS+ MK L+ I C+ + DV T LR
Sbjct: 879 -PEKDVETSSLLNKEVVI-PNLEKLDISY---MKDLKEIWPCELGMSQEVDVST----LR 929
Query: 609 LLDLSECG-----MERNPFEVIARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQ 663
++ +S C NP +I EL+ +F + +E
Sbjct: 930 VIKVSSCDNLVNLFPCNPMPLIHHLEELQVIFCGSIEVLFNIEL--------------DS 975
Query: 664 LGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIP----- 718
+G + G + L R + L L + + L I+G +G I+
Sbjct: 976 IGQIGEGINNSSL---RIIQLQNLGKLSEVWRIKGADNSSLLISGFQGVESIIVNKCKMF 1032
Query: 719 -DVFQ------SMNHLKELLIRD---SKGIECLVDTC-----LIEVGTLFFCKLHWLRIE 763
+VF + L E+ I+D + LV++ + G +F+ + R
Sbjct: 1033 RNVFTPTTTNFDLGALMEIRIQDCGEKRRNNELVESSQEQEQFYQAGGVFWTLCQYSREI 1092
Query: 764 HMKHLGALYNGQMPL--SGHFENLEDLYISHCPKLTRLF-TLAVAQNLAQLEKLQVLSC- 819
+++ AL + +P +G +N++ L I C + LF T + N + C
Sbjct: 1093 NIRECYAL-SSVIPCYAAGQMQNVQVLNIYRCNSMKELFETQGMNNNNGDSGCDEGNGCI 1151
Query: 820 ---PELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECL 876
P L +++ + P LK + +CG LE++ + L QLE L
Sbjct: 1152 PAIPRLNNVI----------------MLPNLKILKIEDCGHLEHVFTFSALGSLRQLEEL 1195
Query: 877 EIVCNENLKYVFGQSTHNDGQ--NQNELKIIELSALEELTLVNLPNINSICPEDCYLMWP 934
I + +K + + Q + +++ L+ + L NL + + WP
Sbjct: 1196 TIEKCKAMKVIVKEEDEYGEQTTKASSKEVVVFPRLKSIELENLQELMGFYLGKNEIQWP 1255
Query: 935 SLLQFNLQNCGEFFMVS-----------INTCMALHNNPRINEASHQTLQNITEVRVNNC 983
SL + ++NC E + + INT ++ + E Q + +NC
Sbjct: 1256 SLDKVMIKNCPEMMVFAPGESTVPKRKYINTSFGIYGMEEVLET-----QGMNNNNDDNC 1310
Query: 984 ELEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISG 1043
+G N G +P+L + ++F N++ ++IS
Sbjct: 1311 CDDG--------NGG----------------IPRLNNV---------IMFPNIKILQISN 1337
Query: 1044 CRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIVED 1079
C L+ IF+ L QLK L I C + IV++
Sbjct: 1338 CGSLEHIFTFSALESLMQLKELTIADCKAMKVIVKE 1373
Score = 49.7 bits (117), Expect = 5e-04
Identities = 60/256 (23%), Positives = 113/256 (43%), Gaps = 39/256 (15%)
Query: 784 NLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDD---RDEISAYDYR 840
NL+ L I C L +FT + +L QLE+L + C ++ I+ ++D A
Sbjct: 1165 NLKILKIEDCGHLEHVFTFSALGSLRQLEELTIEKCKAMKVIVKEEDEYGEQTTKASSKE 1224
Query: 841 LLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIV----CNENLKYVFGQSTHNDG 896
+++FP+LK + L+ ++ L + +Q L+ V C E + + G+ST
Sbjct: 1225 VVVFPRLKSIELEN---LQELMGFYLGKNEIQWPSLDKVMIKNCPEMMVFAPGESTVPKR 1281
Query: 897 QNQN-ELKIIELSALEELTLVNLPNINSICPE--------DCYLMWPSLLQFNLQNCGEF 947
+ N I + + E +N N ++ C + + +M+P++ + NCG
Sbjct: 1282 KYINTSFGIYGMEEVLETQGMNNNNDDNCCDDGNGGIPRLNNVIMFPNIKILQISNCGS- 1340
Query: 948 FMVSINTCMALHNNPRINEASHQTLQNITEVRVNNCELEGIFQLVGLTNDGE-----KDP 1002
+ I T AL ++L + E+ + +C+ + +V D E K
Sbjct: 1341 -LEHIFTFSAL-----------ESLMQLKELTIADCKAMKV--IVKEEYDVEQTRVLKAV 1386
Query: 1003 LTSCLEMLYLENLPQL 1018
+ SCL+ + L +LP+L
Sbjct: 1387 VFSCLKSITLCHLPEL 1402
Score = 46.2 bits (108), Expect = 0.005
Identities = 83/391 (21%), Positives = 151/391 (38%), Gaps = 74/391 (18%)
Query: 601 VTQLTNLRLLDLSECGMERNPFEVIARHT--ELEELFFADCRSKWEVEFLKEFSVPQVLQ 658
V N+++L +S CG + F A + +L+EL ADC++ +V +E+ V Q
Sbjct: 1324 VIMFPNIKILQISNCGSLEHIFTFSALESLMQLKELTIADCKAM-KVIVKEEYDVEQT-- 1380
Query: 659 RYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAEKAEVLC--IAGIEGGAKNI 716
++ +FS + L H L +L + L + + C + G G
Sbjct: 1381 --RVLKAVVFSCLKSITLCHLPELVGFFLGKNEFWWPSLDKVTIIDCPQMMGFTPGGS-- 1436
Query: 717 IPDVFQSMNHLKELLIRDSKG---IEC-----LVDTCLIEVGTLFFCK-----LHWLRIE 763
+ +HLK I S G +EC + T + L C + W
Sbjct: 1437 ------TTSHLK--YIHSSLGKHTLECGLNFQVTTTAYHQTPFLSSCPATSEGMPW-SFH 1487
Query: 764 HMKHLGALYNGQMPLSG-----HFENLEDLYISHCPKLTRLF------------------ 800
++ + ++N + H + LE +++ HC + +F
Sbjct: 1488 NLIEISLMFNDVEKIIPSNELLHLQKLEKVHVRHCNGVEEVFEALEAGANSSNGFDESLQ 1547
Query: 801 --TLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVL 858
TL NL Q+E L+ L C L++I + FP L +REC L
Sbjct: 1548 TTTLVKLPNLTQVE-LEYLDC--LRYIW--------KTNQWTTFEFPNLTTVTIRECHGL 1596
Query: 859 EYIIPITLAQGLVQLECLEIVCNENLKYVFGQST-----HNDGQNQNELKIIELSALEEL 913
E++ ++ L+QL+ L I + ++ V + + + ++ K I L L+ +
Sbjct: 1597 EHVFTSSMVGSLLQLQELHIYNCKYMEEVIARDADVVEEEEEDDDDDKRKDITLPFLKTV 1656
Query: 914 TLVNLPNINSICPEDCYLMWPSLLQFNLQNC 944
TL +LP + +P L +++ C
Sbjct: 1657 TLASLPRLKGFWLGKEDFSFPLLDTLSIEEC 1687
>UniRef100_Q9ZT67 Resistance protein candidate RGC20 [Lactuca sativa]
Length = 1758
Score = 251 bits (642), Expect = 7e-65
Identities = 311/1292 (24%), Positives = 544/1292 (42%), Gaps = 248/1292 (19%)
Query: 1 MEYLYGFASAISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQ 60
M+ + ++ L+ V L Y V+D++ + L A R + +D + R +
Sbjct: 1 MDVVNAILKPVAETLMEPVKKHLGYIISSTKHVRDMSNKMRELNAARHAEEDHLDRNIRT 60
Query: 61 TRKTAEVVEKWLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNL 120
+ + V WL++ ++ +D ++ S+ +C C I ++ VGR+ L
Sbjct: 61 RLEISNQVRSWLEE----VEKIDAKVKALPSDVTAC---CSLKI-KHEVGREA------L 106
Query: 121 KLYIE---EGRQY---------IEIERPASLSAGYFSAERCW-EFDSRKPAYEELMCALK 167
KL +E RQ+ I + + S+ A +A + +F SR+ + + + AL+
Sbjct: 107 KLIVEIESATRQHSLITWTDHPIPLGKVDSMKASMSTASTDYNDFQSREKTFTQALKALE 166
Query: 168 DDDVT-MIGLYGMGGCGKTMLAMEVGK--RCGNLFDQVLFVPISSTVEVERIQEKIAGSL 224
++ + MI L GMGG GKT + + K + +F ++ I + IQ+ +A L
Sbjct: 167 PNNASHMIALCGMGGVGKTTMMQRLKKVAKQNRMFSYMVEAVIGEKTDPIAIQQAVADYL 226
Query: 225 EFEFQEKDEMDRSKRLCMRLTQ-----EDRVLVILDDVWQMLDFDAIGI-PSIEHHKGCK 278
E +E + R+ +L +++ LVILDDVWQ +D + IG+ P K
Sbjct: 227 RIELKESTKPARADKLREWFKANSGEGKNKFLVILDDVWQSVDLEDIGLSPFPNQGVDFK 286
Query: 279 ILITSRSEAVCTLMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNE 336
+L+TSR E VCT+M + + L E LFQ+ SE + + +I +
Sbjct: 287 VLLTSRDEHVCTVMGVGSNSILNVGLLIEAEAQSLFQQFVETSEP---ELHKIGEDIVRK 343
Query: 337 CKGLPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEE 396
C GLP+A +A +L+ K + WK AL R+ N+ K + SY NL +E
Sbjct: 344 CCGLPIAIKTMACTLRNKRKDAWKDALSRIEHYDLRNVAP------KVFETSYHNLHDKE 397
Query: 397 AKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLD 456
KS+FL+C +FPED IP E L R GL I V+++ ARN + +L+ + LL++
Sbjct: 398 TKSVFLMCGLFPEDFNIPTEELMRYGWGLKIFDRVYTFIEARNRINTCIERLVQTNLLIE 457
Query: 457 VNEGKCVKMHDLVR-------------------NVAHWIAENEIKCASEKDIMTLEHTSL 497
++ CVKMHDLVR N+ W EN+ + + +T E S
Sbjct: 458 SDDVGCVKMHDLVRAFVLGMYSEVEHASVVNHGNIPGW-TENDPTDSCKAISLTCESMSG 516
Query: 498 RYLWCEKFPNSLDCSNLDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLK 557
KFPN + L + + + ++GM L+V+ + + + P+L S +
Sbjct: 517 NIPGDFKFPN---LTILKLMHGDKSLRFPQDFYEGMEKLQVI---SYDKMKYPMLPLSPQ 570
Query: 558 SLTNLRCILFSKWDL--VDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSEC 615
TNLR + + L D S +G+M +E ++ + LP + L LRLLDL++C
Sbjct: 571 CSTNLRVLHLHECSLKMFDCSCIGNMANVEVLSFANSGIEMLPSTIGNLKKLRLLDLTDC 630
Query: 616 GMERNPFEVIARHTELEELF--FADC--------------------RSK----WEVEFLK 649
V +LEEL+ F+D RSK E +F +
Sbjct: 631 HGLHITHGVFNNLVKLEELYMGFSDRPDQTRGNISMTDVSYNELAERSKGLSALEFQFFE 690
Query: 650 EFSVPQ-----VLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKD-----LAE 699
+ P L+R++I +G G D F + L T+ + D L
Sbjct: 691 NNAQPNNMSFGKLKRFKISMGCTLYGGSDYFKKTYAVQNTLKLVTNKGELLDSRMNELFV 750
Query: 700 KAEVLCIAGIE---------GGAKNIIPDVF-----------------------QSMNHL 727
+ E+LC++ + +++ P VF + +++L
Sbjct: 751 ETEMLCLSVDDMNDLGDVCVKSSRSPQPSVFKILRVFVVSKCVELRYLFTIGVAKDLSNL 810
Query: 728 KELLIRDSKGIECLVDTCLIEVG--TLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENL 785
+ L + +E L+ C+ G T+ F KL L + +G LSG +N+
Sbjct: 811 EHLEVDSCNNMEQLI--CIENAGKETITFLKLKILSL----------SGLPKLSGLCQNV 858
Query: 786 EDLYISHCPKLTRL-------FTLAVAQNLAQLEKL--QVLSCPELQHILIDDDRDEISA 836
L + P+L L FT QN + L + + P+L+ + ID+ +
Sbjct: 859 NKLEL---PQLIELKLKGIPGFTCIYPQNKLETSSLLKEEVVIPKLETLQIDEMENLKEI 915
Query: 837 YDYRLLLFP--KLKKFHVRECGVLEYII---PITLAQGLVQLE-----CLEIVCNENLKY 886
+ Y++ KL+K V C L + P++L L +LE +E + N +L
Sbjct: 916 WHYKVSNGERVKLRKIEVSNCDKLVNLFPHNPMSLLHHLEELEVKKCGSIESLFNIDLDC 975
Query: 887 VFGQSTHNDGQNQNELKIIELSALEELTLVNLPN-----------INSICPEDC----YL 931
V ++ ++ +K+ L E+ + N + SI E C +
Sbjct: 976 VDAIGEEDNMRSLRNIKVKNSWKLREVWCIKGENNSCPLVSGFQAVESISIESCKRFRNV 1035
Query: 932 MWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQ-----------TLQNITE--- 977
P+ FN+ G +SI+ C N + ++S + LQ +T+
Sbjct: 1036 FTPTTTNFNM---GALLEISIDDCGEYMENEKSEKSSQEQEQTDILSEEVKLQEVTDTIS 1092
Query: 978 ---------------VRVNNCE----LEGIFQLVGLTN----------DGEKDPLTSCLE 1008
+R N E +E +F++ T+ ++ P+ LE
Sbjct: 1093 NVVFTSCLIHSFYNNLRKLNLEKYGGVEVVFEIESSTSRELVTTYHKQQQQQQPIFPNLE 1152
Query: 1009 MLYLENLPQLRYLCKSS-----VESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLK 1063
LYL + + ++ K + ++ + F NL + +S C+ +K +FS MA L LK
Sbjct: 1153 ELYLYYMDNMSHVWKCNNWNKFLQQSESPFHNLTTIHMSDCKSIKYLFSPLMAELLSNLK 1212
Query: 1064 ALKIEKCNQLDQIV---EDIGTAFPSGTQTST 1092
+ I++C+ +++IV +D+ + T +ST
Sbjct: 1213 RINIDECDGIEEIVSKRDDVDEEMTTSTHSST 1244
Score = 75.1 bits (183), Expect = 1e-11
Identities = 84/364 (23%), Positives = 143/364 (39%), Gaps = 82/364 (22%)
Query: 779 SGHFENLEDLYISHCPKLTRLF-TLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAY 837
SG + L L I C + +F T + N K C D+ DEI
Sbjct: 1310 SGQMQKLRVLKIERCKGVKEVFETQGICSN-----KNNKSGC--------DEGNDEIPRV 1356
Query: 838 DYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQ 897
+ +++ P L + +CG LE+I + + L QLE L I+ ++K + + +
Sbjct: 1357 N-SIIMLPNLMILEISKCGSLEHIFTFSALESLRQLEELMILDCGSMKVIVKEEHASSSS 1415
Query: 898 NQNELKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEF---------- 947
+ + +++ L+ + L NLP + WPSL ++NC +
Sbjct: 1416 SSSSKEVVVFPRLKSIKLFNLPELEGFFLGMNEFQWPSLAYVVIKNCPQMTVFAPGGSTA 1475
Query: 948 -FMVSINTCMALHNNPRI-----NEASHQT------------------------------ 971
+ I+T + H+ N A HQT
Sbjct: 1476 PMLKHIHTTLGKHSLGESGLNFHNVAHHQTPFPSLHGAISCPVTTEGMRWSFHNLIELDV 1535
Query: 972 -----------------LQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCLEMLYLE 1013
LQ + ++ V C LE +F+ L + L + L + L+
Sbjct: 1536 GCNRDVKKIIPSSEMLQLQKLEKIHVRYCHGLEEVFE-TALESATTVFNLPN-LRHVELK 1593
Query: 1014 NLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQL 1073
+ LRY+ KS+ + T F NL +++I GC RL+ +F+S M G L QL+ L I C +
Sbjct: 1594 VVSALRYIWKSN-QWTVFDFPNLTRVDIRGCERLEHVFTSSMVGSLLQLQELHIRDCYHM 1652
Query: 1074 DQIV 1077
++I+
Sbjct: 1653 EEII 1656
Score = 45.8 bits (107), Expect = 0.007
Identities = 78/412 (18%), Positives = 160/412 (37%), Gaps = 81/412 (19%)
Query: 724 MNHLKELLIRDSKGIECLVDT---CLIEVGTLFFCK-LHWLRIEHMKHLGALY-----NG 774
++HL+EL ++ IE L + C+ +G + L +++++ L ++ N
Sbjct: 951 LHHLEELEVKKCGSIESLFNIDLDCVDAIGEEDNMRSLRNIKVKNSWKLREVWCIKGENN 1010
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQ-NLAQLEKLQVLSCPEL----------- 822
PL F+ +E + I C + +FT N+ L ++ + C E
Sbjct: 1011 SCPLVSGFQAVESISIESCKRFRNVFTPTTTNFNMGALLEISIDDCGEYMENEKSEKSSQ 1070
Query: 823 ---------QHILIDDDRDEISAYDYRLLL----FPKLKKFHVRECGVLEYIIPITLAQG 869
+ + + + D IS + L + L+K ++ + G +E + I +
Sbjct: 1071 EQEQTDILSEEVKLQEVTDTISNVVFTSCLIHSFYNNLRKLNLEKYGGVEVVFEIESSTS 1130
Query: 870 LVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNLPNINSICPEDC 929
E+V + H Q Q + LEEL L + N++ + +
Sbjct: 1131 R------ELVT----------TYHKQQQQQQPI----FPNLEELYLYYMDNMSHVWKCN- 1169
Query: 930 YLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRVNNCELEGIF 989
W LQ + + ++ C ++ + + L N+ + ++ C+ GI
Sbjct: 1170 --NWNKFLQQSESPFHNLTTIHMSDCKSIKY--LFSPLMAELLSNLKRINIDECD--GIE 1223
Query: 990 QLVGLTNDGEKDPLTSC---------LEMLYLENLPQLR-------YLCKSSVESTNL-- 1031
++V +D +++ TS L+ L L L L+ +L + +
Sbjct: 1224 EIVSKRDDVDEEMTTSTHSSTILFPHLDSLTLFRLDNLKCIGGGGAFLDRFKFSQAGVVC 1283
Query: 1032 --LFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIVEDIG 1081
L Q +++EI C L + +G + +L+ LKIE+C + ++ E G
Sbjct: 1284 WSLCQYSREIEIRSCHALSSVIPCYASGQMQKLRVLKIERCKGVKEVFETQG 1335
Score = 45.1 bits (105), Expect = 0.012
Identities = 31/131 (23%), Positives = 61/131 (45%), Gaps = 2/131 (1%)
Query: 708 GIEGGAKNIIPDV-FQSMNHLKELLIRDSKGIECLVDTCLIEVGTLF-FCKLHWLRIEHM 765
G K IIP + L+++ +R G+E + +T L T+F L + ++ +
Sbjct: 1536 GCNRDVKKIIPSSEMLQLQKLEKIHVRYCHGLEEVFETALESATTVFNLPNLRHVELKVV 1595
Query: 766 KHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHI 825
L ++ F NL + I C +L +FT ++ +L QL++L + C ++ I
Sbjct: 1596 SALRYIWKSNQWTVFDFPNLTRVDIRGCERLEHVFTSSMVGSLLQLQELHIRDCYHMEEI 1655
Query: 826 LIDDDRDEISA 836
++ D ++ A
Sbjct: 1656 IVKDANVDVEA 1666
Score = 41.2 bits (95), Expect = 0.17
Identities = 41/173 (23%), Positives = 77/173 (43%), Gaps = 17/173 (9%)
Query: 783 ENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHI---LIDDDRDEISAYDY 839
+ LE +++ +C L +F A+ V + P L+H+ ++ R + +
Sbjct: 1554 QKLEKIHVRYCHGLEEVFETALESATT------VFNLPNLRHVELKVVSALRYIWKSNQW 1607
Query: 840 RLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQ 899
+ FP L + +R C LE++ ++ L+QL+ L I +++ + + + D + +
Sbjct: 1608 TVFDFPNLTRVDIRGCERLEHVFTSSMVGSLLQLQELHIRDCYHMEEIIVKDANVDVEAE 1667
Query: 900 NE----LKIIELSALEELTLVNLPNIN--SICPEDCYLMWPSLLQFNLQNCGE 946
E I L L+ LTL LP + S+ ED +P L + NC E
Sbjct: 1668 EESDGKTNEIVLPCLKSLTLGWLPCLKGFSLGKED--FSFPLLDTLEINNCPE 1718
Score = 40.8 bits (94), Expect = 0.23
Identities = 53/250 (21%), Positives = 107/250 (42%), Gaps = 37/250 (14%)
Query: 844 FPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQ--NQNE 901
F L H+ +C ++Y+ +A+ L L+ + I + ++ + + D +
Sbjct: 1182 FHNLTTIHMSDCKSIKYLFSPLMAELLSNLKRINIDECDGIEEIVSKRDDVDEEMTTSTH 1241
Query: 902 LKIIELSALEELTLVNLPNINSICPEDCYL-----------MWPSLLQFNLQNCGEFFMV 950
I L+ LTL L N+ I +L W SL Q++ + +
Sbjct: 1242 SSTILFPHLDSLTLFRLDNLKCIGGGGAFLDRFKFSQAGVVCW-SLCQYSRE-------I 1293
Query: 951 SINTCMALHNNPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCLEM 1009
I +C AL + I + +Q + +++ C+ ++ +F+ G+ ++ K+ + C E
Sbjct: 1294 EIRSCHALSSV--IPCYASGQMQKLRVLKIERCKGVKEVFETQGICSN--KNNKSGCDEG 1349
Query: 1010 LYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEK 1069
+ +P++ + ++ NL +EIS C L+ IF+ L QL+ L I
Sbjct: 1350 N--DEIPRVNSI---------IMLPNLMILEISKCGSLEHIFTFSALESLRQLEELMILD 1398
Query: 1070 CNQLDQIVED 1079
C + IV++
Sbjct: 1399 CGSMKVIVKE 1408
>UniRef100_Q9ZSC9 Resistance protein candidate RGC2S [Lactuca sativa]
Length = 1813
Score = 247 bits (631), Expect = 1e-63
Identities = 283/1131 (25%), Positives = 484/1131 (42%), Gaps = 151/1131 (13%)
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I++ + V + Y +V+ + + + L R SV++ ++R + + ++
Sbjct: 15 IAQRALVPVTDHVGYMISCRKYVRVMQTKMTELNTSRISVEEHISRNTRNHLQIPSQIKD 74
Query: 71 WLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSK-------KKRNLKLY 123
WL NV+ + C + R+ +G+K K R L L
Sbjct: 75 WLDQVEGIRANVENFPIDVIT--------CCSLRIRHKLGQKAFKITEQIESLTRQLSL- 125
Query: 124 IEEGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALK-DDDVTMIGLYGMGGC 182
I + + R S++A SA +F SR+ + + + AL+ + M+ L GMGG
Sbjct: 126 ISWTDDPVPLGRVGSMNAST-SASSSDDFPSREKTFTQALKALEPNQQFHMVALCGMGGV 184
Query: 183 GKTMLAMEVGKRCGN--LFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRL 240
GKT + + K LF+ ++ I + IQE IA L + EK + R+ +L
Sbjct: 185 GKTRMMQRLKKAAEEKKLFNYIVRAVIGEKTDPFAIQEAIADYLGIQLNEKTKPARADKL 244
Query: 241 CMRLTQED-----RVLVILDDVWQMLDFDAIGI-PSIEHHKGCKILITSRSEAVCTLMDC 294
+ + L++LDDVWQ++D + IG+ P K+L+TSR VCT+M
Sbjct: 245 REWFKKNSDGGKTKFLIVLDDVWQLVDLEDIGLSPFPNQGVDFKVLLTSRDSQVCTMMGV 304
Query: 295 QKK--IQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLK 352
+ I + LT E LFQ+ SE ++ + +I +C GLP+A +A +L+
Sbjct: 305 EANSIINVGLLTEAEAQSLFQQFVETSEP---ELQKIGEDIVRKCCGLPIAIKTMACTLR 361
Query: 353 GKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCE 412
K + WK AL R+ N+ K + SY NL EE KS FL+C +FPED +
Sbjct: 362 NKRKDAWKDALSRIEHYDIHNVAP------KVFETSYHNLQEEETKSTFLMCGLFPEDFD 415
Query: 413 IPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVR-- 470
IP E L R GL + V++ AR + +L+ + LL++ ++ CVKMHDLVR
Sbjct: 416 IPTEELMRYGWGLKLFDRVYTIREARTRLNTCIERLVQTNLLIESDDVGCVKMHDLVRAF 475
Query: 471 -----------------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSN 513
N+ W EN+I + ++ +T + S KFP N
Sbjct: 476 VLGMFSEVEHASIVNHGNMPEW-TENDITDSCKRISLTCKSMS-------KFPGDFKFPN 527
Query: 514 LDFLQI---HTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKW 570
L L++ + + ++GM L V+ + + + PLL + + TN+R + +K
Sbjct: 528 LMILKLMHGDKSLRFPQDFYEGMEKLHVI---SYDKMKYPLLPLAPRCSTNIRVLHLTKC 584
Query: 571 DL--VDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARH 628
L D S +G++ LE ++ + LP V L LRLLDL C R V+
Sbjct: 585 SLKMFDCSCIGNLSNLEVLSFANSRIEWLPSTVRNLKKLRLLDLRFCDGLRIEQGVLKSL 644
Query: 629 TELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNH--HRTLFLSY 686
+LEE + + SGF D+ N R+ LS
Sbjct: 645 VKLEEFYIGNA-----------------------------SGFIDDNCNEMAERSDNLSA 675
Query: 687 LD----TSNAAIKDLA----EKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGI 738
L+ + A +K+++ E+ ++ +G I S ++ +L+ +
Sbjct: 676 LEFAFFNNKAEVKNMSFENLERFKISVGRSFDGN----INMSSHSYENMLQLVTNKGDVL 731
Query: 739 ECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTR 798
+ ++ ++ LF +H + + + + Q S F NL+ L IS C +L
Sbjct: 732 DSKLNGLFLKTKVLFL-SVHGMNDLEDVEVKSTHPTQ---SSSFCNLKVLIISKCVELRY 787
Query: 799 LFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVL 858
LF L +A L++LE L+V C ++ ++ + FPKLK + + L
Sbjct: 788 LFKLNLANTLSRLEHLEVCECENMEELI------HTGICGEETITFPKLKFLSLSQLPKL 841
Query: 859 EYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNL 918
+ GL L L + ++ Q+ E +I LE L + ++
Sbjct: 842 SSLCHNVNIIGLPHLVDLILKGIPGFTVIYPQNKLRTSSLLKEEVVI--PKLETLQIDDM 899
Query: 919 PNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEV 978
N+ I P C L ++ + +++C L N N S L ++ E+
Sbjct: 900 ENLEEIWP--CELSGGEKVKLR--------EIKVSSCDKLVNLFPRNPMS--LLHHLEEL 947
Query: 979 RVNNC-ELEGIFQL----VGLTNDGEKDPLTSCLEMLYLENLPQLRYLCK-SSVESTNLL 1032
+V NC +E +F + VG GE+D S L + +ENL +LR + + ++++L+
Sbjct: 948 KVKNCGSIESLFNIDLDCVGAI--GEEDN-KSLLRSINMENLGKLREVWRIKGADNSHLI 1004
Query: 1033 --FQNLQQMEISGCRRLKCIFSSCMAG-GLPQLKALKIEKCNQLDQIVEDI 1080
FQ ++ ++I C+R IF+ A L L ++IE C + E I
Sbjct: 1005 NGFQAVESIKIEKCKRFSNIFTPITANFYLVALLEIQIEGCGGNHESEEQI 1055
Score = 68.2 bits (165), Expect = 1e-09
Identities = 82/401 (20%), Positives = 151/401 (37%), Gaps = 112/401 (27%)
Query: 787 DLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILI---------------DDDR 831
++ IS C L+ + A + +L+ L+V C ++ + D+
Sbjct: 1298 EIEISKCNVLSSVIPCYAAGQMQKLQVLRVTGCDGMKEVFETQLGTSSNKNRKGGGDEGN 1357
Query: 832 DEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQS 891
I + +++ P LK + CG LE+I + + L QL+ L+IV +K + +
Sbjct: 1358 GGIPRVNNNVIMLPNLKTLKIYMCGGLEHIFTFSALESLTQLQELKIVGCYGMKVIVKKE 1417
Query: 892 THNDGQNQ-----------------NELKIIELSALEELTLVNLPNINSICPEDCYLMWP 934
G+ Q + K++ L+ + L NLP + P
Sbjct: 1418 EDEYGEQQTTTTTTTKGASSSSSSSSSKKVVVFPRLKSIELFNLPELVGFFLGMNEFRLP 1477
Query: 935 SLLQFNLQNCGEFFMVS-----------INTCMALH----------------------NN 961
SL + ++ C + + + I+T + H +
Sbjct: 1478 SLEEVTIKYCSKMMVFAAGGSTAPQLKYIHTRLGKHTLDQESGLNFHQTSFQSLYGDTSG 1537
Query: 962 PRINEASHQTLQNITE--------------------------VRVNNCE-LEGIFQLV-- 992
P +E + + N+ E + V++C +E +F+
Sbjct: 1538 PATSEGTTWSFHNLIELDMELNYDVKKIIPSSELLQLQKLEKIHVSSCYWVEEVFETALE 1597
Query: 993 --------GLTNDGEKDPLTSCLEMLYLENLPQ--------LRYLCKSSVESTNLLFQNL 1036
G+ D E T+ + L NL + LRY+ KS+ + T F NL
Sbjct: 1598 AAGRNGNSGIGFD-ESSQTTTTTTLFNLRNLREMKLHFLRGLRYIWKSN-QWTAFEFPNL 1655
Query: 1037 QQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIV 1077
++ IS CRRL+ +F+S M G L QL+ L I CN +++++
Sbjct: 1656 TRVHISRCRRLEHVFTSSMVGSLLQLQELDISWCNHMEEVI 1696
Score = 45.8 bits (107), Expect = 0.007
Identities = 47/216 (21%), Positives = 92/216 (41%), Gaps = 34/216 (15%)
Query: 672 QDEFLNHHRTLFLS-YLDTSNAAIKDLAEKAEVLCIA---GIEGGAKNIIPDV-FQSMNH 726
Q+ LN H+T F S Y DTS A + + I + K IIP +
Sbjct: 1517 QESGLNFHQTSFQSLYGDTSGPATSEGTTWSFHNLIELDMELNYDVKKIIPSSELLQLQK 1576
Query: 727 LKELLIRDSKGIECLVDTCLIEVG-------------------TLFFCK-LHWLRIEHMK 766
L+++ + +E + +T L G TLF + L +++ ++
Sbjct: 1577 LEKIHVSSCYWVEEVFETALEAAGRNGNSGIGFDESSQTTTTTTLFNLRNLREMKLHFLR 1636
Query: 767 HLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHIL 826
L ++ + F NL ++IS C +L +FT ++ +L QL++L + C ++ ++
Sbjct: 1637 GLRYIWKSNQWTAFEFPNLTRVHISRCRRLEHVFTSSMVGSLLQLQELDISWCNHMEEVI 1696
Query: 827 I---------DDDRDEISAYDYRLLLFPKLKKFHVR 853
+ D +R+ + +L+ P+LK ++
Sbjct: 1697 VKDADVSVEEDKERESDGKTNKEILVLPRLKSLKLK 1732
Score = 42.7 bits (99), Expect = 0.060
Identities = 121/608 (19%), Positives = 231/608 (37%), Gaps = 117/608 (19%)
Query: 503 EKFPNSLDCSNLDFLQIHTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNL 562
E S + S L+F + +V + F+ + ++ + GR + S S N+
Sbjct: 665 EMAERSDNLSALEFAFFNNKAEVKNMSFENLERFKI----SVGRSFDGNINMSSHSYENM 720
Query: 563 RCILFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLT---------NLRLLDLS 613
++ +K D++D G K + + L +L DV + T NL++L +S
Sbjct: 721 LQLVTNKGDVLDSKLNGLFLKTKVLFLSVHGMNDLEDVEVKSTHPTQSSSFCNLKVLIIS 780
Query: 614 ECGMERNPFEVIARHT--ELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGF 671
+C R F++ +T LE L +C + E+ I G
Sbjct: 781 KCVELRYLFKLNLANTLSRLEHLEVCECENMEEL----------------IHTGIC---- 820
Query: 672 QDEFLNHHRTLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELL 731
+E + + FLS K LC NII + HL +L+
Sbjct: 821 GEETITFPKLKFLSLSQLP---------KLSSLC------HNVNII-----GLPHLVDLI 860
Query: 732 IRDSKGIECLVDTCLIEVGTLF-----FCKLHWLRIEHMKHLGALYNGQMPLSGHFENLE 786
++ G + + +L KL L+I+ M++L ++ ++ G L
Sbjct: 861 LKGIPGFTVIYPQNKLRTSSLLKEEVVIPKLETLQIDDMENLEEIWPCELS-GGEKVKLR 919
Query: 787 DLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAY---DYRLLL 843
++ +S C KL LF L LE+L+V +C ++ L + D D + A D + LL
Sbjct: 920 EIKVSSCDKLVNLFPRNPMSLLHHLEELKVKNCGSIES-LFNIDLDCVGAIGEEDNKSLL 978
Query: 844 FPKLKKFHVRECGVLEYIIPIT------LAQGLVQLECLEIVCNENLKYVFGQSTHN--- 894
+ ++ G L + I L G +E ++I + +F T N
Sbjct: 979 ----RSINMENLGKLREVWRIKGADNSHLINGFQAVESIKIEKCKRFSNIFTPITANFYL 1034
Query: 895 -------------DGQNQNELKII-ELSALEELTLVNLPNINSICPEDCYLMWPSLLQFN 940
+ +++ +++I+ E L+E+T N+ N + P + +L +
Sbjct: 1035 VALLEIQIEGCGGNHESEEQIEILSEKETLQEVTDTNISNDVVLFPSCLMHSFHNLHKLK 1094
Query: 941 LQNCGEFFMV--------SINTCMALHNN-------PRINEASHQTLQNITEV-RVNNCE 984
L+ +V + + H+N P + E + N++ V + +N
Sbjct: 1095 LERVKGVEVVFEIESESPTSRELVTTHHNQQHPIILPNLQELDLSFMDNMSHVWKCSNWN 1154
Query: 985 LEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGC 1044
+ L + P + L +++ + ++YL + L NL+ + ISGC
Sbjct: 1155 -----KFFTLPKQQSESPFHN-LTTIHMFSCRSIKYLFSPLMAE---LLSNLKDIWISGC 1205
Query: 1045 RRLKCIFS 1052
+K + S
Sbjct: 1206 NGIKEVVS 1213
>UniRef100_Q9ZSD1 Resistance protein candidate RGC2B [Lactuca sativa]
Length = 1810
Score = 243 bits (621), Expect = 2e-62
Identities = 286/1128 (25%), Positives = 482/1128 (42%), Gaps = 141/1128 (12%)
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I++ + V + Y +V+ + + + L R SV++ ++R + + ++
Sbjct: 15 IAQTALVPVTDHVGYMISCRKYVRVMQMKMTELNTSRISVEEHISRNTRNHLQIPSQTKE 74
Query: 71 WLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSK-------KKRNLKLY 123
WL NV+ + C + R+ +G+K K R L L
Sbjct: 75 WLDQVEGIRANVENFPIDVIT--------CCSLRIRHKLGQKAFKITEQIESLTRQLSL- 125
Query: 124 IEEGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDD-VTMIGLYGMGGC 182
I + + R S++A SA +F SR+ + + + AL+ + M+ L GMGG
Sbjct: 126 ISWTDDPVPLGRVGSMNAST-SASLSDDFPSREKTFTQALKALEPNQKFHMVALCGMGGV 184
Query: 183 GKTMLAMEVGKRCGN--LFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRL 240
GKT + + K LF+ ++ I + IQE IA L + EK + R+ +L
Sbjct: 185 GKTRMMQRLKKAAEEKKLFNYIVGAVIGEKTDPFAIQEAIADYLGIQLNEKTKPARADKL 244
Query: 241 CMRLTQED-----RVLVILDDVWQMLDFDAIGI-PSIEHHKGCKILITSRSEAVCTLMDC 294
+ + L++LDDVWQ++D + IG+ P K+L+TSR VCT+M
Sbjct: 245 REWFKKNSDGGKTKFLIVLDDVWQLVDLEDIGLSPFPNQGVDFKVLLTSRDSQVCTMMGV 304
Query: 295 QKK--IQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLK 352
+ I + LT E LFQ+ SE ++ + +I +C GLP+A +A +L+
Sbjct: 305 EANSIINVGLLTEAEAQSLFQQFVETSEP---ELQKIGEDIVRKCCGLPIAIKTMACTLR 361
Query: 353 GKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCE 412
K + WK AL R+ N+ K + SY NL EE KS FL+C +FPED +
Sbjct: 362 NKRKDAWKDALSRIEHYDIHNVAP------KVFETSYHNLQEEETKSTFLMCGLFPEDFD 415
Query: 413 IPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVR-- 470
IP E L R GL + V++ AR + +L+ + LL++ ++ CVKMHDLVR
Sbjct: 416 IPTEELMRYGWGLKLFDRVYTIREARTRLNTCIERLVQTNLLIESDDVGCVKMHDLVRAF 475
Query: 471 -----------------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSN 513
N+ W EN++ S K I SL + P L
Sbjct: 476 VLGMFSEVEHASIVNHGNMPGWPDENDMIVHSCKRI------SLTCKGMIEIPVDLKFPK 529
Query: 514 LDFLQI---HTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKW 570
L L++ + + ++GM L V+ + + + PLL + + TN+R + ++
Sbjct: 530 LTILKLMHGDKSLRFPQDFYEGMEKLHVI---SYDKMKYPLLPLAPRCSTNIRVLHLTEC 586
Query: 571 DL--VDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARH 628
L D S +G++ LE ++ + LP V L LRLLDL C R V+
Sbjct: 587 SLKMFDCSSIGNLSNLEVLSFANSHIEWLPSTVRNLKKLRLLDLRFCDGLRIEQGVLKSF 646
Query: 629 TELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNH--HRTLFLSY 686
+LEE + D SGF D+ N R+ LS
Sbjct: 647 VKLEEFYIGDA-----------------------------SGFIDDNCNEMAERSYNLSA 677
Query: 687 LD----TSNAAIKDLA-EKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECL 741
L+ + A +K+++ E E I+ +NI S ++ +L+ ++
Sbjct: 678 LEFAFFNNKAEVKNMSFENLERFKISVGCSFDENINMS-SHSYENMLQLVTNKGDVLDSK 736
Query: 742 VDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFT 801
++ ++ LF +H + + + + Q S F NL+ L IS C +L LF
Sbjct: 737 LNGLFLKTEVLFL-SVHGMNDLEDVEVKSTHPTQ---SSSFCNLKVLIISKCVELRYLFK 792
Query: 802 LAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYI 861
L +A L++LE L+V C ++ ++ I + FPKLK + + L +
Sbjct: 793 LNLANTLSRLEHLEVCECENMEELI----HTGIGGCGEETITFPKLKFLSLSQLPKLSSL 848
Query: 862 IPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNLPNI 921
GL L L + ++ Q+ E +I LE L + ++ N+
Sbjct: 849 CHNVNIIGLPHLVDLILKGIPGFTVIYPQNKLRTSSLLKEGVVI--PKLETLQIDDMENL 906
Query: 922 NSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRVN 981
I P C L ++ + +++C L N N S L ++ E+ V
Sbjct: 907 EEIWP--CELSGGEKVKLR--------AIKVSSCDKLVNLFPRNPMS--LLHHLEELTVE 954
Query: 982 NC-ELEGIFQL----VGLTNDGEKDPLTSCLEMLYLENLPQLRYLCK-SSVESTNLL--F 1033
NC +E +F + VG GE+D S L + +ENL +LR + + ++++L+ F
Sbjct: 955 NCGSIESLFNIDLDCVGAI--GEEDN-KSLLRSINVENLGKLREVWRIKGADNSHLINGF 1011
Query: 1034 QNLQQMEISGCRRLKCIFSSCMAG-GLPQLKALKIEKCNQLDQIVEDI 1080
Q ++ ++I C+R + IF+ A L L ++IE C + E I
Sbjct: 1012 QAVESIKIEKCKRFRNIFTPITANFYLVALLEIQIEGCGGNHESEEQI 1059
Score = 59.7 bits (143), Expect = 5e-07
Identities = 78/399 (19%), Positives = 147/399 (36%), Gaps = 109/399 (27%)
Query: 787 DLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD-------------DRDE 833
++ I C L+ + A + +L+ L++ SC ++ + +
Sbjct: 1299 EIEIVGCYALSSVIPCYAAGQMQKLQVLRIESCDGMKEVFETQLGTSSNKNNEKSGCEEG 1358
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
I + +++ P LK + CG LE+I + + L QL+ L+I +K + +
Sbjct: 1359 IPRVNNNVIMLPNLKILSIGNCGGLEHIFTFSALESLRQLQELKIKFCYGMKVIVKKEED 1418
Query: 894 NDGQNQ-------------------NELKIIELSALEELTLVNLPNINSICPEDCYLMWP 934
G+ Q + K++ L+ + LVNLP + P
Sbjct: 1419 EYGEQQTTTTTTKGASSSSSSSSSSSSKKVVVFPCLKSIVLVNLPELVGFFLGMNEFRLP 1478
Query: 935 SLLQFNLQNCGEFFM--------------------------------------------- 949
SL + ++ C + +
Sbjct: 1479 SLDKLKIKKCPKMMVFTAGGSTAPQLKYIHTRLGKHTLDQESGLNFHQTSFQSLYGDTLG 1538
Query: 950 --VSINTCMALHNNPRIN-EASHQT-----------LQNITEVRVNNCE-LEGIFQLV-- 992
S T + HN ++ E +H LQ + ++ V C+ +E +F+
Sbjct: 1539 PATSEGTTWSFHNFIELDVEGNHDVKKIIPSSELLQLQKLEKINVRWCKRVEEVFETALE 1598
Query: 993 --------GLTNDGEKDPLTSCLEML------YLENLPQLRYLCKSSVESTNLLFQNLQQ 1038
G+ D T+ L L L L LRY+ KS+ + T F NL +
Sbjct: 1599 AAGRNGNSGIGFDESSQTTTTTLVNLPNLREMNLWGLDCLRYIWKSN-QWTAFEFPNLTR 1657
Query: 1039 MEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIV 1077
++I C+RL+ +F+S M G L QL+ L I C+++++++
Sbjct: 1658 VDIYKCKRLEHVFTSSMVGSLSQLQELHISNCSEMEEVI 1696
Score = 45.4 bits (106), Expect = 0.009
Identities = 62/307 (20%), Positives = 123/307 (39%), Gaps = 71/307 (23%)
Query: 782 FENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILID-DDRDE----ISA 836
F NL + I C + LF+ +A+ L+ L+ +++ C ++ ++ + DD DE ++
Sbjct: 1169 FHNLTTINILKCKSIKYLFSPLMAELLSNLKDIRISECDGIKEVVSNRDDEDEEMTTFTS 1228
Query: 837 YDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDG 896
LFP L + L + ENLK + G ++G
Sbjct: 1229 THTTTTLFPSL--------------------------DSLTLSFLENLKCIGGGGAKDEG 1262
Query: 897 QNQNELK--IIELSALEELTLVNLPNINSICPEDCYLMWPSLLQF--NLQNCGEFFMVSI 952
N+ + L++ L ++ W SL Q+ ++ G + + S+
Sbjct: 1263 SNEISFNNTTATTAVLDQFELSEAGGVS----------W-SLCQYAREIEIVGCYALSSV 1311
Query: 953 NTCMALHNNPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCLEMLY 1011
C A +Q + +R+ +C+ ++ +F+ T+ + + + C E
Sbjct: 1312 IPCYAAGQ-----------MQKLQVLRIESCDGMKEVFETQLGTSSNKNNEKSGCEE--- 1357
Query: 1012 LENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCN 1071
+P+ V + ++ NL+ + I C L+ IF+ L QL+ LKI+ C
Sbjct: 1358 --GIPR--------VNNNVIMLPNLKILSIGNCGGLEHIFTFSALESLRQLQELKIKFCY 1407
Query: 1072 QLDQIVE 1078
+ IV+
Sbjct: 1408 GMKVIVK 1414
Score = 44.7 bits (104), Expect = 0.016
Identities = 52/221 (23%), Positives = 91/221 (40%), Gaps = 33/221 (14%)
Query: 672 QDEFLNHHRTLFLS-YLDTSNAAIKD-LAEKAEVLCIAGIEGG--AKNIIPDV-FQSMNH 726
Q+ LN H+T F S Y DT A + +EG K IIP +
Sbjct: 1518 QESGLNFHQTSFQSLYGDTLGPATSEGTTWSFHNFIELDVEGNHDVKKIIPSSELLQLQK 1577
Query: 727 LKELLIRDSKGIECLVDTCLIEVG----------------TLFFCKLHWLRIEHMKHLGA 770
L+++ +R K +E + +T L G T L LR ++ L
Sbjct: 1578 LEKINVRWCKRVEEVFETALEAAGRNGNSGIGFDESSQTTTTTLVNLPNLREMNLWGLDC 1637
Query: 771 L---YNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILI 827
L + + F NL + I C +L +FT ++ +L+QL++L + +C E++ +++
Sbjct: 1638 LRYIWKSNQWTAFEFPNLTRVDIYKCKRLEHVFTSSMVGSLSQLQELHISNCSEMEEVIV 1697
Query: 828 DDDRDEI---------SAYDYRLLLFPKLKKFHVRECGVLE 859
D D + + +L+ P+L +RE L+
Sbjct: 1698 KDADDSVEEDKEKESDGETNKEILVLPRLNSLILRELPCLK 1738
>UniRef100_Q9ZSD0 Resistance protein candidate RGC2C [Lactuca sativa]
Length = 1804
Score = 243 bits (619), Expect = 3e-62
Identities = 280/1110 (25%), Positives = 473/1110 (42%), Gaps = 129/1110 (11%)
Query: 23 LSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTR-AKKQTRKTAEVVEKWLKDANIAMDN 81
L Y ++ D+ + L +D+V++ + A V+ WL+D ++
Sbjct: 25 LRYLVSCRKYISDMDLKMKELKEAKDNVEEHKNHNISNRLEVPAAQVQSWLED----VEK 80
Query: 82 VDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASL-- 139
++ ++ + CF N RY GR + + + P L
Sbjct: 81 INAKVETVPKDVGCCF----NLKIRYRAGRDAFNIIEEIDSVMRRHSLITWTDHPIPLGR 136
Query: 140 ------SAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGK 193
S S E +F SR+ + E + AL+ + MI L GMGG GKT + + K
Sbjct: 137 VDSVMASTSTLSTEHN-DFQSREVRFSEALKALEANH--MIALCGMGGVGKTHMMQRLKK 193
Query: 194 --RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQED--- 248
+ F ++ I + IQ+ +A L E +E D+ R+++L +
Sbjct: 194 VAKEKRKFGYIIEAVIGEISDPIAIQQVVADYLCIELKESDKKTRAEKLRQGFKAKSDGG 253
Query: 249 --RVLVILDDVWQMLDFDAIGI-PSIEHHKGCKILITSRSEAVCTLMDCQKK--IQLSTL 303
+ L+ILDDVWQ +D + IG+ PS K+L+TSR E VC++M + I + L
Sbjct: 254 NTKFLIILDDVWQSVDLEDIGLSPSPNQGVDFKVLLTSRDEHVCSVMGVEANSIINVGLL 313
Query: 304 TNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVAL 363
E LFQ+ SE + + +I C GLP+A +A +L+ K + WK AL
Sbjct: 314 IEAEAQRLFQQFVETSEP---ELHKIGEDIVRRCCGLPIAIKTMACTLRNKRKDAWKDAL 370
Query: 364 DRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAI 423
RL+ N+ + + SY+NL +E KS+FL+C +FPED IP E L R
Sbjct: 371 SRLQHHDIGNVATAV------FRTSYENLPDKETKSVFLMCGLFPEDFNIPTEELMRYGW 424
Query: 424 GLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVR------------- 470
GL + V++ ARN + ++L+ + LL+ + G VKMHDLVR
Sbjct: 425 GLKLFDRVYTIIEARNRLNTCIDRLVQTNLLIGSDNGVHVKMHDLVRAFVLGMYSEVEQA 484
Query: 471 ------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQI---HT 521
N+ W EN++ S K I SL +FP L L L++
Sbjct: 485 SIVNHGNMPGWPDENDMIVHSCKRI------SLTCKGMIEFPVDLKFPKLTILKLMHGDK 538
Query: 522 YTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDL--VDISFVG 579
+ E ++GM LRV+ + + + PLL + + TN+R + ++ L D S +G
Sbjct: 539 SLKFPQEFYEGMEKLRVISYH---KMKYPLLPLAPQCSTNIRVLHLTECSLKMFDCSCIG 595
Query: 580 DMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADC 639
++ LE ++ + LP V L LRLLDL C R V+ +LEE + +
Sbjct: 596 NLSNLEVLSFANSCIEWLPSTVRNLKKLRLLDLRLCYGLRIEQGVLKSLVKLEEFYIGNA 655
Query: 640 RSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDTSNAAIKDLAE 699
F+ + + Y + S + F N+ + N + ++L E
Sbjct: 656 YG-----FIDDNCKEMAERSYNL------SALEFAFFNNK-------AEVKNMSFENL-E 696
Query: 700 KAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHW 759
+ ++ +G I S ++ L+ ++ ++ ++ LF +H
Sbjct: 697 RFKISVGCSFDGN----INMSSHSYENMLRLVTNKGDVLDSKLNGLFLKTEVLFL-SVHG 751
Query: 760 LRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSC 819
+ + + + Q S F NL+ L IS C +L LF L VA L++LE L+V C
Sbjct: 752 MNDLEDVEVKSTHPTQ---SSSFCNLKVLIISKCVELRYLFKLNVANTLSRLEHLEVCKC 808
Query: 820 PELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIV 879
++ ++ I + FPKLK + + L + GL L L++
Sbjct: 809 KNMEELI----HTGIGGCGEETITFPKLKFLSLSQLPKLSGLCHNVNIIGLPHLVDLKLK 864
Query: 880 CNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQF 939
++ Q+ E +I LE L + ++ N+ I P C L ++
Sbjct: 865 GIPGFTVIYPQNKLRTSSLLKEEVVI--PKLETLQIDDMENLEEIWP--CELSGGEKVKL 920
Query: 940 NLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRVNNC-ELEGIFQL----VGL 994
+ +++C L N N S L ++ E+ V NC +E +F + VG
Sbjct: 921 R--------EIKVSSCDKLVNLFPRNPMS--LLHHLEELTVENCGSIESLFNIDLDCVGA 970
Query: 995 TNDGEKDPLTSCLEMLYLENLPQLRYLCK-SSVESTNLL--FQNLQQMEISGCRRLKCIF 1051
GE+D S L + +ENL +LR + + ++++L+ FQ ++ ++I C+R + IF
Sbjct: 971 I--GEEDN-KSLLRSINVENLGKLREVWRIKGADNSHLINGFQAVESIKIEKCKRFRNIF 1027
Query: 1052 SSCMAG-GLPQLKALKIEKCNQLDQIVEDI 1080
+ A L L ++IE C + E I
Sbjct: 1028 TPITANFYLVALLEIQIEGCGGNHESEEQI 1057
Score = 57.0 bits (136), Expect = 3e-06
Identities = 81/393 (20%), Positives = 148/393 (37%), Gaps = 104/393 (26%)
Query: 787 DLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD-------------DRDE 833
++ I +C L+ + A + +L+ L+V++C ++ + +
Sbjct: 1297 EIKIGNCHALSSVIPCYAAGQMQKLQVLRVMACNGMKEVFETQLGTSSNKNNEKSGCEEG 1356
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
I + +++ P LK + CG LE+I + + L QL+ L I +K + +
Sbjct: 1357 IPRVNNNVIMLPNLKILSIGNCGGLEHIFTFSALESLRQLQELTIKGCYRMKVIVKKEED 1416
Query: 894 NDGQNQ--------------NELKIIELSALEELTLVNL---------------PNINSI 924
G+ Q + K++ L+ + LVNL P+++ +
Sbjct: 1417 EYGEQQTTTTTTKGASSSSSSSKKVVVFPCLKSIVLVNLPELVGFFLGMNEFRLPSLDKL 1476
Query: 925 CPEDCYLMW---------PSLLQFNLQ------------NCGEFFMVSIN---------- 953
E C M P L + + N + + S N
Sbjct: 1477 IIEKCPKMMVFTAGGSTAPQLKYIHTRLGKHTLDQESGLNFHQVHIYSFNGDTLGPATSE 1536
Query: 954 -TCMALHNNPRINEASHQT------------LQNITEVRVNNCE-LEGIFQLV------- 992
T + HN ++ S+ LQ + ++ V C+ +E +F+
Sbjct: 1537 GTTWSFHNFIELDVKSNHDVKKIIPSSELLQLQKLVKINVMWCKRVEEVFETALEAAGRN 1596
Query: 993 ---GLTNDGEKDPLTSCLEML------YLENLPQLRYLCKSSVESTNLLFQNLQQMEISG 1043
G+ D T+ L L L L LRY+ KS+ + T F NL ++EI
Sbjct: 1597 GNSGIGFDESSQTTTTTLVNLPNLGEMKLRGLDCLRYIWKSN-QWTAFEFPNLTRVEIYE 1655
Query: 1044 CRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQI 1076
C L+ +F+S M G L QL+ L+I CN ++ +
Sbjct: 1656 CNSLEHVFTSSMVGSLLQLQELEIGLCNHMEVV 1688
>UniRef100_Q6Y140 Resistance protein RGC2 [Lactuca sativa]
Length = 1821
Score = 242 bits (618), Expect = 4e-62
Identities = 275/1112 (24%), Positives = 485/1112 (42%), Gaps = 160/1112 (14%)
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I++ + V + Y +V+ + + + L R SV++ ++R + + ++
Sbjct: 15 IAQTALVPVTEHVGYMISCRKYVRVMQMKMTELNTSRISVEEHISRNTRNHLQIPSQIKD 74
Query: 71 WLKD-----ANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIE 125
WL AN+A +D + C + R+ +G+K K ++
Sbjct: 75 WLDQVEGIRANVANFPIDVI-------------SCCSLRIRHKLGQKAFKITEQIESLTR 121
Query: 126 EGRQYIEIERPASL--------SAGYFSAERCWEFDSRKPAYEELMCALKDDDVT-MIGL 176
+ I + P L S S++ F SR+ + + + AL+ + +I L
Sbjct: 122 QNSLIIWTDEPVPLGRVGSMIASTSAASSDHHDVFPSREQIFRKALEALEPVQKSHIIAL 181
Query: 177 YGMGGCGKTMLAMEVGK--RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEM 234
+GMGG GKT + ++ + +F+ ++ V I IQ+ +A L E +E +
Sbjct: 182 WGMGGVGKTTMMKKLKEVVEQKKMFNIIVQVVIGEKTNPIAIQQAVADYLSIELKENTKE 241
Query: 235 DRSKRLCMRLTQE---DRVLVILDDVWQMLDFDAIGIPSIEHHKGC--KILITSRSEAVC 289
R+ +L + ++ LVILDDVWQ +D + IG+ + + KG K+L+TSR VC
Sbjct: 242 ARADKLRKWFEDDGGKNKFLVILDDVWQFVDLEDIGLSPLPN-KGVNFKVLLTSRDSHVC 300
Query: 290 TLMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWI--SIKNMAREISNECKGLPVATV 345
TLM + + + L + E LF++ A + + + +A I++ C+GLP+A
Sbjct: 301 TLMGAEANSILNIKVLKDVEGQSLFRQFAKNAGDDDLDPAFNGIADSIASRCQGLPIAIK 360
Query: 346 AVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCS 405
+A SLKG+++ W AL RL + K + E+ ++ +K +SYDNL E KS+FLLC+
Sbjct: 361 TIALSLKGRSKPAWDHALSRLENHK-IGSEEVVREVFK---ISYDNLQDEVTKSIFLLCA 416
Query: 406 VFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKM 465
+FPED +IP+E L R GL + E + ARN + +L + LL ++ CVKM
Sbjct: 417 LFPEDFDIPIEELVRYGWGLKLFIEAKTIREARNRLNTCTERLRETNLLFGSDDFGCVKM 476
Query: 466 HDLVR-------------------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWCEKFP 506
HD+VR NV+ W+ N I + + SL +FP
Sbjct: 477 HDVVRDFVLYXXXXVQXASIXNHGNVSEWLEXNH-------SIYSCKRISLTXKGMSEFP 529
Query: 507 NSLDCSNLDFLQI-HTYTQVS--DEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLR 563
L NL L++ H +S ++ + M ++V+ + + PLL +SL+ TN+R
Sbjct: 530 KDLXFPNLSILKLXHGDKSLSFPEDFYGKMEKVQVI---SYDKLMYPLLPSSLECSTNVR 586
Query: 564 C--ILFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNP 621
+ + + D S +G++ +E ++ + + LP + L LRLLDL+ C R
Sbjct: 587 VLHLHYCSLRMFDCSSIGNLLNMEVLSFANSNIEWLPSTIGNLKKLRLLDLTNCKGLRID 646
Query: 622 FEVIARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRT 681
V+ +LEEL+ R + L + + ++ +R L
Sbjct: 647 NGVLKNLVKLEELYMGVNRPYGQAVSLTDENCNEMAER------------SKNLLALESE 694
Query: 682 LFLSYLDTSNAAIKDLAEKAEVLCIAGIEGG---AKNIIPDVFQSMNHLKELLIRDSKGI 738
LF N + ++L E+ ++ ++G +++ + + ELL G+
Sbjct: 695 LFKYNAQVKNISFENL-ERFKISVGRSLDGSFSKSRHSYGNTLKLAIDKGELLESRMNGL 753
Query: 739 ECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTR 798
+ + VG ++ HL + S F NL L +S C +L
Sbjct: 754 FEKTEVLCLSVGDMY-------------HLSDV----KVKSSSFYNLRVLVVSECAELKH 796
Query: 799 LFTLAVAQNLAQLEKLQVLSCPELQHILI--DDDRDEISAYDYRLLLFPKLKKFHVRECG 856
LFTL VA L++LE LQV C ++ ++ +RD I+ FPKLK +
Sbjct: 797 LFTLGVANTLSKLEYLQVYKCDNMEELIHTGGSERDTIT--------FPKLKLLSLNALP 848
Query: 857 VLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLV 916
L + L + +E E+V E Y T +N+ E EE+ +
Sbjct: 849 KL-----LGLCLNVNTIELPELV--EMKLYSIPGFTSIYPRNKLEASSF---LKEEVVIP 898
Query: 917 NLPNINSICPEDCYLMWPS---------LLQFNLQNCGEFFMVSINTCMALHNNPRINEA 967
L + E+ +WPS L + ++NC + + + M+L
Sbjct: 899 KLDILEIHDMENLKEIWPSELSRGEKVKLREIKVRNCDKLVNLFPHNPMSL--------- 949
Query: 968 SHQTLQNITEVRVNNC-ELEGIF--QLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCK- 1023
L ++ E+ V C +E +F L + GE+D S L + +EN +LR + +
Sbjct: 950 ----LHHLEELIVEKCGSIEELFNIDLDCASVIGEEDN-NSSLRNINVENSMKLREVWRI 1004
Query: 1024 SSVESTNLLFQNLQQME---ISGCRRLKCIFS 1052
+++ LF+ Q +E I+ C+R +F+
Sbjct: 1005 KGADNSRPLFRGFQVVEKIIITRCKRFTNVFT 1036
Score = 64.3 bits (155), Expect = 2e-08
Identities = 78/394 (19%), Positives = 146/394 (36%), Gaps = 104/394 (26%)
Query: 787 DLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD-------------DRDE 833
++ I C L+ + A + +L+ L+V+ C ++ + +
Sbjct: 1312 EIEIYECHALSSVIPCYAAGQMQKLQVLRVMGCDGMKEVFETQLGTSSNKNNEKSGCEEG 1371
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
I + +++ P LK +R CG LE+I + + L QL+ L+I+ +K + +
Sbjct: 1372 IPRVNNNVIMLPNLKILEIRGCGGLEHIFTFSALESLRQLQELKIIFCYGMKVIVKKEED 1431
Query: 894 NDGQNQ--------------NELKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQF 939
G+ Q + K++ L+ + LVNLP + PSL +
Sbjct: 1432 EYGEQQTTTTTTKGASSSSSSSKKVVVFPCLKSIVLVNLPELVGFFLGMNEFRLPSLDKL 1491
Query: 940 NLQNCGEFFM-----------------------------------------------VSI 952
++ C + + S
Sbjct: 1492 IIKKCPKMMVFTAGGSTAPQLKYIHTRLGKHTLDQESGLNFHQTSFQSLYGDTLGPATSE 1551
Query: 953 NTCMALHNNPRINEASHQ------------TLQNITEVRVNNC-ELEGIFQLV------- 992
T + HN ++ S+ LQ + ++ +N+C +E +F+
Sbjct: 1552 GTTWSFHNLIELDVKSNHDVKKIIPSSELLQLQKLEKININSCVGVEEVFETALEAAGRN 1611
Query: 993 ---GLTNDGEKDPLTSC------LEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISG 1043
G+ D T+ L + L L LRY+ KS+ + T F NL ++EI
Sbjct: 1612 GNSGIGFDESSQTTTTTLVNLPNLREMNLHYLRGLRYIWKSN-QWTAFEFPNLTRVEIYE 1670
Query: 1044 CRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIV 1077
C L+ +F+S M G L QL+ L I C+Q++ ++
Sbjct: 1671 CNSLEHVFTSSMVGSLLQLQELLIWNCSQIEVVI 1704
Score = 47.0 bits (110), Expect = 0.003
Identities = 77/365 (21%), Positives = 143/365 (39%), Gaps = 76/365 (20%)
Query: 756 KLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQ 815
KL L I M++L ++ ++ G L ++ + +C KL LF L LE+L
Sbjct: 899 KLDILEIHDMENLKEIWPSELS-RGEKVKLREIKVRNCDKLVNLFPHNPMSLLHHLEELI 957
Query: 816 VLSCPELQHI---------LIDDDRDEISAYDYRLLLFPKLKKFH-----------VREC 855
V C ++ + +I ++ + S + + KL++ R
Sbjct: 958 VEKCGSIEELFNIDLDCASVIGEEDNNSSLRNINVENSMKLREVWRIKGADNSRPLFRGF 1017
Query: 856 GVLEYII------------PITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELK 903
V+E II PIT L L + + C N + QS Q Q ++
Sbjct: 1018 QVVEKIIITRCKRFTNVFTPITTNFDLGALLEISVDCRGNDES--DQSNQEQEQEQTDI- 1074
Query: 904 IIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPR 963
+ E L+E T V++ N+ ++P L + N
Sbjct: 1075 LSEEETLQEAT-VSISNV----------VFPPCLMHSFHN-------------------- 1103
Query: 964 INEASHQTLQNITEVRVNNCELEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCK 1023
+++ + ++ + V E +LV N+ ++ + L+ LYL N+ ++ K
Sbjct: 1104 LHKLKLERVRGVEVVFEIESESPTCRELVTTHNNQQQPIILPYLQELYLRNMDNTSHVWK 1163
Query: 1024 SS---------VESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLD 1074
S + + F NL + I CR +K +FS MA L LK ++I+ C+ ++
Sbjct: 1164 CSNWNNFFTLPKQQSESPFHNLTTITIMFCRSIKHLFSPLMAELLSNLKKVRIDDCDGIE 1223
Query: 1075 QIVED 1079
++V +
Sbjct: 1224 EVVSN 1228
Score = 42.4 bits (98), Expect = 0.079
Identities = 76/354 (21%), Positives = 150/354 (41%), Gaps = 59/354 (16%)
Query: 759 WLRIEHM----KHLGALYNG--QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNL-AQL 811
WL H K + G + P F NL L + H K +L+ ++ ++
Sbjct: 505 WLEXNHSIYSCKRISLTXKGMSEFPKDLXFPNLSILKLXHGDK-----SLSFPEDFYGKM 559
Query: 812 EKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLV 871
EK+QV+S +L + L+ + + L + L+ F G L+
Sbjct: 560 EKVQVISYDKLMYPLLPSSLECSTNVRVLHLHYCSLRMFDCSSIG------------NLL 607
Query: 872 QLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYL 931
+E L N N++++ ST N +L++++L+ + L + N N + E+ Y+
Sbjct: 608 NMEVLSFA-NSNIEWL--PSTIG---NLKKLRLLDLTNCKGLRIDNGVLKNLVKLEELYM 661
Query: 932 ----MWPSLLQFNLQNCGEFFMVSINTCMALHN-----NPRINEASHQTLQNITEVRVNN 982
+ + +NC E S N +AL + N ++ S + L+ ++ V
Sbjct: 662 GVNRPYGQAVSLTDENCNEMAERSKNL-LALESELFKYNAQVKNISFENLERF-KISVGR 719
Query: 983 CELEGIFQLV--------------GLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVES 1028
L+G F G + + L E+L L ++ + +L V+S
Sbjct: 720 -SLDGSFSKSRHSYGNTLKLAIDKGELLESRMNGLFEKTEVLCL-SVGDMYHLSDVKVKS 777
Query: 1029 TNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLDQIVEDIGT 1082
++ F NL+ + +S C LK +F+ +A L +L+ L++ KC+ +++++ G+
Sbjct: 778 SS--FYNLRVLVVSECAELKHLFTLGVANTLSKLEYLQVYKCDNMEELIHTGGS 829
Score = 38.9 bits (89), Expect = 0.87
Identities = 28/120 (23%), Positives = 58/120 (48%), Gaps = 15/120 (12%)
Query: 744 TCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLA 803
T L+ + L LH+LR L ++ + F NL + I C L +FT +
Sbjct: 1627 TTLVNLPNLREMNLHYLR-----GLRYIWKSNQWTAFEFPNLTRVEIYECNSLEHVFTSS 1681
Query: 804 VAQNLAQLEKLQVLSCPELQHILIDD-------DRDEIS---AYDYRLLLFPKLKKFHVR 853
+ +L QL++L + +C +++ +++ D D+++ S + +L+ P+LK ++
Sbjct: 1682 MVGSLLQLQELLIWNCSQIEVVIVKDADVSVEEDKEKESDGKTTNKEILVLPRLKSLKLQ 1741
>UniRef100_Q6Y134 Resistance protein RGC2 [Lactuca sativa]
Length = 1070
Score = 242 bits (618), Expect = 4e-62
Identities = 279/1096 (25%), Positives = 478/1096 (43%), Gaps = 127/1096 (11%)
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I++ + V + Y +V+ + + L R SV++ ++R + + ++
Sbjct: 15 IAQTALVPVTEHVGYIISCRKYVRVMQMKMRELNTSRISVEEHISRNTRNHLQIPSQIKD 74
Query: 71 WLKD-----ANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIE 125
WL AN+A +D + C + R+ +G+K K ++
Sbjct: 75 WLDQVEGIKANVANFPIDVI-------------SCCSLRIRHKLGQKAFKITEQIESLTR 121
Query: 126 EGRQYIEIERPASL--------SAGYFSAERCWEFDSRKPAYEELMCALKDDDVT-MIGL 176
+ I + P L S S++ F SR+ + + + AL+ + MI L
Sbjct: 122 QNSLIIWTDEPVPLGRVGSMIASTSAASSDHHDVFPSREQIFRKALEALEPVQKSHMIAL 181
Query: 177 YGMGGCGKTMLAMEVGK--RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEM 234
+GMGG GKTM+ ++ + F+ ++ V I IQ+ +A SL E +E +
Sbjct: 182 WGMGGVGKTMMMKKLKEVVEQKKTFNIIVQVVIGEKTNPIAIQQAVADSLSIELKENTKE 241
Query: 235 DRSKRLCMRLTQE---DRVLVILDDVWQMLDFDAIGI-PSIEHHKGCKILITSRSEAVCT 290
R+ +L + ++ LVILDDVWQ +D + IG+ P K+L+TSR VCT
Sbjct: 242 ARADKLRKWFEADGGKNKFLVILDDVWQFVDLEDIGLSPHPNXGVXFKVLLTSRDSHVCT 301
Query: 291 LMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWI--SIKNMAREISNECKGLPVATVA 346
LM + + + L + E LF++ A + + + +A I++ C+GLP+A
Sbjct: 302 LMGAEANSILNIKVLKDVEGKSLFRQFAKNAGDDDLDPAFIGIADSIASRCQGLPIAIKT 361
Query: 347 VASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSV 406
+A SLKG+++ W VAL RL + K + E+ ++ +K +SYDNL E KS+FLLC++
Sbjct: 362 IALSLKGRSKSAWDVALSRLENHK-IGSEEVVREVFK---ISYDNLQDEVTKSIFLLCAL 417
Query: 407 FPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMH 466
FPED +IP E L R GL + E + ARN + +L + LL ++ CVKMH
Sbjct: 418 FPEDFDIPTEELVRYGWGLKLFIEAKTIREARNRLNTCTERLRETNLLFGSDDIGCVKMH 477
Query: 467 DLVRN--------VAHWIAENEIKCASE-----KDIMTLEHTSLRYLWCEKFPNSLDCSN 513
D+VR+ V H N SE I + + SL +FP L N
Sbjct: 478 DVVRDFVLHIFSEVQHASIVNHGNXXSEWLEENHSIYSCKRISLTCKGMSEFPKDLKFPN 537
Query: 514 LDFLQI-HTYTQVS-DEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWD 571
L L++ H +S E F G + Y+K PLL +SL+ TNLR + +
Sbjct: 538 LSILKLMHGDKSLSFPENFYGKMEKVQVISYDK--LMYPLLPSSLECSTNLRVLHLHECS 595
Query: 572 L--VDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHT 629
L D S +G++ +E ++ + LP + L LRLLDL++CG V+
Sbjct: 596 LRMFDCSSIGNLLNMEVLSFANSGIEWLPSTIGNLKKLRLLDLTDCGGLHIDNGVLKNLV 655
Query: 630 ELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLFLSYLDT 689
+LEEL+ R L + + ++ +R + L S L
Sbjct: 656 KLEELYMGANRLFGNAISLTDENCNEMAERSKNLLA-----------------LESELFK 698
Query: 690 SNAAIKDLA-EKAE--VLCIAGIEGG----AKNIIPDVFQSMNHLKELLIRDSKGIECLV 742
SNA +K+L+ E E + + GG +++ + + + + ELL G+
Sbjct: 699 SNAQLKNLSFENLERFKISVGHFSGGYFSKSRHSYENTLKLVVNKGELLESRMNGLFEKT 758
Query: 743 DTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTL 802
+ + VG M L + M S F NL L +S C +L LF L
Sbjct: 759 EVLCLSVG-------------DMNDLSDV----MVKSSSFYNLRVLVVSECAELKHLFKL 801
Query: 803 AVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLEYII 862
VA L++LE L+V C ++ ++ + + FPKLK ++ L ++
Sbjct: 802 GVANTLSKLEHLEVYKCDNMEELI------HTGGSEGDTITFPKLKLLYLHG---LPNLL 852
Query: 863 PITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELK-IIELSALEELTLVNLPNI 921
+ L ++L L + ++ N + LK + + L+ L + ++ N+
Sbjct: 853 GLCLNVNTIELPELVQMKLYSIPGFTSIYPRNKLETSTLLKEEVVIPKLDILEIDDMENL 912
Query: 922 NSICPEDCYL-MWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRV 980
I P + L + ++NC + + + M+L ++ + E + +I E+
Sbjct: 913 KEIWPSELSRGEKVKLREIKVRNCDKLVNLFPHNPMSLLHH--LEELIVEKCGSIEELFN 970
Query: 981 NNCELEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLR--YLCKSSVESTNLL--FQNL 1036
N + G+ GE+D S L + +EN +LR + K + S L FQ +
Sbjct: 971 INLDCAGVI--------GEEDN-NSSLRNIKVENSVKLREVWRIKGADNSCPLFRGFQAV 1021
Query: 1037 QQMEISGCRRLKCIFS 1052
+ + I C R + +F+
Sbjct: 1022 ESISIRWCDRFRNVFT 1037
Score = 42.7 bits (99), Expect = 0.060
Identities = 72/330 (21%), Positives = 145/330 (43%), Gaps = 49/330 (14%)
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNL-AQLEKLQVLSCPELQHILIDDDRDE 833
+ P F NL L + H K +L+ +N ++EK+QV+S +L + L+
Sbjct: 528 EFPKDLKFPNLSILKLMHGDK-----SLSFPENFYGKMEKVQVISYDKLMYPLLPSS--- 579
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
L L+ H+ EC + + + L+ +E L N ++++ ST
Sbjct: 580 -------LECSTNLRVLHLHECSLRMF--DCSSIGNLLNMEVLSFA-NSGIEWL--PSTI 627
Query: 894 NDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYL----MWPSLLQFNLQNCGEFFM 949
N +L++++L+ L + N N + E+ Y+ ++ + + +NC E
Sbjct: 628 G---NLKKLRLLDLTDCGGLHIDNGVLKNLVKLEELYMGANRLFGNAISLTDENCNEMAE 684
Query: 950 VSINTCMALHNNPRINEASHQTL--QNITEVRVNNCELEGIF----------QLVGLTND 997
S N +AL + + A + L +N+ +++ G + L + N
Sbjct: 685 RSKNL-LALESELFKSNAQLKNLSFENLERFKISVGHFSGGYFSKSRHSYENTLKLVVNK 743
Query: 998 GE-----KDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFS 1052
GE + L E+L L ++ + L V+S++ F NL+ + +S C LK +F
Sbjct: 744 GELLESRMNGLFEKTEVLCL-SVGDMNDLSDVMVKSSS--FYNLRVLVVSECAELKHLFK 800
Query: 1053 SCMAGGLPQLKALKIEKCNQLDQIVEDIGT 1082
+A L +L+ L++ KC+ +++++ G+
Sbjct: 801 LGVANTLSKLEHLEVYKCDNMEELIHTGGS 830
>UniRef100_Q6Y138 Resistance protein RGC2 [Lactuca sativa]
Length = 892
Score = 237 bits (605), Expect = 1e-60
Identities = 250/935 (26%), Positives = 420/935 (44%), Gaps = 131/935 (14%)
Query: 173 MIGLYGMGGCGKTMLAMEVGKRCGNL--FDQVLFVPISSTVEVERIQEKIAGSLEFEFQE 230
MI L+GMGG GKT + ++ + G F+ ++ V I IQ+ +A L E +E
Sbjct: 1 MIALWGMGGVGKTTMMKKLKEVVGQKKSFNIIIQVVIGEKTNPIAIQQAVADYLSIELKE 60
Query: 231 KDEMDRSKRLCMRLTQE---DRVLVILDDVWQMLDFDAIGIPSIEHHKGC--KILITSRS 285
+ R+ +L R + ++ LVILDDVWQ +D + IG+ + + KG K+L+TSR
Sbjct: 61 NTKEARADKLRKRFEADGGKNKFLVILDDVWQFVDLEDIGLSPLPN-KGVNFKVLLTSRD 119
Query: 286 EAVCTLMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWI--SIKNMAREISNECKGLP 341
VCTLM + + + L + E LF++ A + + + +A I++ C+GLP
Sbjct: 120 SHVCTLMGAEANSILNIKVLKDVEGQSLFRQFAKNAGDDDLDPAFNGIADSIASRCQGLP 179
Query: 342 VATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLF 401
+A +A SLKG+++ W VAL RL + K + E+ ++ +K +SYDNL E KS+F
Sbjct: 180 IAIKTIALSLKGRSKSAWDVALSRLENHK-IGSEEVVREVFK---ISYDNLQDEVTKSIF 235
Query: 402 LLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGK 461
LLC++FPED +IP E L R GL + E + ARN + +L + LL ++
Sbjct: 236 LLCALFPEDFDIPTEELVRYGWGLKLFIEAKTIREARNRLNTCTERLRETNLLFGSDDIG 295
Query: 462 CVKMHDLVR-------------------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWC 502
CVKMHD+VR NV+ W+ EN I + + SL
Sbjct: 296 CVKMHDVVRDFVLHIFSEVQHASIVNHGNVSEWLEENH-------SIYSCKRISLTCKGM 348
Query: 503 EKFPNSLDCSNLDFLQI-HTYTQVS-DEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLT 560
+FP L NL L++ H +S E F G + Y+K PLL +SL+ T
Sbjct: 349 SQFPKDLKFPNLSILKLMHGDKSLSFPENFYGKMEKVQVISYDK--LMYPLLPSSLECST 406
Query: 561 NLRC--ILFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGME 618
N+R + + + D S +G++ +E ++ + + LP + L LRLLDL+ C
Sbjct: 407 NVRVLHLHYCSLRMFDCSSIGNLLNMEVLSFANSNIEWLPSTIGNLKKLRLLDLTNCKGL 466
Query: 619 RNPFEVIARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNH 678
R V+ +LEEL+ R + L + + ++ +R L
Sbjct: 467 RIDNGVLKNLVKLEELYMGVNRPYGQAVSLTDENCNEMAER------------SKNLLAL 514
Query: 679 HRTLFLSYLDTSNAAIKDLAEKAEVLCIAGIEGG---AKNIIPDVFQSMNHLKELLIRDS 735
LF N + ++L E+ ++ ++G +++ + + ELL
Sbjct: 515 ESQLFKYNAQVKNISFENL-ERFKISVGRSLDGSFSKSRHSYENTLKLAIDKGELLESRM 573
Query: 736 KGIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPK 795
G+ + + VG ++ HL + S F NL L +S C +
Sbjct: 574 NGLFEKTEVLCLSVGDMY-------------HLSDV----KVKSSSFYNLRVLVVSECAE 616
Query: 796 LTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVREC 855
L LFTL VA L++LE L+V C ++ ++ + + FPKLK ++
Sbjct: 617 LKHLFTLGVANTLSKLEHLEVYKCDNMEELI------HTGGSEGDTITFPKLKLLNLHGL 670
Query: 856 GVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSAL--EEL 913
L + L + +E E+V + Y T +N+ +E S+L EE+
Sbjct: 671 PNL-----LGLCLNVNAIELPELV--QMKLYSIPGFTSIYPRNK-----LEASSLLKEEV 718
Query: 914 TLVNLPNINSICPEDCYLMWPS---------LLQFNLQNCGEFFMVSINTCMALHNNPRI 964
+ L + E+ +WPS L + ++NC + + + M+L
Sbjct: 719 VIPKLDILEIHDMENLKEIWPSELSRGEKVKLREIKVRNCDKLVNLFPHNPMSL------ 772
Query: 965 NEASHQTLQNITEVRVNNC-ELEGIF--QLVGLTNDGEKDPLTSCLEMLYLENLPQLRYL 1021
L ++ E+ V C +E +F L + GE+D S L + +EN +LR +
Sbjct: 773 -------LHHLEELIVEKCGSIEELFNIDLDCASVIGEEDN-NSSLRNINVENSMKLREV 824
Query: 1022 CK-SSVESTNLLFQNLQQME---ISGCRRLKCIFS 1052
+ +++ LF+ Q +E I+ C+R +F+
Sbjct: 825 WRIKGADNSRPLFRGFQVVEKIIITRCKRFTNVFT 859
Score = 43.9 bits (102), Expect = 0.027
Identities = 73/332 (21%), Positives = 144/332 (42%), Gaps = 53/332 (15%)
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNL-AQLEKLQVLSCPELQHILIDDDRDE 833
Q P F NL L + H K +L+ +N ++EK+QV+S +L + L+ +
Sbjct: 350 QFPKDLKFPNLSILKLMHGDK-----SLSFPENFYGKMEKVQVISYDKLMYPLLPSSLEC 404
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
+ L + L+ F G L+ +E L N N++++ ST
Sbjct: 405 STNVRVLHLHYCSLRMFDCSSIG------------NLLNMEVLSFA-NSNIEWL--PSTI 449
Query: 894 NDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYL----MWPSLLQFNLQNCGEFFM 949
N +L++++L+ + L + N N + E+ Y+ + + +NC E
Sbjct: 450 G---NLKKLRLLDLTNCKGLRIDNGVLKNLVKLEELYMGVNRPYGQAVSLTDENCNEMAE 506
Query: 950 VSINTCMALHN-----NPRINEASHQTLQNITEVRVNNCELEGIFQLV------------ 992
S N +AL + N ++ S + L+ ++ V L+G F
Sbjct: 507 RSKNL-LALESQLFKYNAQVKNISFENLERF-KISVGR-SLDGSFSKSRHSYENTLKLAI 563
Query: 993 --GLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCI 1050
G + + L E+L L ++ + +L V+S++ F NL+ + +S C LK +
Sbjct: 564 DKGELLESRMNGLFEKTEVLCL-SVGDMYHLSDVKVKSSS--FYNLRVLVVSECAELKHL 620
Query: 1051 FSSCMAGGLPQLKALKIEKCNQLDQIVEDIGT 1082
F+ +A L +L+ L++ KC+ +++++ G+
Sbjct: 621 FTLGVANTLSKLEHLEVYKCDNMEELIHTGGS 652
>UniRef100_Q9ZT69 Resistance protein candidate RGC2J [Lactuca sativa]
Length = 1847
Score = 237 bits (604), Expect = 2e-60
Identities = 280/1107 (25%), Positives = 483/1107 (43%), Gaps = 150/1107 (13%)
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I++ + + + Y +V+D+ + + L R S ++ ++R + + ++
Sbjct: 15 IAQTALVPLTDHVGYMISCRKYVRDMQMKMTELNTSRISAEEHISRNTRNHLQIPSQIKD 74
Query: 71 WLKD-----ANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIE 125
WL AN+A +D + C + R+ +G+K K ++
Sbjct: 75 WLDQVEGIRANVANFPIDVI-------------SCCSLRIRHKLGQKAFKITEQIESLTR 121
Query: 126 EGRQYIEIERPASL--------SAGYFSAERCWEFDSRKPAYEELMCALKDDDVT-MIGL 176
+ I + P L S S++ F SR+ + + + AL+ + +I L
Sbjct: 122 QNSLIIWTDEPVPLGRVGSMIASTSAASSDHHDVFPSREQIFRKALEALEPVQKSHIIAL 181
Query: 177 YGMGGCGKTML------AMEVGKRCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQE 230
+GMGG GKT + +E K C N+ QV+ I IQ+ +A L E +E
Sbjct: 182 WGMGGVGKTTMMKKLKEVVEQKKTC-NIIVQVV---IGEKTNPIAIQQAVADYLSIELKE 237
Query: 231 KDEMDRSKRLCMRLTQE---DRVLVILDDVWQMLDFDAIGIPSIEHHKGC--KILITSRS 285
+ R+ +L R + ++ LVILDDVWQ D + IG+ + + KG K+L+TSR
Sbjct: 238 NTKEARADKLRKRFEADGGKNKFLVILDDVWQFFDLEDIGLSPLPN-KGVNFKVLLTSRD 296
Query: 286 EAVCTLMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWI--SIKNMAREISNECKGLP 341
VCTLM + + + L + E LF++ A + + + +A I++ C+GLP
Sbjct: 297 SHVCTLMGAEANSILNIKVLKDVEGKSLFRQFAKNAGDDDLDPAFIGIADSIASRCQGLP 356
Query: 342 VATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLF 401
+A +A SLKG+++ W VAL RL + K + E+ ++ +K +SYDNL E KS+F
Sbjct: 357 IAIKTIALSLKGRSKSAWDVALSRLENHK-IGSEEVVREVFK---ISYDNLQDEVTKSIF 412
Query: 402 LLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGK 461
LLC++FPED +IP+E L R GL + E + ARN + +L + LL ++
Sbjct: 413 LLCALFPEDFDIPIEELVRYGWGLKLFIEAKTIREARNRLNNCTERLRETNLLFGSHDFG 472
Query: 462 CVKMHDLVR-------------------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWC 502
CVKMHD+VR N++ W +N+ + ++ +T + S
Sbjct: 473 CVKMHDVVRDFVLHMFSEVKHASIVNHGNMSEWPEKNDTSNSCKRISLTCKGMS------ 526
Query: 503 EKFPNSLDCSNLDFLQI--HTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLT 560
KFP ++ NL L++ + E F G + Y+K PLL +SL+ T
Sbjct: 527 -KFPKDINYPNLLILKLMHGDKSLCFPENFYGKMEKVQVISYDK--LMYPLLPSSLECST 583
Query: 561 NLRC--ILFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGME 618
N+R + + + D S +G++ +E ++ + + LP + L LRLLDL+ C
Sbjct: 584 NVRVLHLHYCSLRMFDCSSIGNLLNMEVLSFANSNIEWLPSTIGNLKKLRLLDLTNCKGL 643
Query: 619 RNPFEVIARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNH 678
R V+ +LEEL+ R + L + + ++++ GS + L
Sbjct: 644 RIDNGVLKNLVKLEELYMGVNRPYGQAVSLTDENCNEMVE------GS------KKLLAL 691
Query: 679 HRTLFLSYLDTSNAAIKDLAE-KAEVLC-IAGIEGGAKNIIPDVFQSMNHLKELLIRDSK 736
LF N + ++L K V C + G +++ + + ELL
Sbjct: 692 EYELFKYNAQVKNISFENLKRFKISVGCSLHGSFSKSRHSYENTLKLAIDKGELLESRMN 751
Query: 737 GIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKL 796
G+ + + VG ++ HL + S F NL L +S C +L
Sbjct: 752 GLFEKTEVLCLSVGDMY-------------HLSDV----KVKSSSFYNLRVLVVSECAEL 794
Query: 797 TRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECG 856
LFTL VA L++LE L+V C ++ ++ + + FPKLK
Sbjct: 795 KHLFTLGVANTLSKLEHLKVYKCDNMEELI------HTGGSEGDTITFPKLK-------- 840
Query: 857 VLEYIIPITLAQGLVQLECLEIVCNENLKYV----FGQSTHNDGQNQNELKIIELSALEE 912
L Y+ + GL CL + E K V + +N+L+ L EE
Sbjct: 841 -LLYLHGLPNLLGL----CLNVNAIELPKLVQMKLYSIPGFTSIYPRNKLEASSL-LKEE 894
Query: 913 LTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTL 972
+ + L + E+ +WPS + + + + + C L N N S L
Sbjct: 895 VVIPKLDILEIHDMENLKEIWPS--ELSRGEKVKLRKIKVRNCDKLVNLFPHNPMS--LL 950
Query: 973 QNITEVRVNNC-ELEGIF--QLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCK-SSVES 1028
++ E+ V C +E +F L + GE+D S L + +EN +LR + + ++
Sbjct: 951 HHLEELIVEKCGSIEELFNIDLDCASVIGEEDN-NSSLRNINVENSMKLREVWRIKGADN 1009
Query: 1029 TNLLFQNLQQME---ISGCRRLKCIFS 1052
+ LF+ Q +E I+ C+R +F+
Sbjct: 1010 SRPLFRGFQVVEKIIITRCKRFTNVFT 1036
Score = 49.7 bits (117), Expect = 5e-04
Identities = 36/123 (29%), Positives = 61/123 (49%), Gaps = 18/123 (14%)
Query: 972 LQNITEVRVNNCE-LEGIFQLV----------GLTNDGEKDPLTSCLEML------YLEN 1014
LQ + ++ +N+C +E +F+ G+ D T+ L L L
Sbjct: 1611 LQKLEKININSCVGVEEVFETALEAAGRNGNSGIGFDESSQTTTTTLVNLPNLREMNLWG 1670
Query: 1015 LPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKCNQLD 1074
L LRY+ KS+ + T F L ++EIS C L+ +F+S M G L QL+ L I +C ++
Sbjct: 1671 LDCLRYIWKSN-QWTAFEFPKLTRVEISNCNSLEHVFTSSMVGSLSQLQELHISQCKLME 1729
Query: 1075 QIV 1077
+++
Sbjct: 1730 EVI 1732
Score = 43.5 bits (101), Expect = 0.035
Identities = 68/329 (20%), Positives = 141/329 (42%), Gaps = 47/329 (14%)
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNL-AQLEKLQVLSCPELQHILIDDDRDE 833
+ P ++ NL L + H K +L +N ++EK+QV+S +L + L+ +
Sbjct: 527 KFPKDINYPNLLILKLMHGDK-----SLCFPENFYGKMEKVQVISYDKLMYPLLPSSLEC 581
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
+ L + L+ F G L+ +E L N N++++ ST
Sbjct: 582 STNVRVLHLHYCSLRMFDCSSIG------------NLLNMEVLSFA-NSNIEWL--PSTI 626
Query: 894 NDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYL----MWPSLLQFNLQNCGEFFM 949
N +L++++L+ + L + N N + E+ Y+ + + +NC E
Sbjct: 627 G---NLKKLRLLDLTNCKGLRIDNGVLKNLVKLEELYMGVNRPYGQAVSLTDENCNEMVE 683
Query: 950 VSINTCMALHNNPRIN-EASHQTLQNITEVRVN-NCELEGIFQLV--------------G 993
S + + N + + + +N+ +++ C L G F G
Sbjct: 684 GSKKLLALEYELFKYNAQVKNISFENLKRFKISVGCSLHGSFSKSRHSYENTLKLAIDKG 743
Query: 994 LTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSS 1053
+ + L E+L L ++ + +L V+S++ F NL+ + +S C LK +F+
Sbjct: 744 ELLESRMNGLFEKTEVLCL-SVGDMYHLSDVKVKSSS--FYNLRVLVVSECAELKHLFTL 800
Query: 1054 CMAGGLPQLKALKIEKCNQLDQIVEDIGT 1082
+A L +L+ LK+ KC+ +++++ G+
Sbjct: 801 GVANTLSKLEHLKVYKCDNMEELIHTGGS 829
Score = 41.6 bits (96), Expect = 0.13
Identities = 73/330 (22%), Positives = 129/330 (38%), Gaps = 74/330 (22%)
Query: 782 FENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILID-DDRDE----ISA 836
F NL + I C + LF+ +A+ L+ L+K+ + C ++ ++ + DD DE ++
Sbjct: 1180 FHNLTTINIDFCRSIKYLFSPLMAELLSNLKKVNIKWCYGIEEVVSNRDDEDEEMTTFTS 1239
Query: 837 YDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDG 896
+LFP L+ L + ENLK + G ++G
Sbjct: 1240 THTTTILFP--------------------------HLDSLTLSFLENLKCIGGGGAKDEG 1273
Query: 897 QNQNELK--IIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINT 954
N+ + L++ L ++ W SL Q+ + +SI
Sbjct: 1274 SNEISFNNTTATTAVLDQFELSEAGGVS----------W-SLCQYARE-------ISIEF 1315
Query: 955 CMALHN-NPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCLEMLYL 1012
C AL + P Q LQ +T V++C L+ +F+ + + + + C E
Sbjct: 1316 CNALSSVIPCYAAGQMQKLQVLT---VSSCNGLKEVFETQLRRSSNKNNEKSGCDE--GN 1370
Query: 1013 ENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALKIEKC-- 1070
+P+ V + ++ L+ +EIS C L+ IF+ L QL+ L I C
Sbjct: 1371 GGIPR--------VNNNVIMLSGLKILEISFCGGLEHIFTFSALESLRQLEELTIMNCWS 1422
Query: 1071 ------NQLDQIVEDIGTAFPSGTQTSTYS 1094
+ D+ E T GT +S+ S
Sbjct: 1423 MKVIVKKEEDEYGEQQTTTTTKGTSSSSSS 1452
Score = 37.0 bits (84), Expect = 3.3
Identities = 20/87 (22%), Positives = 41/87 (46%), Gaps = 9/87 (10%)
Query: 782 FENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILI---------DDDRD 832
F L + IS+C L +FT ++ +L+QL++L + C ++ +++ D +++
Sbjct: 1688 FPKLTRVEISNCNSLEHVFTSSMVGSLSQLQELHISQCKLMEEVIVKDADVSVEEDKEKE 1747
Query: 833 EISAYDYRLLLFPKLKKFHVRECGVLE 859
+ +L P LK + LE
Sbjct: 1748 SDGKMNKEILALPSLKSLKLESLPSLE 1774
>UniRef100_Q6Y136 Resistance protein RGC2 [Lactuca sativa]
Length = 1066
Score = 236 bits (601), Expect = 4e-60
Identities = 285/1130 (25%), Positives = 481/1130 (42%), Gaps = 194/1130 (17%)
Query: 11 ISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEK 70
I++ + V + Y +V+ + + L R SV++ ++R + + ++
Sbjct: 15 IAQTALVPVTDHVGYMISCRKYVRVMQMKMRELNTSRISVEEHISRNTRNHLQIPSQTKE 74
Query: 71 WLKD-----ANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSK-------KKR 118
WL AN+A +D + C + R+ +G+K K R
Sbjct: 75 WLDQVEGIRANVANFPIDVI-------------SCCSLRIRHKLGQKAFKITEQIESLTR 121
Query: 119 NLKLYIEEGRQYIEIERPASLSAGYFSAERCWE--FDSRKPAYEELMCALKDDDVT-MIG 175
L L I + + + S++A + + F SR+ + + + AL+ + MI
Sbjct: 122 QLSL-ISWTDDPVPLGKVGSMNASTSAPSSVYHDVFPSREQIFRKALEALEPVQKSHMIA 180
Query: 176 LYGMGGCGKTMLAM---EVGKRCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKD 232
L+GMGG GKT + EV +R +F ++ V I IQ+ +A L E +E
Sbjct: 181 LWGMGGVGKTTMMKKLKEVVER-KKMFSIIVQVVIGEKTNPIAIQQAVADYLSIELKENT 239
Query: 233 EMDRSKRLCMRLTQE---DRVLVILDDVWQMLDFDAIGIPSIEHHKGC--KILITSRSEA 287
+ R+ +L + ++ LVILDDVWQ +D + IG+ + + KG K+L+TSR
Sbjct: 240 KEARADKLRKWFEADGGKNKFLVILDDVWQFVDLEDIGLSPLPN-KGVNFKVLLTSRDSH 298
Query: 288 VCTLMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWI--SIKNMAREISNECKGLPVA 343
VCTLM + + + LT E LF++ A + + + +A I++ C+GLP+A
Sbjct: 299 VCTLMGAEANSILNIKVLTAVEGQSLFRQFAKNAGDDDLDPAFNRIADSIASRCQGLPIA 358
Query: 344 TVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLL 403
+A SLKG+++ W AL RL + K + E+ ++ +K +SYDNL E KS+FLL
Sbjct: 359 IKTIALSLKGRSKPAWDHALSRLENHK-IGSEEVVREVFK---ISYDNLQDEITKSIFLL 414
Query: 404 CSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCV 463
C++FPED +IP E L R GL + E + ARN + +L + LL ++ CV
Sbjct: 415 CALFPEDFDIPTEELMRYGWGLKLFIEAKTIREARNRLNTCTERLRETNLLFGSDDIGCV 474
Query: 464 KMHDLVR-------------------NVAHWIAENEIKCASEKDIMTLEHTSLRYLWCEK 504
KMHD+VR NV+ W+ EN I + + SL +
Sbjct: 475 KMHDVVRDFVLHIFSEVQHASIVNHGNVSEWLEENH-------SIYSCKRISLTCKGMSE 527
Query: 505 FPNSLDCSNLDFLQI-HTYTQVS-DEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNL 562
FP L NL L++ H +S E F G + Y+K PLL +SL+ TN+
Sbjct: 528 FPKDLKFPNLSILKLMHGDKSLSFPENFYGKMEKVQVISYDK--LMYPLLPSSLECSTNV 585
Query: 563 RC--ILFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERN 620
R + + + D S +G++ +E ++ + + LP + L LRLLDL+ C R
Sbjct: 586 RVLHLHYCSLRMFDCSSIGNLLNMEVLSFANSNIEWLPSTIGNLKKLRLLDLTNCKGLRI 645
Query: 621 PFEVIARHTELEELFFA--------------DC-----RSK----WEVEFLK-EFSVPQV 656
V+ +LEEL+ +C RSK E E K V +
Sbjct: 646 DNGVLKNLVKLEELYMGVNHPYGQAVSLTDENCDEMAERSKNLLALESELFKYNAQVKNI 705
Query: 657 ----LQRYQIQLGSMFSGFQDEFLNHHRT---LFLSYLDTSNAAIKDLAEKAEVLCIAGI 709
L+R++I +G G+ + ++ ++ L ++ + + + L EK EVLC++
Sbjct: 706 SFENLERFKISVGRSLDGYFSKNMHSYKNTLKLGINKGELLESRMNGLFEKTEVLCLS-- 763
Query: 710 EGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLG 769
+ ++D +EV + F
Sbjct: 764 ----------------------------VGDMIDLSDVEVKSSSF--------------- 780
Query: 770 ALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD 829
YN L L +S C +L LFTL VA L LE L+V C ++ ++
Sbjct: 781 --YN-----------LRVLVVSECAELKHLFTLGVANTLKMLEHLEVHKCKNMEELI--- 824
Query: 830 DRDEISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFG 889
+ + FPKLK + L + L L L+ ++
Sbjct: 825 ---HTGGSEGDTITFPKLKFLSLSGLPKLSGLCHNVNIIELPHLVDLKFKGIPGFTVIYP 881
Query: 890 QSTHNDGQNQNELKIIELSALEELTLVNLPNINSICP-EDCYLMWPSLLQFNLQNCGEFF 948
Q+ E + + LE L + ++ N+ I P E L + + NC +
Sbjct: 882 QNKLGTSSLLKEELQVVIPKLETLQIDDMENLEEIWPCERSGGEKVKLREITVSNCDK-- 939
Query: 949 MVSINTCMALHNNPRINEASHQTLQNITEVRVNNC-ELEGIFQL-VGLTNDGEKDPLTSC 1006
+V++ C NP L ++ E+ V NC +E +F + + ++ S
Sbjct: 940 LVNLFPC-----NPM------SLLHHLEELTVENCGSIESLFNIDLDCVGGIGEEYNKSI 988
Query: 1007 LEMLYLENLPQLRYL--CKSSVESTNLL--FQNLQQMEISGCRRLKCIFS 1052
L + +ENL +LR + K + S L+ F+ ++ + I GC+R + IF+
Sbjct: 989 LRSIKVENLGKLREVWGIKGADNSRPLIHGFKAVESISIWGCKRFRNIFT 1038
Score = 43.9 bits (102), Expect = 0.027
Identities = 73/330 (22%), Positives = 140/330 (42%), Gaps = 49/330 (14%)
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNL-AQLEKLQVLSCPELQHILIDDDRDE 833
+ P F NL L + H K +L+ +N ++EK+QV+S +L + L+ +
Sbjct: 527 EFPKDLKFPNLSILKLMHGDK-----SLSFPENFYGKMEKVQVISYDKLMYPLLPSSLEC 581
Query: 834 ISAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTH 893
+ L + L+ F G L+ +E L N N++++ ST
Sbjct: 582 STNVRVLHLHYCSLRMFDCSSIG------------NLLNMEVLSFA-NSNIEWL--PSTI 626
Query: 894 NDGQNQNELKIIELSALEELTLVNLPNINSICPEDCYL----MWPSLLQFNLQNCGEFFM 949
N +L++++L+ + L + N N + E+ Y+ + + +NC E
Sbjct: 627 G---NLKKLRLLDLTNCKGLRIDNGVLKNLVKLEELYMGVNHPYGQAVSLTDENCDEMAE 683
Query: 950 VSINTCMALHN-----NPRINEASHQTLQNITEVRVNNCELEGIFQL----------VGL 994
S N +AL + N ++ S + L+ ++ V L+G F +G+
Sbjct: 684 RSKNL-LALESELFKYNAQVKNISFENLERF-KISVGR-SLDGYFSKNMHSYKNTLKLGI 740
Query: 995 TNDGE--KDPLTSCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFS 1052
N GE + + E + L + S VE + F NL+ + +S C LK +F+
Sbjct: 741 -NKGELLESRMNGLFEKTEVLCLSVGDMIDLSDVEVKSSSFYNLRVLVVSECAELKHLFT 799
Query: 1053 SCMAGGLPQLKALKIEKCNQLDQIVEDIGT 1082
+A L L+ L++ KC +++++ G+
Sbjct: 800 LGVANTLKMLEHLEVHKCKNMEELIHTGGS 829
>UniRef100_Q6Y130 Resistance protein candidate RGC2 [Lactuca sativa]
Length = 1923
Score = 232 bits (591), Expect = 5e-59
Identities = 268/1098 (24%), Positives = 470/1098 (42%), Gaps = 106/1098 (9%)
Query: 1 MEYLYGFASAISRDLVCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQ 60
M+ + + L+ V L Y +V+D+ + S L + + V+D + +
Sbjct: 1 MDVINAIIKPVVDTLMVPVKRHLGYMINCTKYVRDMHNKLSELNSAKTGVEDHIKQNTSS 60
Query: 61 TRKTAEVVEKWLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSK----- 115
+ V WL+D V+ + S+ +SCF + R+ VGRK K
Sbjct: 61 LLEVPAQVRGWLEDVGKINAKVEDI----PSDVSSCF----SLKLRHKVGRKAFKIIEEV 112
Query: 116 ----KKRNLKLYIEEGRQYIEIERPASLSAGYFSAERCWE-FDSRKPAYEELMCALKDDD 170
+K +L ++ + I + + S+ A + + F SR+ + E + AL +
Sbjct: 113 ESVTRKHSLIIWTDHP---IPLGKVDSMKASVSTPSTYHDDFKSREQIFTEALQALHPNH 169
Query: 171 VT-MIGLYGMGGCGKTMLAMEVGK--RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFE 227
+ MI L GMGG GKT + + K + +FD ++ I + IQE +A L E
Sbjct: 170 KSHMIALCGMGGVGKTTMMQRLKKIVQEKKMFDFIIEAVIGHKTDPIAIQEAVADYLSIE 229
Query: 228 FQEKDEMDRSKRLCMRLTQE-----DRVLVILDDVWQMLDFDAIGIPSIEHHK-GCKILI 281
+EK + R+ L L + ++ LVILDDVWQ +D + IG+ + + K+L+
Sbjct: 230 LKEKTKSARADMLRKMLVAKSDGGKNKFLVILDDVWQFVDLEDIGLSPLPNQGVNFKVLL 289
Query: 282 TSRSEAVCTLMDCQKK--IQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKG 339
TSR VCT+M + + + L ++E LF + IS + + +I +C G
Sbjct: 290 TSRDVDVCTMMGVEANSILNMKILLDEEAQSLFMEFVQISSDVDPKLHKIGEDIVRKCCG 349
Query: 340 LPVATVAVASSLKGKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKS 399
LP+A +A +L+ K++ W AL RL N + +SYD L +E K
Sbjct: 350 LPIAIKTMALTLRNKSKDAWSDALSRLEHHDLHNFVN------EVFGISYDYLQDQETKY 403
Query: 400 LFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNE 459
+FLLC +FPED IP E L R GL + +V++ AR + +LI + LL++ +
Sbjct: 404 IFLLCGLFPEDYNIPPEELMRYGWGLNLFKKVYTIREARARLNTCIERLIHTNLLMEGDV 463
Query: 460 GKCVKMHDLVR----------NVAHWIAENEIKCASEKDIM-TLEHTSLRYLWCEKFPNS 508
CVKMHDL A + + E D+ + + SL FP
Sbjct: 464 VGCVKMHDLALAFVMDMFSKVQDASIVNHGSMSGWPENDVSGSCQRISLTCKGMSGFPID 523
Query: 509 LDCSNLDFLQI---HTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLK-SLTNLRC 564
L+ NL L++ + + + ++ M L+V+ + + P L +S + TNLR
Sbjct: 524 LNFPNLTILKLMHGDKFLKFPPDFYEQMEKLQVVSFHEM---KYPFLPSSPQYCSTNLRV 580
Query: 565 ILFSKWDLV-DISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFE 623
+ + L+ D S +G++ LE ++ + LP + L LRLLDL++C R
Sbjct: 581 LHLHQCSLMFDCSCIGNLFNLEVLSFANSGIEWLPSRIGNLKKLRLLDLTDCFGLRIDKG 640
Query: 624 VIARHTELEELFFADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLF 683
V+ +LEE++ + K S +L + EF
Sbjct: 641 VLKNLVKLEEVYMRVAVRSKKAGNRKAISFTDDNCNEMAELSKNLFALEFEFFE------ 694
Query: 684 LSYLDTSNAAIKDLA-EKAEVLCIA-GIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECL 741
NA K+++ EK E I+ G E ++I N L+ L+ + + +E
Sbjct: 695 ------INAQPKNMSFEKLERFKISMGSELRVDHLISSSHSFENTLR-LVTKKGELLESK 747
Query: 742 VDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQM--PLSGHFENLEDLYISHCPKLTRL 799
++ + L+ L + M L + + P S F NL L +S C +L L
Sbjct: 748 MNELFQKTDVLY------LSVGDMNDLEDIEVKSLHPPQSSSFYNLRVLVVSRCAELRYL 801
Query: 800 FTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVLE 859
FT++V + L++LE L+V C ++ ++ + E + FPKLK ++ L
Sbjct: 802 FTVSVVRALSKLEHLRVSYCKNMEELIHTGGKGE------EKITFPKLKFLYLHTLSKLS 855
Query: 860 YIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNELKIIELSALEELTLVNLP 919
+ + QL LE+ N+ ++ ++ N K + + LE+L++ +
Sbjct: 856 GLCHNVNIIEIPQLLELELFYIPNITNIYHKNNSETSCLLN--KEVMIPKLEKLSVRGMD 913
Query: 920 NINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVR 979
N+ I WP ++ + + + ++ C L N N + + E+
Sbjct: 914 NLKEI--------WP--CEYRMSGEVKVREIKVDYCNNLVNLFPCNPM--PLIHYLEELE 961
Query: 980 VNNC-ELEGIFQL-VGLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVESTNLL----F 1033
V NC +E +F + + +D +S L + + L L + + E+ + L F
Sbjct: 962 VKNCGSIEMLFNIDLDCVGGVGEDCGSSNLRSIVVFQLWNLSEVWRVKGENNSHLLVSGF 1021
Query: 1034 QNLQQMEISGCRRLKCIF 1051
Q ++ + I C R + IF
Sbjct: 1022 QAVESITIGSCVRFRHIF 1039
Score = 78.6 bits (192), Expect = 1e-12
Identities = 81/327 (24%), Positives = 144/327 (43%), Gaps = 42/327 (12%)
Query: 782 FENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRD-------EI 834
F NL+ L I C +L +FT + +L QLE+L+V C ++ I+ ++ D
Sbjct: 1507 FPNLKILIIRDCDRLEHIFTFSAVASLKQLEELRVWDCKAMKVIVKKEEEDASSSSSSSS 1566
Query: 835 SAYDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIV----CNENLKYVFGQ 890
S+ ++++FP+LK + G L+ ++ L Q L+ V C + + + GQ
Sbjct: 1567 SSSSKKVVVFPRLKSITL---GNLQNLVGFFLGMNDFQFPLLDDVVINICPQMVVFTSGQ 1623
Query: 891 STH---NDGQNQNELKIIEL-------SALEELTLVNLPNINSICPEDCYLMWPSLLQFN 940
T Q I+E + L NI S P P ++
Sbjct: 1624 LTALKLKHVQTGVGTYILECGLNFHVSTTAHHQNLFQSSNITSSSPATTKGGVP----WS 1679
Query: 941 LQNCGEFFMVSINTCMALHNNPRINEASH-QTLQNITEVRVNNCEL-EGIFQLVGLTNDG 998
QN ++ ++ + ++ + Q LQN+ +R+ C L E +F+ + TN G
Sbjct: 1680 YQN-----LIKLHVSSYMETPKKLFPCNELQQLQNLEMIRLWRCNLVEEVFEALQGTNSG 1734
Query: 999 EKDPLT------SCLEMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFS 1052
S L + LE L LRY+ +S+ + T NL ++EI C RL+ +F+
Sbjct: 1735 SASASQTTLVKLSNLRQVELEGLMNLRYIWRSN-QWTVFELANLTRVEIKECARLEYVFT 1793
Query: 1053 SCMAGGLPQLKALKIEKCNQLDQIVED 1079
M G L QL+ L + C ++++++ +
Sbjct: 1794 IPMVGSLLQLQDLTVRSCKRMEEVISN 1820
Score = 42.7 bits (99), Expect = 0.060
Identities = 63/270 (23%), Positives = 111/270 (40%), Gaps = 36/270 (13%)
Query: 848 KKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVF---GQSTHNDGQNQNELKI 904
++ +R C L +IP A+ + +LE L I +K +F G + +N G +
Sbjct: 1312 REITIRMCYKLSSLIPSYTARQMQKLEKLTIENCGGMKELFETQGINNNNIGCEEGNFDT 1371
Query: 905 IELSALEE---LTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNN 961
+ L LVNL +N ++P L++ G+ + I C A+
Sbjct: 1372 PAIPRRNNGSMLQLVNLKELNIKSANHLEYVFPYSA---LESLGKLEELWIRNCSAMKVI 1428
Query: 962 PRINEASHQTLQNITEVRVNNCELEGIFQLVGLTNDGEKDPLTSCLEMLYL--------- 1012
+ ++ QT++ N + + + L+N CL +L
Sbjct: 1429 VKEDDGEQQTIRT-KGASSNEVVVFPPIKSIILSN-------LPCLMGFFLGMKEFTHGW 1480
Query: 1013 ENLPQLRY----LCKSSVES--TNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLKALK 1066
PQ++Y L K S+E N+ F NL+ + I C RL+ IF+ L QL+ L+
Sbjct: 1481 STAPQIKYIDTSLGKHSLEYGLINIQFPNLKILIIRDCDRLEHIFTFSAVASLKQLEELR 1540
Query: 1067 IEKCNQLDQIV----EDIGTAFPSGTQTST 1092
+ C + IV ED ++ S + +S+
Sbjct: 1541 VWDCKAMKVIVKKEEEDASSSSSSSSSSSS 1570
Score = 37.0 bits (84), Expect = 3.3
Identities = 66/334 (19%), Positives = 132/334 (38%), Gaps = 51/334 (15%)
Query: 777 PLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISA 836
P+ +F NL L + H K + Q+EKLQV+S E+++ + S
Sbjct: 521 PIDLNFPNLTILKLMHGDKFLKF----PPDFYEQMEKLQVVSFHEMKYPFLPSSPQYCST 576
Query: 837 YDYRLLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDG 896
L+ H+ +C ++ + L LE L N ++++ +
Sbjct: 577 ---------NLRVLHLHQCSLM---FDCSCIGNLFNLEVLSFA-NSGIEWLPSRIG---- 619
Query: 897 QNQNELKIIELSALEELTLVNLPNINSICPEDCYLMWP---------SLLQFNLQNCGEF 947
N +L++++L+ L + N + E+ Y+ + F NC E
Sbjct: 620 -NLKKLRLLDLTDCFGLRIDKGVLKNLVKLEEVYMRVAVRSKKAGNRKAISFTDDNCNEM 678
Query: 948 FMVSINTCMA----LHNNPRINEASHQTLQNI-----TEVRVNNC-----ELEGIFQLV- 992
+S N N + S + L+ +E+RV++ E +LV
Sbjct: 679 AELSKNLFALEFEFFEINAQPKNMSFEKLERFKISMGSELRVDHLISSSHSFENTLRLVT 738
Query: 993 --GLTNDGEKDPLTSCLEMLYLE--NLPQLRYL-CKSSVESTNLLFQNLQQMEISGCRRL 1047
G + + + L ++LYL ++ L + KS + F NL+ + +S C L
Sbjct: 739 KKGELLESKMNELFQKTDVLYLSVGDMNDLEDIEVKSLHPPQSSSFYNLRVLVVSRCAEL 798
Query: 1048 KCIFSSCMAGGLPQLKALKIEKCNQLDQIVEDIG 1081
+ +F+ + L +L+ L++ C +++++ G
Sbjct: 799 RYLFTVSVVRALSKLEHLRVSYCKNMEELIHTGG 832
Score = 37.0 bits (84), Expect = 3.3
Identities = 24/115 (20%), Positives = 52/115 (44%), Gaps = 12/115 (10%)
Query: 715 NIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNG 774
N++ +VF+++ + G T L+++ L + +E + +L ++
Sbjct: 1719 NLVEEVFEALQGT-------NSGSASASQTTLVKLSNL-----RQVELEGLMNLRYIWRS 1766
Query: 775 QMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDD 829
NL + I C +L +FT+ + +L QL+ L V SC ++ ++ +D
Sbjct: 1767 NQWTVFELANLTRVEIKECARLEYVFTIPMVGSLLQLQDLTVRSCKRMEEVISND 1821
Score = 36.6 bits (83), Expect = 4.3
Identities = 34/154 (22%), Positives = 66/154 (42%), Gaps = 23/154 (14%)
Query: 787 DLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHIL-----------IDDDRDEIS 835
++ I C KL+ L A+ + +LEKL + +C ++ + ++ +
Sbjct: 1313 EITIRMCYKLSSLIPSYTARQMQKLEKLTIENCGGMKELFETQGINNNNIGCEEGNFDTP 1372
Query: 836 AYDYR----LLLFPKLKKFHVRECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVF--- 888
A R +L LK+ +++ LEY+ P + + L +LE L I +K +
Sbjct: 1373 AIPRRNNGSMLQLVNLKELNIKSANHLEYVFPYSALESLGKLEELWIRNCSAMKVIVKED 1432
Query: 889 ---GQSTHNDGQNQNELKIIELSALEELTLVNLP 919
Q+ G + NE ++ ++ + L NLP
Sbjct: 1433 DGEQQTIRTKGASSNE--VVVFPPIKSIILSNLP 1464
>UniRef100_Q6Y132 Resistance protein RGC2 [Lactuca sativa]
Length = 1285
Score = 226 bits (577), Expect = 2e-57
Identities = 280/1109 (25%), Positives = 486/1109 (43%), Gaps = 136/1109 (12%)
Query: 23 LSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNV 82
L Y +++D+ + L A + V+++ + V+ WL D+V
Sbjct: 33 LRYLILCTKYMRDMGIKIIELNAAKVGVEEKTRHNISNNLEVPAQVKGWL-------DDV 85
Query: 83 DQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERP------ 136
++ ++ N+ G C N R++ GR + + + ++ + P
Sbjct: 86 GKINAQVENVPNN-IGSCFNLKIRHTAGRSAVEISEEIDSVMRRYKEINWADHPIPPGRV 144
Query: 137 -ASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVT-MIGLYGMGGCGKTMLAMEVGK- 193
+ S+ + + +F SR+ + + + AL + + MI L GMGG GKT + + K
Sbjct: 145 HSMKSSTSTLSTKHNDFQSRELTFTKALKALDLNHKSHMIALCGMGGVGKTTMMQRLKKV 204
Query: 194 -RCGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQE----- 247
+ +F ++ I + IQE I+ L E + R+ L +
Sbjct: 205 AKEKRMFSYIIEAVIGEKTDPISIQEAISYYLGVELNANTKSVRADMLRQGFKAKSDVGK 264
Query: 248 DRVLVILDDVWQMLDFDAIGI-PSIEHHKGCKILITSRSEAVCTLMDCQKK--IQLSTLT 304
D+ L+ILDDVWQ +D + IG+ P K+L+TSR +CT+M + + LT
Sbjct: 265 DKFLIILDDVWQSVDLEDIGLSPFPNQGVNFKVLLTSRDRHICTVMGVEGHSIFNVGLLT 324
Query: 305 NDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVEWKVALD 364
E+ LF + EG+ + + +I ++C GLP+A +A +L+ K+ WK AL
Sbjct: 325 EAESKRLFWQ---FVEGSDPELHKIGEDIVSKCCGLPIAIKTMACTLRDKSTDAWKDALS 381
Query: 365 RLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIG 424
RL N+ K + SYDNL EE KS F LC +FPED IP+E L R G
Sbjct: 382 RLEHHDIENVAS------KVFKASYDNLQDEETKSTFFLCGLFPEDSNIPMEELVRYGWG 435
Query: 425 LGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENEIKCA 484
L + +V++ AR + +LI + LL+ V++ +C+KMHDL+R+ + +
Sbjct: 436 LKLFKKVYTIREARTRLNTCIERLIYTNLLIKVDDVQCIKMHDLIRSFVLDMFSKVEHAS 495
Query: 485 SEKDIMTLE------HTSLRYL------WCE-----KFPNSLDCSNLDFLQIHTYTQVSD 527
TLE H S + L CE KFPN + L + +
Sbjct: 496 IVNHGNTLEWPADDMHDSCKGLSLTCKGICEFCGDLKFPNLM---ILKLMHGDKSLRFPQ 552
Query: 528 EIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDL--VDISFVGDMKKLE 585
++GM+ L+V+ + + + PLL S + TNLR + + L D S +G++ LE
Sbjct: 553 NFYEGMQKLQVI---SYDKMKYPLLPLSSECSTNLRVLHLHECSLQMFDFSSIGNLLNLE 609
Query: 586 SITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWEV 645
++ D LP + L LR+LDL ++ +LEEL+
Sbjct: 610 VLSFADSCIQMLPSTIGNLKKLRVLDLRGSDDLHIEQGILKNLVKLEELYMG-------- 661
Query: 646 EFLKEFSVPQVLQRYQIQLGSMFSGFQDEFLNH--HRTLFLSYLD----TSNAAIKDLA- 698
F EF G D+ N R+ LS L+ +NA K+++
Sbjct: 662 -FYDEFR----------HRGKGIYNMTDDNYNEIAERSKGLSALEIEFFRNNAQPKNMSF 710
Query: 699 EKAEVLCIAGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVDTCLIE--VGTLFFCK 756
EK E I G + + D + M ++ L +K E L+D+ L E V T C
Sbjct: 711 EKLEKF---KISVGRRYLYGDYMKHMYAVQNTLKLVTKKGE-LLDSRLNELFVKTEMLC- 765
Query: 757 LHWLRIEHMKHLGAL--YNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKL 814
L ++ M LG L + + P F+ L L +S C +L LFT+ VA++L+ LE L
Sbjct: 766 ---LSVDDMNDLGDLDVKSSRFPQPSSFKILRVLVVSMCAELRYLFTIGVAKDLSNLEHL 822
Query: 815 QVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHV----RECGVLEYIIPITLAQGL 870
+V SC ++ ++ ++ + + + F KLK + + G+ + I L Q L
Sbjct: 823 EVDSCDNMEELICSENAGK------KTITFLKLKVLCLFGLPKLSGLCHNVNRIELLQ-L 875
Query: 871 VQLECLEIVCNENLKYVFGQSTHNDGQNQNELKI-IELSALEELTLVNLPNINSICPEDC 929
V+L+ I N+ ++ + N + LK + + LE+L+++++ N+ I P D
Sbjct: 876 VELKLSRI---GNITSIYPK---NKLETSCFLKAEVLVPKLEKLSIIHMDNLKEIWPCD- 928
Query: 930 YLMWPSLLQFNLQNCGEFFMVSINTCMALHNNPRINEASHQTLQNITEVRVNNCELEGIF 989
+ + + NL+ + +N+C L N N L ++ E++V C +
Sbjct: 929 ---FRTSDEVNLRE------IYVNSCDKLMNLFPCNPM--PLLHHLQELQVKWCGSIEVL 977
Query: 990 QLVGLTNDGE--KDPLTSCLEMLYLENLPQLRYLCKSSVESTN-----LLFQNLQQMEIS 1042
+ L GE + + + L + ++ L +LR + + + N FQ ++++ +
Sbjct: 978 FNIDLDCAGEIGEGGIKTNLRSIEVDCLGKLREVWRIKGDQVNSGVNIRSFQAVEKIMVK 1037
Query: 1043 GCRRLKCIFSSCMAG-GLPQLKALKIEKC 1070
C+R + +F+ A L L + IE C
Sbjct: 1038 RCKRFRNLFTPTGANFDLGALMEISIEDC 1066
Score = 40.4 bits (93), Expect = 0.30
Identities = 23/83 (27%), Positives = 46/83 (54%), Gaps = 3/83 (3%)
Query: 782 FENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILID-DDRDE--ISAYD 838
F NL +Y+ C ++ LF+ +A+ L+ L+K+ + C ++ ++ + DD+DE + +
Sbjct: 1175 FYNLTTIYMYGCRRIKYLFSPLMAKLLSNLKKVHIEFCDGIEEVVSNRDDKDEEMTTFTN 1234
Query: 839 YRLLLFPKLKKFHVRECGVLEYI 861
+LFP L H+ L++I
Sbjct: 1235 TSTILFPHLDSLHLSSLKTLKHI 1257
>UniRef100_Q6Y142 Resistance protein RGC2 [Lactuca sativa]
Length = 892
Score = 221 bits (564), Expect = 7e-56
Identities = 248/959 (25%), Positives = 409/959 (41%), Gaps = 157/959 (16%)
Query: 183 GKTMLAMEVGKRCGN--LFDQVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRL 240
GKT + + K +F ++ V I + IQE IA L E EK++ R+ +L
Sbjct: 2 GKTTIMQRLKKVAEEKKMFKFIVEVVIGEKTDPISIQEAIAYYLSVELSEKNKSVRANKL 61
Query: 241 CMRLTQE-----DRVLVILDDVWQMLDFDAIGIPSIEHH-KGCKILITSRSEAVCTLMDC 294
+ ++ L++LDDVWQ +D + IGI + + K+L+TSR VCT+M
Sbjct: 62 RRGFKAKSDGGKNKFLIVLDDVWQSVDLEDIGISPLPNQCVDFKVLLTSRDRNVCTMMGV 121
Query: 295 QKK--IQLSTLTNDETWDLFQKQALISEGTWISIKNMAREISNECKGLPVATVAVASSLK 352
+ + + L + E LF + S+ + M +I +C GLP+A +A +L+
Sbjct: 122 EGNSILHVGLLIDSEAQRLFWQFVETSDH---ELHKMGEDIVKKCCGLPIAIKTMACTLR 178
Query: 353 GKAEVEWKVALDRLRSSKPVNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCE 412
K++ WK AL RL N+ K + SYDNL +E KS FLLC +F ED
Sbjct: 179 DKSKDAWKDALFRLEHHDIENVAS------KVFKTSYDNLQDDETKSTFLLCGLFSEDFN 232
Query: 413 IPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVR-- 470
IP E L R GL + +V++ AR + +LI + LLL+ + + VKMHDLVR
Sbjct: 233 IPTEELVRYGWGLKLFKKVYNIREARTRLNTYIERLIHTNLLLESVDVRWVKMHDLVRAF 292
Query: 471 ------NVAHWIA---ENEIKCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLDFLQI-- 519
V H N ++ + + + SL +FP L NL L++
Sbjct: 293 VLGMYSEVEHASIINHGNTLEWHVDDTDDSYKRLSLTCKSMSEFPRDLKFPNLMILKLIH 352
Query: 520 -HTYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDL--VDIS 576
+ + + ++GM L+V+ + + + PLL +S + TNLR + + L D S
Sbjct: 353 GDKFLRFPQDFYEGMGKLQVI---SYDKMKYPLLPSSFQCSTNLRVLHLHECSLRMFDCS 409
Query: 577 FVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFF 636
+G++ LE ++ D LP + L +RLLDL+ C V+ + +LEEL+
Sbjct: 410 CIGNLLNLEVLSFADSGIEWLPSTIGNLKKIRLLDLTNCHGLCIANGVLKKLVKLEELYM 469
Query: 637 ADCR-----------------------SKWEVEFLKEFSVP-----QVLQRYQIQLGSMF 668
R S E+E K P + LQR+QI +G
Sbjct: 470 RGVRQHRKAVNLTEDNCNEMAERSKDLSALELEVYKNSVQPKNMSFEKLQRFQISVGRYL 529
Query: 669 SGFQDEFLNHHRT---LFLSYLDTSNAAIKDLAEKAEVLCIAGIEGGAKNIIPDVFQSMN 725
G + + + L + + + + +L +K EVLC++ MN
Sbjct: 530 YGASIKSRHSYENTLKLVVQKGELLESRMNELFKKTEVLCLS-------------VGDMN 576
Query: 726 HLKELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENL 785
L+++ ++ S Q S F +L
Sbjct: 577 DLEDIEVKSS--------------------------------------SQPFQSSSFYHL 598
Query: 786 EDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFP 845
L +S C +L LFT V L +LE L+V C ++ ++ D +E + FP
Sbjct: 599 RVLVVSKCAELKHLFTPGVTNTLKKLEHLEVYKCDNMEELIHTGDSEE------ETITFP 652
Query: 846 KLKKFHV----RECGVLEYIIPITLAQGLVQLECLEIVCNENLKYVFGQSTHNDGQNQNE 901
KLK + + G+ + + I L Q L++LE L+ + Y +S + E
Sbjct: 653 KLKFLSLCGLPKLLGLCDNVKIIELPQ-LMELE-LDNIPGFTSIYPMKKS-ETSSLLKEE 709
Query: 902 LKIIELSALEELTLVNLPNINSICPEDCYLMWPSLLQFNLQNCGEFFMVSINTCMALHNN 961
+ I +L L ++ NL I WP +FN +F + ++ C L N
Sbjct: 710 VLIPKLEKLHVSSMWNLKEI-----------WP--CEFNTSEEVKFREIEVSNCDKLVNL 756
Query: 962 PRINEASHQTLQNITEVRVNNC-ELEGIFQLVGLTNDG--EKDPLTSCLEMLYLENLPQL 1018
N S L ++ E+ V NC +E +F + L DG E++ + L + +ENL +L
Sbjct: 757 FPHNPMS--MLHHLEELEVENCGSIESLFN-IDLDCDGAIEQEDNSISLRNIEVENLGKL 813
Query: 1019 R--YLCKSSVESTNLL--FQNLQQMEISGCRRLKCIFSSCMAG-GLPQLKALKIEKCNQ 1072
R + K S L+ FQ ++ + + C+R + +F+ L L + I+ C +
Sbjct: 814 REVWRIKGGDNSRPLVHGFQAVESIRVRKCKRFRNVFTPTTTNFDLGALLEISIDDCGE 872
>UniRef100_Q9T048 Putative disease resistance protein At4g27190 [Arabidopsis
thaliana]
Length = 985
Score = 219 bits (557), Expect = 5e-55
Identities = 236/900 (26%), Positives = 397/900 (43%), Gaps = 118/900 (13%)
Query: 16 VCGVIGQLSYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVV------- 68
V G I +L Y F+ V + K +SN+ A+ +S++ R+T K + E +
Sbjct: 7 VIGEILRLMYESTFSR-VANAIKFKSNVKALNESLE-RLTELKGNMSEDHETLLTKDKPL 64
Query: 69 ----EKWLKDANIAMDNVDQLLQMAKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYI 124
+W ++A + V ++ E+ SC R + RKL K +K+
Sbjct: 65 RLKLMRWQREA----EEVISKARLKLEERVSC-----GMSLRPRMSRKLVKILDEVKMLE 115
Query: 125 EEGRQYIEIERPASLSAGYFSAERCWEFDSRKPAYEELMC--------ALKDDDVTMIGL 176
++G +++++ S + ER ++ + L + IG+
Sbjct: 116 KDGIEFVDMLSVES------TPERVEHVPGVSVVHQTMASNMLAKIRDGLTSEKAQKIGV 169
Query: 177 YGMGGCGKTMLAMEVGKR-----CGNLFDQVLFVPISSTVEVERIQEKIAGSLEFEFQ-E 230
+GMGG GKT L + + F V+FV +S + +Q++IA L+ + Q E
Sbjct: 170 WGMGGVGKTTLVRTLNNKLREEGATQPFGLVIFVIVSKEFDPREVQKQIAERLDIDTQME 229
Query: 231 KDEMDRSKRLCMRLTQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCT 290
+ E ++R+ + L +E + L+ILDDVW+ +D D +GIP E +KG K+++TSR VC
Sbjct: 230 ESEEKLARRIYVGLMKERKFLLILDDVWKPIDLDLLGIPRTEENKGSKVILTSRFLEVCR 289
Query: 291 LMDCQKKIQLSTLTNDETWDLFQKQALISEGTWI---SIKNMAREISNECKGLPVATVAV 347
M +++ L ++ W+LF K A G + ++ +A+ +S EC GLP+A + V
Sbjct: 290 SMKTDLDVRVDCLLEEDAWELFCKNA----GDVVRSDHVRKIAKAVSQECGGLPLAIITV 345
Query: 348 ASSLKGKAEVE-WKVALDRLRSSKP--VNIEKGLQNPYKCLQLSYDNLDTEEAKSLFLLC 404
++++GK V+ W L +L S P +IE+ + P L+LSYD L+ ++AK FLLC
Sbjct: 346 GTAMRGKKNVKLWNHVLSKLSKSVPWIKSIEEKIFQP---LKLSYDFLE-DKAKFCFLLC 401
Query: 405 SVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVK 464
++FPED I V + R + G + E+ S E + NE L CLL D + VK
Sbjct: 402 ALFPEDYSIEVTEVVRYWMAEGFMEELGSQEDSMNEGITTVESLKDYCLLEDGDRRDTVK 461
Query: 465 MHDLVRNVAHWIAENE--------------IKCASEKDIMTLEHTSLRYLWCEKFPNSLD 510
MHD+VR+ A WI + +K +L SL E P+ ++
Sbjct: 462 MHDVVRDFAIWIMSSSQDDSHSLVMSGTGLQDIRQDKLAPSLRRVSLMNNKLESLPDLVE 521
Query: 511 --CSNLDFLQIH---TYTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCI 565
C L + +V + LR+L L G + + SL L +L +
Sbjct: 522 EFCVKTSVLLLQGNFLLKEVPIGFLQAFPTLRILNL--SGTRIKSFPSCSLLRLFSLHSL 579
Query: 566 -LFSKWDLVDISFVGDMKKLESITLCDCSFVELPDVVTQLTNLRLLDLSE-CGMERNPFE 623
L + LV + + + KLE + LC +E P + +L R LDLS +E P
Sbjct: 580 FLRDCFKLVKLPSLETLAKLELLDLCGTHILEFPRGLEELKRFRHLDLSRTLHLESIPAR 639
Query: 624 VIARHTELEELFFADCRSKWEVE--------FLKEFSVPQVLQRYQIQLGS--------- 666
V++R + LE L +W V+ ++E Q LQ I+L S
Sbjct: 640 VVSRLSSLETLDMTSSHYRWSVQGETQKGQATVEEIGCLQRLQVLSIRLHSSPFLLNKRN 699
Query: 667 ----------MFSGFQDEFLNHH--RTLFLSYLDTSNAAIKDLAEKAEVLCI---AGIEG 711
+ G + H R L +S+L+ S +I L L + GIE
Sbjct: 700 TWIKRLKKFQLVVGSRYILRTRHDKRRLTISHLNVSQVSIGWLLAYTTSLALNHCQGIEA 759
Query: 712 GAKNIIPD--VFQSMNHL--KELLIRDSKGIECLVDTCLIEVGTLFFCKLHWLRIEHMKH 767
K ++ D F+++ L + ++I + +E +V T + + L L H++
Sbjct: 760 MMKKLVSDNKGFKNLKSLTIENVIINTNSWVE-MVSTNTSKQSSDILDLLPNLEELHLRR 818
Query: 768 --LGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHI 825
L Q L E L+ + I+ C KL L + LE++++ C LQ++
Sbjct: 819 VDLETFSELQTHLGLKLETLKIIEITMCRKLRTLLDKRNFLTIPNLEEIEISYCDSLQNL 878
Score = 37.4 bits (85), Expect = 2.5
Identities = 26/116 (22%), Positives = 56/116 (47%), Gaps = 13/116 (11%)
Query: 949 MVSINTCMALHNNPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCL 1007
++ I C L +++ + T+ N+ E+ ++ C+ L+ + + + P L
Sbjct: 840 IIEITMCRKLRT--LLDKRNFLTIPNLEEIEISYCDSLQNLHEALLY-----HQPFVPNL 892
Query: 1008 EMLYLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFSSCMAGGLPQLK 1063
+L L NLP L +C + +++ L+Q+E+ C +L C+ S G + ++K
Sbjct: 893 RVLKLRNLPNLVSIC-----NWGEVWECLEQVEVIHCNQLNCLPISSTCGRIKKIK 943
>UniRef100_O81825 Putative disease resistance protein At4g27220 [Arabidopsis
thaliana]
Length = 919
Score = 208 bits (529), Expect = 8e-52
Identities = 226/865 (26%), Positives = 396/865 (45%), Gaps = 96/865 (11%)
Query: 29 FNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQM 88
F + + L + L ++ V + + R+ Q + + WL+ +NV L ++
Sbjct: 2 FRSNARALNRALERLKNVQTKVNEALKRSGIQEKSLERKLRIWLRKVE---ENVP-LGEL 57
Query: 89 AKSEKNSCFGHCPNWIWRYSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLSAGYFSAER 148
+++SC IW ++ +K + L+ E+G+ I+ S
Sbjct: 58 ILEKRSSCA------IWLSDKDVEILEKVKRLE---EQGQDLIKKISVNKSSREIVERVL 108
Query: 149 CWEFDSRKPAYE---ELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGK-----RCGNLFD 200
F +K A E +L LK +V IG++GMGG GKT L + F
Sbjct: 109 GPSFHPQKTALEMLDKLKDCLKKKNVQKIGVWGMGGVGKTTLVRTLNNDLLKYAATQQFA 168
Query: 201 QVLFVPISSTVEVERIQEKIAGSLEFEFQEKDEMDRSKRLCMRLTQEDRVLVILDDVWQM 260
V++V +S +++R+Q IA L F + +C RL L+ILDDVW
Sbjct: 169 LVIWVTVSKDFDLKRVQMDIAKRLGKRFTREQMNQLGLTICERLIDLKNFLLILDDVWHP 228
Query: 261 LDFDAIGIP-SIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQALIS 319
+D D +GIP ++E K K+++TSR VC M + I+++ L E W+LF +
Sbjct: 229 IDLDQLGIPLALERSKDSKVVLTSRRLEVCQQMMTNENIKVACLQEKEAWELFCHN--VG 286
Query: 320 E-GTWISIKNMAREISNECKGLPVATVAVASSLKGKAEVE-WKVALDRLRSSKP-VNIEK 376
E ++K +A+++S+EC GLP+A + + +L+GK +VE WK L+ L+ S P ++ E+
Sbjct: 287 EVANSDNVKPIAKDVSHECCGLPLAIITIGRTLRGKPQVEVWKHTLNLLKRSAPSIDTEE 346
Query: 377 GLQNPYKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEVHSYEG 436
+ + L+LSYD L + KS FL C++FPED I V L + G++ H YE
Sbjct: 347 KI---FGTLKLSYDFLQ-DNMKSCFLFCALFPEDYSIKVSELIMYWVAEGLLDGQHHYED 402
Query: 437 ARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIAENE--------------IK 482
NE +L SCLL D + VKMHD+VR+ A W ++ I+
Sbjct: 403 MMNEGVTLVERLKDSCLLEDGDSCDTVKMHDVVRDFAIWFMSSQGEGFHSLVMAGRGLIE 462
Query: 483 CASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD----FLQIHTYT-QVSDEIFKGMRMLR 537
+K + +++ SL E+ PN++ ++ LQ +++ +V + + LR
Sbjct: 463 FPQDKFVSSVQRVSLMANKLERLPNNV-IEGVETLVLLLQGNSHVKEVPNGFLQAFPNLR 521
Query: 538 VLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDMKKLESITLCDCSFVEL 597
+L L P ++L SL +L +L + L ++ + + KL+ + L + + EL
Sbjct: 522 ILDLSGVRIRTLPDSFSNLHSLRSL--VLRNCKKLRNLPSLESLVKLQFLDLHESAIREL 579
Query: 598 PDVVTQLTNLRLLDLSEC-GMERNPFEVIARHTELEELFFADCRSKWEVE--------FL 648
P + L++LR + +S ++ P I + + LE L A W ++ L
Sbjct: 580 PRGLEALSSLRYICVSNTYQLQSIPAGTILQLSSLEVLDMAGSAYSWGIKGEEREGQATL 639
Query: 649 KEFSVPQVLQRYQIQLGSMFS-GFQDEFLNHHRTLF---------------------LSY 686
E + LQ I+L + S ++ + L T F +S
Sbjct: 640 DEVTCLPHLQFLAIKLLDVLSFSYEFDSLTKRLTKFQFLFSPIRSVSPPGTGEGCLAISD 699
Query: 687 LDTSNAAIKDLAEKAEVLCI---AGIEGGAKNIIPDVFQSMNHLKELLIRDSKGIECLVD 743
++ SNA+I L + L + G+ G +N++ S +K L I + L
Sbjct: 700 VNVSNASIGWLLQHVTSLDLNYCEGLNGMFENLVTKSKSSFVAMKALSIHYFPSLS-LAS 758
Query: 744 TCLIEVGTLFFCKLHWLRIE--HMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFT 801
C ++ F L L ++ +++ +G L NG + + + L+ L +S C +L RLF+
Sbjct: 759 GCESQLD--LFPNLEELSLDNVNLESIGEL-NGFLGM--RLQKLKLLQVSGCRQLKRLFS 813
Query: 802 -LAVAQNLAQLEKLQVLSCPELQHI 825
+A L L++++V+SC L+ +
Sbjct: 814 DQILAGTLPNLQEIKVVSCLRLEEL 838
Score = 47.0 bits (110), Expect = 0.003
Identities = 40/147 (27%), Positives = 72/147 (48%), Gaps = 27/147 (18%)
Query: 955 CMALHNNPRINEASHQTLQNITEVRVNNCE-LEGIFQLVGLTNDGEKDPLTSCLEMLYLE 1013
C+A+ + N + LQ++T + +N CE L G+F+ + +T L + Y
Sbjct: 694 CLAISDVNVSNASIGWLLQHVTSLDLNYCEGLNGMFENL-VTKSKSSFVAMKALSIHYFP 752
Query: 1014 NLPQLRYLCKS-----------SVESTNL------------LFQNLQQMEISGCRRLKCI 1050
+L L C+S S+++ NL Q L+ +++SGCR+LK +
Sbjct: 753 SL-SLASGCESQLDLFPNLEELSLDNVNLESIGELNGFLGMRLQKLKLLQVSGCRQLKRL 811
Query: 1051 FS-SCMAGGLPQLKALKIEKCNQLDQI 1076
FS +AG LP L+ +K+ C +L+++
Sbjct: 812 FSDQILAGTLPNLQEIKVVSCLRLEEL 838
Score = 42.7 bits (99), Expect = 0.060
Identities = 26/79 (32%), Positives = 42/79 (52%), Gaps = 6/79 (7%)
Query: 971 TLQNITEVRVNNC-ELEGIFQLVGLTNDGEKDPLTSCLEMLYLENLPQLRYLCKSSVEST 1029
TL N+ E++V +C LE +F + D + L L ++ L+ LPQLR LC V
Sbjct: 820 TLPNLQEIKVVSCLRLEELFNFSSVPVDFCAESLLPKLTVIKLKYLPQLRSLCNDRV--- 876
Query: 1030 NLLFQNLQQMEISGCRRLK 1048
+ ++L+ +E+ C LK
Sbjct: 877 --VLESLEHLEVESCESLK 893
Score = 36.6 bits (83), Expect = 4.3
Identities = 34/136 (25%), Positives = 63/136 (46%), Gaps = 19/136 (13%)
Query: 956 MALHNNPRINEASHQTLQ-----NITEVRVNNCELEGIFQLVGLTNDGEKDPLTSCLEML 1010
+++H P ++ AS Q N+ E+ ++N LE I +L G + L++L
Sbjct: 746 LSIHYFPSLSLASGCESQLDLFPNLEELSLDNVNLESIGELNGFLGMRLQK-----LKLL 800
Query: 1011 YLENLPQLRYLCKSSVESTNLLFQNLQQMEISGCRRLKCIFS-------SCMAGGLPQLK 1063
+ QL+ L + + L NLQ++++ C RL+ +F+ C LP+L
Sbjct: 801 QVSGCRQLKRLFSDQILAGTL--PNLQEIKVVSCLRLEELFNFSSVPVDFCAESLLPKLT 858
Query: 1064 ALKIEKCNQLDQIVED 1079
+K++ QL + D
Sbjct: 859 VIKLKYLPQLRSLCND 874
>UniRef100_O22727 Putative disease resistance protein At1g61190 [Arabidopsis
thaliana]
Length = 967
Score = 206 bits (524), Expect = 3e-51
Identities = 236/892 (26%), Positives = 400/892 (44%), Gaps = 102/892 (11%)
Query: 36 LAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVDQLLQMAKSE--K 93
L +E +L A + VQ++V R + + ++ E V+ WL N LL ++ E K
Sbjct: 38 LQREMEDLRATQHEVQNKVAREESRHQQRLEAVQVWLDRVNSIDIECKDLLSVSPVELQK 97
Query: 94 NSCFGHCPNWIWR-YSVGRKLSKKKRNLKLYIEEG----------RQYIEIERPASLSAG 142
G C ++ Y G+++ + EG R +E ERP + G
Sbjct: 98 LCLCGLCSKYVCSSYKYGKRVFLLLEEVTKLKSEGNFDEVSQPPPRSEVE-ERPTQPTIG 156
Query: 143 YFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKR---CGNLF 199
+ + K A+ LM +D V ++GL+GMGG GKT L ++ + G F
Sbjct: 157 --------QEEMLKKAWNRLM----EDGVGIMGLHGMGGVGKTTLFKKIHNKFAETGGTF 204
Query: 200 DQVLFVPISSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRLTQEDRVLVILDD 256
D V+++ +S ++ ++QE IA L ++ K+E D++ + R+ + R +++LDD
Sbjct: 205 DIVIWIVVSQGAKLSKLQEDIAEKLHLCDDLWKNKNESDKATDI-HRVLKGKRFVLMLDD 263
Query: 257 VWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQKQA 316
+W+ +D +AIGIP CK+ T+R + VC M K +Q+ L ++ W+LF+ +
Sbjct: 264 IWEKVDLEAIGIPYPSEVNKCKVAFTTRDQKVCGQMGDHKPMQVKCLEPEDAWELFKNK- 322
Query: 317 LISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSKPV 372
+ + T S I +ARE++ +C+GLP+A + ++ K V EW+ A+D L S
Sbjct: 323 -VGDNTLRSDPVIVGLAREVAQKCRGLPLALSCIGETMASKTMVQEWEHAIDVLTRSAAE 381
Query: 373 NIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVGEV 431
+ +QN L+ SYD+L+ E KS FL C++FPED +I + L I G +GE
Sbjct: 382 FSD--MQNKILPILKYSYDSLEDEHIKSCFLYCALFPEDDKIDTKTLINKWICEGFIGED 439
Query: 432 HSYEGARNEVTVAKNKLISSCLLLDVNEGKCVK----MHDLVRNVAHWIA-------ENE 480
+ ARN+ LI + LL N+ VK MHD+VR +A WIA EN
Sbjct: 440 QVIKRARNKGYEMLGTLIRANLL--TNDRGFVKWHVVMHDVVREMALWIASDFGKQKENY 497
Query: 481 I--------KCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHTYTQVSDEIF 530
+ + KD + SL E+ CS L FLQ + +S E
Sbjct: 498 VVRARVGLHEIPKVKDWGAVRRMSLMMNEIEEITCESKCSELTTLFLQSNQLKNLSGEFI 557
Query: 531 KGMRMLRVLFL-YNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVG--DMKKLESI 587
+ M+ L VL L +N P + L SL L W ++ VG ++KKL +
Sbjct: 558 RYMQKLVVLDLSHNPDFNELPEQISGLVSLQYLDL----SWTRIEQLPVGLKELKKLIFL 613
Query: 588 TLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRSKWEVEF 647
LC + +++L +LR L L E + + V+ +LE L D R E
Sbjct: 614 NLCFTERLCSISGISRLLSLRWLSLRESNVHGDA-SVLKELQQLENL--QDLRITESAEL 670
Query: 648 LK-EFSVPQVLQRYQIQLGSMFSGFQDEFLNHHRTLF-----LSYLDTSNAAIKDLAEKA 701
+ + + +++ +I+ G + F FL L+ SY N ++ ++
Sbjct: 671 ISLDQRLAKLISVLRIE-GFLQKPFDLSFLASMENLYGLLVENSYFSEINIKCRESETES 729
Query: 702 EVLCIAGIEGGAKNIIPDVFQSMNHLKELL------------IRDSKGI-ECLVDTCLIE 748
L I N+ + + +K+L IRDS+ + E + I
Sbjct: 730 SYLHINPKIPCFTNLTGLIIMKCHSMKDLTWILFAPNLVNLDIRDSREVGEIINKEKAIN 789
Query: 749 VGTLF--FCKLHWLRIEHMKHLGALYNGQMPLSGHFENLEDLYISHCPKLTRLFTLAVAQ 806
+ ++ F KL L + + L ++Y +P F L ++ + +CPKL +L A +
Sbjct: 790 LTSIITPFQKLERLFLYGLPKLESIYWSPLP----FPLLSNIVVKYCPKLRKLPLNATSV 845
Query: 807 NLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLLFPKLKKFHVRECGVL 858
L + +++ + PE ++ L +D D + + + + K H G L
Sbjct: 846 PLVEEFEIR-MDPPEQENELEWEDEDTKNRFLPSIKPLVRRLKIHYSGMGFL 896
>UniRef100_Q940K0 Probable disease resistance protein At1g61180 [Arabidopsis
thaliana]
Length = 889
Score = 204 bits (520), Expect = 9e-51
Identities = 237/903 (26%), Positives = 408/903 (44%), Gaps = 126/903 (13%)
Query: 24 SYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVD 83
SY ++ L +E +L AI+ VQ++V R + + ++ E V+ WL N
Sbjct: 25 SYIRTLEKNLRALQREMEDLRAIQHEVQNKVARDEARHQRRLEAVQVWLDRVNSVDIECK 84
Query: 84 QLLQMAKSE--KNSCFGHCPNWIWR-YSVGRKLSKKKRNLKLYIEEG----------RQY 130
LL + E K G C ++ Y G+K+ +K EG R
Sbjct: 85 DLLSVTPVELQKLCLCGLCSKYVCSSYKYGKKVFLLLEEVKKLNSEGNFDEVSQPPPRSE 144
Query: 131 IEIERPASLSAGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAME 190
+E ERP + G + D + A+ LM +D V ++GL+GMGG GKT L +
Sbjct: 145 VE-ERPTQPTIG--------QEDMLEKAWNRLM----EDGVGIMGLHGMGGVGKTTLFKK 191
Query: 191 VGKR---CGNLFDQVLFVPISSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRL 244
+ + G FD V+++ +S V + ++QE IA L ++ K+E D++ + R+
Sbjct: 192 IHNKFAEIGGTFDIVIWIVVSKGVMISKLQEDIAEKLHLCDDLWKNKNESDKATDI-HRV 250
Query: 245 TQEDRVLVILDDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLT 304
+ R +++LDD+W+ +D +AIGIP CK+ T+RS VC M K +Q++ L
Sbjct: 251 LKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCKVAFTTRSREVCGEMGDHKPMQVNCLE 310
Query: 305 NDETWDLFQKQALISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWK 360
++ W+LF+ + + + T S I +ARE++ +C+GLP+A + ++ K V EW+
Sbjct: 311 PEDAWELFKNK--VGDNTLSSDPVIVELAREVAQKCRGLPLALNVIGETMSSKTMVQEWE 368
Query: 361 VALDRLRSSKPVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLT 419
A+ +S + +QN L+ SYD+L E KS FL C++FPED EI E L
Sbjct: 369 HAIHVFNTSAAEFSD--MQNKILPILKYSYDSLGDEHIKSCFLYCALFPEDGEIYNEKLI 426
Query: 420 RSAIGLGIVGEVHSYEGARNEVTVAKNKLISSCLLLDVNEGKCVKMHDLVRNVAHWIA-- 477
I G +GE + ARN+ L + LL V CV MHD+VR +A WIA
Sbjct: 427 DYWICEGFIGEDQVIKRARNKGYAMLGTLTRANLLTKVGTYYCV-MHDVVREMALWIASD 485
Query: 478 -----ENEI--------KCASEKDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHTY 522
EN + + KD + SL E+ CS L FLQ +
Sbjct: 486 FGKQKENFVVQAGVGLHEIPKVKDWGAVRKMSLMDNDIEEITCESKCSELTTLFLQSNKL 545
Query: 523 TQVSDEIFKGMRMLRVLFL-YNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDM 581
+ + M+ L VL L YN+ + P + L +L+ + S + + +
Sbjct: 546 KNLPGAFIRYMQKLVVLDLSYNRDFNKLP---EQISGLVSLQFLDLSNTSIEHMPI--GL 600
Query: 582 KKLESITLCDCSFVE-LPDV--VTQLTNLRLLDL--SECGMERNPFEVIARHTELEELFF 636
K+L+ +T D ++ + L + +++L +LRLL L S+ + + + + + L+EL
Sbjct: 601 KELKKLTFLDLTYTDRLCSISGISRLLSLRLLRLLGSKVHGDASVLKELQQLQNLQEL-- 658
Query: 637 ADCRSKWEVEFLKEFSVPQVLQRYQIQLGSMFSGFQDE-FLNHHRTL-FLSYLDTSNAAI 694
+V L +L + S E FL L FL+ ++ ++
Sbjct: 659 -------------AITVSAELISLDQRLAKLISNLCIEGFLQKPFDLSFLASMENLSSLR 705
Query: 695 KDLAEKAEVLC-IAGIEGGAKNIIPDV--FQSMNHLK--------------------ELL 731
+ + +E+ C + E I P + F +++ L+ LL
Sbjct: 706 VENSYFSEIKCRESETESSYLRINPKIPCFTNLSRLEIMKCHSMKDLTWILFAPNLVVLL 765
Query: 732 IRDSKGIECLVD----TCLIEVGTLFFCKLHWLRIEHMKHLGALYNGQMPLSGHFENLED 787
I DS+ + +++ T L + F KL WL + ++ L ++Y +P F L
Sbjct: 766 IEDSREVGEIINKEKATNLTSITP--FLKLEWLILYNLPKLESIYWSPLP----FPVLLT 819
Query: 788 LYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAY-----DYRLL 842
+ +S+CPKL +L A + + + ++ + PE ++ L +D D + + Y+
Sbjct: 820 MDVSNCPKLRKLPLNATSVSKVEEFEIHMYPPPEQENELEWEDDDTKNRFLPSIKPYKYF 879
Query: 843 LFP 845
++P
Sbjct: 880 VYP 882
>UniRef100_O64789 Putative disease resistance protein At1g61310 [Arabidopsis
thaliana]
Length = 925
Score = 202 bits (515), Expect = 3e-50
Identities = 238/902 (26%), Positives = 391/902 (42%), Gaps = 145/902 (16%)
Query: 24 SYPCCFNNFVQDLAKEESNLAAIRDSVQDRVTRAKKQTRKTAEVVEKWLKDANIAMDNVD 83
SY ++ L +E +L A + VQ++V R + + ++ E V+ WL N
Sbjct: 27 SYIRTLEKNLRALQREMEDLRATQHEVQNKVAREESRHQQRLEAVQVWLDRVNSIDIECK 86
Query: 84 QLLQMAKSE--KNSCFGHCPNWIWR-YSVGRKLSKKKRNLKLYIEEGRQYIEIERPASLS 140
LL ++ E K G C ++ Y G+K+ +K+ EG + E+ +P S
Sbjct: 87 DLLSVSPVELQKLCLCGLCTKYVCSSYKYGKKVFLLLEEVKILKSEGN-FDEVSQPPPRS 145
Query: 141 AGYFSAERCWEFDSRKPAYEELMCALKDDDVTMIGLYGMGGCGKTMLAMEVGKR---CGN 197
++ E+ L +D V ++GL+GMGG GKT L ++ + G
Sbjct: 146 E--VEERPTQPTIGQEEMLEKAWNRLMEDGVGIMGLHGMGGVGKTTLFKKIHNKFAEIGG 203
Query: 198 LFDQVLFVPISSTVEVERIQEKIAGSLEF---EFQEKDEMDRSKRLCMRLTQEDRVLVIL 254
FD V+++ +S ++ ++QE IA L ++ K+E D++ + R+ + R +++L
Sbjct: 204 TFDIVIWIVVSQGAKLSKLQEDIAEKLHLCDDLWKNKNESDKATDI-HRVLKGKRFVLML 262
Query: 255 DDVWQMLDFDAIGIPSIEHHKGCKILITSRSEAVCTLMDCQKKIQLSTLTNDETWDLFQK 314
DD+W+ +D +AIGIP CK+ T+RS VC M K +Q++ L ++ W+LF+
Sbjct: 263 DDIWEKVDLEAIGIPYPSEVNKCKVAFTTRSREVCGEMGDHKPMQVNCLEPEDAWELFKN 322
Query: 315 QALISEGTWIS---IKNMAREISNECKGLPVATVAVASSLKGKAEV-EWKVALDRLRSSK 370
+ + + T S I +ARE++ +C+GLP+A + ++ K V EW+ A+D L S
Sbjct: 323 K--VGDNTLSSDPVIVGLAREVAQKCRGLPLALNVIGETMASKTMVQEWEYAIDVLTRSA 380
Query: 371 PVNIEKGLQNP-YKCLQLSYDNLDTEEAKSLFLLCSVFPEDCEIPVEFLTRSAIGLGIVG 429
G++N L+ SYD+L E KS FL C++FPED +I E L I G +G
Sbjct: 381 AE--FSGMENKILPILKYSYDSLGDEHIKSCFLYCALFPEDGQIYTETLIDKLICEGFIG 438
Query: 430 EVHSYEGARNE-----VTVAKNKLISSC------LLLDVNEGKCVKMHDLVRNVAHWIA- 477
E + ARN+ T+ + L++ LL V+ CV MHD+VR +A WIA
Sbjct: 439 EDQVIKRARNKGYAMLGTLTRANLLTKVGTELANLLTKVSIYHCV-MHDVVREMALWIAS 497
Query: 478 ------ENEIKCASE--------KDIMTLEHTSLRYLWCEKFPNSLDCSNLD--FLQIHT 521
EN + AS KD + SL E+ CS L FLQ +
Sbjct: 498 DFGKQKENFVVQASAGLHEIPEVKDWGAVRRMSLMRNEIEEITCESKCSELTTLFLQSNQ 557
Query: 522 YTQVSDEIFKGMRMLRVLFLYNKGRERRPLLTTSLKSLTNLRCILFSKWDLVDISFVGDM 581
+S E + M+ L VL DL D
Sbjct: 558 LKNLSGEFIRYMQKLVVL-------------------------------DLSD------- 579
Query: 582 KKLESITLCDCSFVELPDVVTQLTNLRLLDLSECGMERNPFEVIARHTELEELFFADCRS 641
+ F ELP+ ++ L +L+ LDLS +E+ P + EL++L F D
Sbjct: 580 ---------NRDFNELPEQISGLVSLQYLDLSFTRIEQLPVGL----KELKKLTFLDL-- 624
Query: 642 KWEVEFLKEFSVPQVLQ-RYQIQLGSMFSGFQDEFLNHHRTLFLSYLD-TSNAAIKDLAE 699
+ + ++L R LGS G + L L T +A + L +
Sbjct: 625 AYTARLCSISGISRLLSLRVLSLLGSKVHGDASVLKELQQLENLQDLAITLSAELISLDQ 684
Query: 700 K-AEVLCIAGIEGGAKNIIPDVF-QSMNHLKELLIRDS--KGIECLVD------------ 743
+ A+V+ I GIEG + F SM +L L +++S I+C
Sbjct: 685 RLAKVISILGIEGFLQKPFDLSFLASMENLSSLWVKNSYFSEIKCRESETDSSYLHINPK 744
Query: 744 -TCLIEVGTLFFCKLH------W---------LRIEHMKHLGALYNGQMPLS----GHFE 783
C + L K H W L IE + +G + N + + F
Sbjct: 745 IPCFTNLSRLDIVKCHSMKDLTWILFAPNLVVLFIEDSREVGEIINKEKATNLTSITPFL 804
Query: 784 NLEDLYISHCPKLTRLFTLAVAQNLAQLEKLQVLSCPELQHILIDDDRDEISAYDYRLLL 843
LE L + + PKL ++ + L L + V CP+L+ + + + ++R+L+
Sbjct: 805 KLERLILCYLPKLESIYWSPLPFPL--LLNIDVEECPKLRKLPL-NATSAPKVEEFRILM 861
Query: 844 FP 845
+P
Sbjct: 862 YP 863
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,766,873,187
Number of Sequences: 2790947
Number of extensions: 73190836
Number of successful extensions: 201492
Number of sequences better than 10.0: 2051
Number of HSP's better than 10.0 without gapping: 629
Number of HSP's successfully gapped in prelim test: 1426
Number of HSP's that attempted gapping in prelim test: 193636
Number of HSP's gapped (non-prelim): 5305
length of query: 1094
length of database: 848,049,833
effective HSP length: 138
effective length of query: 956
effective length of database: 462,899,147
effective search space: 442531584532
effective search space used: 442531584532
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 80 (35.4 bits)
Medicago: description of AC137995.11


