

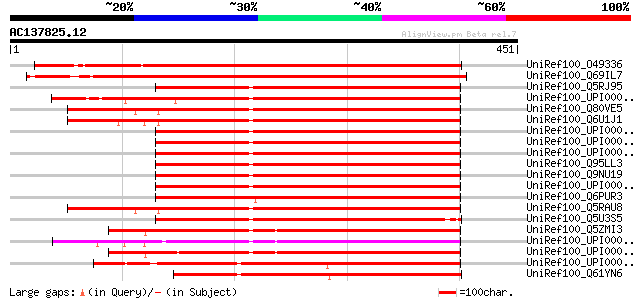
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.12 - phase: 0 /pseudo
(451 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O49336 Expressed protein [Arabidopsis thaliana] 568 e-160
UniRef100_Q69IL7 Putative tbc1 domain family protein [Oryza sativa] 537 e-151
UniRef100_Q5RJ95 Novel protein [Brachydanio rerio] 293 7e-78
UniRef100_UPI00003AD2E8 UPI00003AD2E8 UniRef100 entry 293 9e-78
UniRef100_Q80VE5 Hypothetical protein BC045600 [Mus musculus] 291 3e-77
UniRef100_Q6U1J1 TBC domain-containing protein [Rattus norvegicus] 290 8e-77
UniRef100_UPI000020DC43 UPI000020DC43 UniRef100 entry 289 1e-76
UniRef100_UPI000036D72A UPI000036D72A UniRef100 entry 289 1e-76
UniRef100_UPI000036D729 UPI000036D729 UniRef100 entry 289 1e-76
UniRef100_Q95LL3 TBC1 domain family protein C6orf197 homolog [Ma... 289 1e-76
UniRef100_Q9NU19 TBC1 domain family protein C6orf197 [Homo sapiens] 289 1e-76
UniRef100_UPI00003AD2E9 UPI00003AD2E9 UniRef100 entry 288 2e-76
UniRef100_Q6PUR3 C22orf4-like protein [Brachydanio rerio] 288 2e-76
UniRef100_Q5RAU8 Hypothetical protein DKFZp469J1514 [Pongo pygma... 286 6e-76
UniRef100_Q5U3S5 Hypothetical protein [Brachydanio rerio] 284 3e-75
UniRef100_Q5ZMI3 Hypothetical protein [Gallus gallus] 282 1e-74
UniRef100_UPI00003A952B UPI00003A952B UniRef100 entry 280 8e-74
UniRef100_UPI00003A952A UPI00003A952A UniRef100 entry 278 3e-73
UniRef100_UPI00004311DD UPI00004311DD UniRef100 entry 277 5e-73
UniRef100_Q61YN6 Hypothetical protein CBG03461 [Caenorhabditis b... 273 6e-72
>UniRef100_O49336 Expressed protein [Arabidopsis thaliana]
Length = 440
Score = 568 bits (1464), Expect = e-160
Identities = 277/380 (72%), Positives = 325/380 (84%), Gaps = 5/380 (1%)
Query: 23 DSRFSQTLRNVQGLLKGRSMPGKVLMSRRSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQI 82
DSRF+QTL+NVQG LKGRS+PGKVL++RRS+P +S T Y+RS S NDAG ++
Sbjct: 13 DSRFNQTLKNVQGFLKGRSIPGKVLLTRRSDPPPY---PISPT-YQRSLSENDAGRNELF 68
Query: 83 SGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKVLS 142
VE E ++SK KL+S++S ++ E ++ IG R++DSARV KFNKVLS
Sbjct: 69 ESPVEVEDHNSSKKHDNTYAGKLRSNSSAERSVKE-VQNLKIGVRSSDSARVMKFNKVLS 127
Query: 143 GTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYD 202
T VIL+ LRELAW+GVP YMRP VWRLLLGY PPNSDR+E VL RKR EYL+ + Q+YD
Sbjct: 128 ETTVILEKLRELAWNGVPHYMRPDVWRLLLGYAPPNSDRREAVLRRKRLEYLESVGQFYD 187
Query: 203 IPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGI 262
+PDSERSDDE+NMLRQIAVDCPRTVPDV+FFQQ+QVQKSLERILY WAIRHPASGYVQGI
Sbjct: 188 LPDSERSDDEINMLRQIAVDCPRTVPDVSFFQQEQVQKSLERILYTWAIRHPASGYVQGI 247
Query: 263 NDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQP 322
NDLVTPFLV+F+SE+L+GG+D WSM DLS++K+S+VEADCYWCL+KLLDGMQDHYTFAQP
Sbjct: 248 NDLVTPFLVIFLSEYLDGGVDSWSMDDLSAEKVSDVEADCYWCLTKLLDGMQDHYTFAQP 307
Query: 323 GIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYL 382
GIQRLVFKLKELVRRIDEP+S+H+E+ GLEFLQFAFRW+NCLLIREIPF+LI RLWDTYL
Sbjct: 308 GIQRLVFKLKELVRRIDEPVSRHMEEHGLEFLQFAFRWYNCLLIREIPFNLINRLWDTYL 367
Query: 383 AEGDALPDFLVYIFASFLLT 402
AEGDALPDFLVYI+ASFLLT
Sbjct: 368 AEGDALPDFLVYIYASFLLT 387
>UniRef100_Q69IL7 Putative tbc1 domain family protein [Oryza sativa]
Length = 444
Score = 537 bits (1384), Expect = e-151
Identities = 264/391 (67%), Positives = 309/391 (78%), Gaps = 12/391 (3%)
Query: 16 NNTVTTLDSRFSQTLRNVQGLLKGRSMPGKVLMSRRSEPSDNLNSKVSSTFYKRSFSHND 75
NNT + RF+QTLRNVQG+LKGRS PGKVL++RRSEP +S Y + ND
Sbjct: 17 NNT----EWRFNQTLRNVQGMLKGRSFPGKVLLTRRSEP-------LSPPEYSPRYE-ND 64
Query: 76 AGTSDQISGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARVT 135
+Q G+ E + Q++ + + + K ++ N D + GARATDSAR+
Sbjct: 65 RDEYEQNEGSQEGKGQASGNTADSMSAKKSNPPSTSSTNSLPDAQGLVSGARATDSARIA 124
Query: 136 KFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLD 195
KF LS VILD LREL+WSGVP YMRP +WRLLLGY PPN+DR+EGVL RKR EY++
Sbjct: 125 KFTNELSRPAVILDKLRELSWSGVPPYMRPNIWRLLLGYAPPNADRREGVLTRKRLEYVE 184
Query: 196 CISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPA 255
C+SQYYDIPD+ERSD+E+NMLRQIAVDCPRTVPDVTFFQ Q+QKSLERILY WAIRHPA
Sbjct: 185 CVSQYYDIPDTERSDEEINMLRQIAVDCPRTVPDVTFFQHPQIQKSLERILYTWAIRHPA 244
Query: 256 SGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQD 315
SGYVQGINDL+TPFLVVF+SEHLEG +D WSM LS +SN+EADCYWCLSK LDGMQD
Sbjct: 245 SGYVQGINDLLTPFLVVFLSEHLEGNMDTWSMEKLSPQDVSNIEADCYWCLSKFLDGMQD 304
Query: 316 HYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLIT 375
HYTFAQPGIQRLVF+LKELV RIDEP+S+H+E+QGL+FLQFAFRWFNCL+IREIPF L+T
Sbjct: 305 HYTFAQPGIQRLVFRLKELVHRIDEPVSKHMEEQGLDFLQFAFRWFNCLMIREIPFHLVT 364
Query: 376 RLWDTYLAEGDALPDFLVYIFASFLLTTNSR 406
RLWDTYLAEGD LPDFLVYI ASFLLT + +
Sbjct: 365 RLWDTYLAEGDYLPDFLVYISASFLLTWSDK 395
>UniRef100_Q5RJ95 Novel protein [Brachydanio rerio]
Length = 364
Score = 293 bits (750), Expect = 7e-78
Identities = 142/272 (52%), Positives = 193/272 (70%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+++R+ KF +VL+G L+ LR+L+WSG+P +RP W+LL GY P N++R+E L RK
Sbjct: 41 EASRLDKFRQVLAGPNTDLEELRKLSWSGIPRQVRPITWKLLSGYLPANAERRESTLQRK 100
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 101 RQEYFGFIEQYYDSRNDEHHQDTY---RQIHIDIPRTNPLIPLFQQASVQEIFERILFIW 157
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VV++ E++E ++++++S L + + N+EAD +WC+SKL
Sbjct: 158 AIRHPASGYVQGINDLVTPFFVVYVFEYIEEEVENFNVSSLQEEVLRNIEADSFWCMSKL 217
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQR V L+ELV RIDE + + ++ +E+LQFAFRW N LL+RE+
Sbjct: 218 LDGIQDNYTFAQPGIQRKVKALEELVSRIDETVHRQMQLYEVEYLQFAFRWMNNLLMREL 277
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY AE + F +Y+ A+FL+
Sbjct: 278 PLRCTIRLWDTYQAEPEGFSRFHLYVCAAFLV 309
>UniRef100_UPI00003AD2E8 UPI00003AD2E8 UniRef100 entry
Length = 428
Score = 293 bits (749), Expect = 9e-78
Identities = 160/368 (43%), Positives = 225/368 (60%), Gaps = 10/368 (2%)
Query: 38 KGRSMPGKVLMSRRSEPSDNLNSKVSSTFYKRSFSHNDAGTSDQISGAVEEEVQSTSKSV 97
+ +PG + R + SK +S+F++ F+ N + D I +E+ S+S S+
Sbjct: 12 RSAKLPGSFIKDRSKANVLPMKSKKASSFHE--FARNTSDAWD-IGDDEDEDFTSSSSSL 68
Query: 98 SAAN--VSKLKSSTSLGENLSEDIRKYTIGARATDSARVTKFNKVLSGTVV--ILDNLRE 153
N V+K ++ L + ++ + +D K NK S + + D LR+
Sbjct: 69 QTLNSKVAKATAAQVLENHSKLRVKPERVQPTLSDMPTNCKVNKSSSEAQLSRMSDELRK 128
Query: 154 LAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEV 213
+W GVP +RP WRLL GY P N +R++ L RKR EY I QYYD + E D
Sbjct: 129 CSWPGVPREVRPVTWRLLSGYLPANMERRKLTLQRKREEYFGFIQQYYDSRNEEHHQDTY 188
Query: 214 NMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVF 273
RQI +D PRT P + FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTPF VVF
Sbjct: 189 ---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIWAIRHPASGYVQGINDLVTPFFVVF 245
Query: 274 ISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKE 333
+SE++E ++++ +++LS D + ++EAD +WC+SKLLDG+QD+YTFAQPGIQ+ V L+E
Sbjct: 246 LSEYVEEDVENFDVTNLSQDVLRSIEADSFWCMSKLLDGIQDNYTFAQPGIQKKVKALEE 305
Query: 334 LVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALPDFLV 393
LV RIDE + H +E+LQFAFRW N LL+RE+P RLWDTY +E + F +
Sbjct: 306 LVSRIDEQVHNHFRKYEVEYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSEPEGFSHFHL 365
Query: 394 YIFASFLL 401
Y+ A+FL+
Sbjct: 366 YVCAAFLI 373
>UniRef100_Q80VE5 Hypothetical protein BC045600 [Mus musculus]
Length = 505
Score = 291 bits (744), Expect = 3e-77
Identities = 162/372 (43%), Positives = 228/372 (60%), Gaps = 25/372 (6%)
Query: 52 SEPSDNLNSKVS-STFYKRSFSHNDAGTSDQISGAVEEEVQSTSKSVSAANVSKLKSSTS 110
S P LNSKV+ +T + +H+ + S + +V + K + +++ ++L ++S
Sbjct: 82 SPPFQTLNSKVALATAAQVLENHSKLRVKPERSQSTTSDVPANYKVIKSSSDAQLSRNSS 141
Query: 111 -------LGENLSEDIRKYT-IGARATDS-------------ARVTKFNKVLSGTVVILD 149
L + S +R + AR +D R+ KF ++LS LD
Sbjct: 142 DTCLRNPLHKQQSLPLRPIIPLVARISDQNASGAPPMTVREKTRLEKFRQLLSSHNTDLD 201
Query: 150 NLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERS 209
LR+ +W GVP +RP WRLL GY P N++R++ L RKR EY I QYYD + E
Sbjct: 202 ELRKWSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQRKREEYFGFIEQYYDSRNEEHH 261
Query: 210 DDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 269
D RQI +D PRT P + FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTPF
Sbjct: 262 QDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIWAIRHPASGYVQGINDLVTPF 318
Query: 270 LVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVF 329
VVF+SE++E ++++ +++LS D + ++EAD +WC+SKLLDG+QD+YTFAQPGIQ+ V
Sbjct: 319 FVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKLLDGIQDNYTFAQPGIQKKVK 378
Query: 330 KLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALP 389
L+ELV RIDE + H +E+LQFAFRW N LL+RE+P RLWDTY +E +
Sbjct: 379 ALEELVSRIDEQVHSHFRRYEVEYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSEPEGFS 438
Query: 390 DFLVYIFASFLL 401
F +Y+ A+FL+
Sbjct: 439 HFHLYVCAAFLI 450
>UniRef100_Q6U1J1 TBC domain-containing protein [Rattus norvegicus]
Length = 505
Score = 290 bits (741), Expect = 8e-77
Identities = 167/373 (44%), Positives = 227/373 (60%), Gaps = 26/373 (6%)
Query: 52 SEPS-DNLNSKVS-STFYKRSFSHNDAGTSDQISGAVEEEVQSTSK---SVSAANVSKLK 106
S PS LNSKV+ +T + +H+ + S + +V + K S S A +S+
Sbjct: 81 SSPSLQTLNSKVALATAAQVLENHSKLRVKPERSQSTTSDVPANYKVIKSSSDAQLSRNS 140
Query: 107 SSTSLGENLSED----IRKYT-IGARATDS-------------ARVTKFNKVLSGTVVIL 148
S T L L + +R + AR +D R+ KF ++LS L
Sbjct: 141 SDTCLRNTLHKQQSLPLRPIIPLVARISDQNASGAPPMTVREKTRLEKFRQLLSSHNTDL 200
Query: 149 DNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSER 208
D LR+ +W GVP +RP WRLL GY P N++R++ L RKR EY I QYYD + E
Sbjct: 201 DELRKWSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQRKREEYFGFIEQYYDSRNEEH 260
Query: 209 SDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTP 268
D RQI +D PRT P + FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTP
Sbjct: 261 HQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIWAIRHPASGYVQGINDLVTP 317
Query: 269 FLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLV 328
F VVF+SE++E ++++ +++LS D + ++EAD +WC+SKLLDG+QD+YTFAQPGIQ+ V
Sbjct: 318 FFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKLLDGIQDNYTFAQPGIQKKV 377
Query: 329 FKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDAL 388
L+ELV RIDE + H +E+LQFAFRW N LL+RE+P RLWDTY +E +
Sbjct: 378 KALEELVSRIDEQVHSHFRRYEVEYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSEPEGF 437
Query: 389 PDFLVYIFASFLL 401
F +Y+ A+FL+
Sbjct: 438 SHFHLYVCAAFLI 450
>UniRef100_UPI000020DC43 UPI000020DC43 UniRef100 entry
Length = 555
Score = 289 bits (740), Expect = 1e-76
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 232 EKTRLEKFRQLLSSQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQRK 291
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 292 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 348
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 349 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 408
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 409 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 468
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 469 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 500
>UniRef100_UPI000036D72A UPI000036D72A UniRef100 entry
Length = 505
Score = 289 bits (740), Expect = 1e-76
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 182 EKTRLEKFRQLLSNQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQRK 241
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 242 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 298
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 299 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 358
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 359 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 418
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 419 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 450
>UniRef100_UPI000036D729 UPI000036D729 UniRef100 entry
Length = 555
Score = 289 bits (740), Expect = 1e-76
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 232 EKTRLEKFRQLLSNQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQRK 291
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 292 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 348
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 349 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 408
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 409 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 468
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 469 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 500
>UniRef100_Q95LL3 TBC1 domain family protein C6orf197 homolog [Macaca fascicularis]
Length = 505
Score = 289 bits (740), Expect = 1e-76
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 182 EKTRLEKFRQLLSSHNTDLDELRKCSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQRK 241
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 242 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 298
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 299 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 358
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 359 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 418
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 419 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 450
>UniRef100_Q9NU19 TBC1 domain family protein C6orf197 [Homo sapiens]
Length = 505
Score = 289 bits (740), Expect = 1e-76
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 182 EKTRLEKFRQLLSSQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQRK 241
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 242 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 298
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 299 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 358
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 359 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 418
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 419 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 450
>UniRef100_UPI00003AD2E9 UPI00003AD2E9 UniRef100 entry
Length = 507
Score = 288 bits (738), Expect = 2e-76
Identities = 142/272 (52%), Positives = 187/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N +R++ L RK
Sbjct: 184 EKTRLEKFRQLLSSHNTDLDELRKCSWPGVPREVRPVTWRLLSGYLPANMERRKLTLQRK 243
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 244 REEYFGFIQQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 300
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 301 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDVLRSIEADSFWCMSKL 360
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 361 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRKYEVEYLQFAFRWMNNLLMREL 420
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 421 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 452
>UniRef100_Q6PUR3 C22orf4-like protein [Brachydanio rerio]
Length = 567
Score = 288 bits (738), Expect = 2e-76
Identities = 141/290 (48%), Positives = 194/290 (66%), Gaps = 18/290 (6%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+++R+ KF +VL+G L+ LR+L+WSG+P +RP W+LL GY P N++R+E L RK
Sbjct: 223 EASRLDKFRQVLAGPNTDLEELRKLSWSGIPRQVRPITWKLLSGYLPANAERRESTLQRK 282
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLR------------------QIAVDCPRTVPDVT 231
R EY I QYYD + E D + +I +D PRT P +
Sbjct: 283 RQEYFGFIEQYYDSRNDEHHQDTYRQIHIDIPRMSPESLVLQPKVTEIHIDIPRTNPLIP 342
Query: 232 FFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLS 291
FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTPF VV++ E++E ++++++S L
Sbjct: 343 LFQQASVQEIFERILFIWAIRHPASGYVQGINDLVTPFFVVYVFEYIEEEVENFNVSSLQ 402
Query: 292 SDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGL 351
+ + N+EAD +WC+SKLLDG+QD+YTFAQPGIQR V L+ELV RIDE + +H++ +
Sbjct: 403 EEVLRNIEADSFWCMSKLLDGIQDNYTFAQPGIQRKVKALEELVSRIDETVHRHMQLYEV 462
Query: 352 EFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
E+LQFAFRW N LL+RE+P RLWDTY AE + F +Y+ A+FL+
Sbjct: 463 EYLQFAFRWMNNLLMRELPLRCTIRLWDTYQAEPEGFSHFHLYVCAAFLV 512
>UniRef100_Q5RAU8 Hypothetical protein DKFZp469J1514 [Pongo pygmaeus]
Length = 505
Score = 286 bits (733), Expect = 6e-76
Identities = 162/373 (43%), Positives = 229/373 (60%), Gaps = 26/373 (6%)
Query: 52 SEPS-DNLNSKVS-STFYKRSFSHNDAGTSDQISGAVEEEVQSTSKSVSAANVSKLKSST 109
S PS LNSKV+ +T + +H+ + S + +V ++ K + +++ ++L ++
Sbjct: 81 SSPSFQTLNSKVALATAAQVLENHSKLRVKPERSQSTTSDVPASYKVIKSSSDAQLSRNS 140
Query: 110 S-------LGENLSEDIRKYT-IGARATDS-------------ARVTKFNKVLSGTVVIL 148
S L + S +R + AR +D R+ KF ++LS L
Sbjct: 141 SDTCLRNPLHKQQSLPLRPIIPLVARISDQNASGAPPMTVREKTRLEKFRQLLSSHNTDL 200
Query: 149 DNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSER 208
D LR+ +W GVP +RP WRLL GY P N++R++ L RKR EY I QYYD + E
Sbjct: 201 DELRKYSWPGVPREVRPITWRLLSGYLPANTERRKLTLQRKREEYFGFIEQYYDSRNEEH 260
Query: 209 SDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTP 268
D RQI +D PRT P + FQQ VQ+ ERIL+ WAIRHPASGYVQGINDLVTP
Sbjct: 261 HQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIWAIRHPASGYVQGINDLVTP 317
Query: 269 FLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLV 328
F VVF+SE++E ++++ +++LS D + ++ AD +WC+SKLLDG+QD+YTFAQPGIQ+ V
Sbjct: 318 FFVVFLSEYVEEDVENFDVTNLSQDMLRSIGADSFWCMSKLLDGIQDNYTFAQPGIQKKV 377
Query: 329 FKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDAL 388
L+ELV RIDE + H +E+LQFAFRW N LL+RE+P RLWDTY +E +
Sbjct: 378 KALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMRELPLRCTIRLWDTYQSEPEGF 437
Query: 389 PDFLVYIFASFLL 401
F +Y+ A+FL+
Sbjct: 438 SHFHLYVCAAFLI 450
>UniRef100_Q5U3S5 Hypothetical protein [Brachydanio rerio]
Length = 491
Score = 284 bits (727), Expect = 3e-75
Identities = 141/273 (51%), Positives = 192/273 (69%), Gaps = 8/273 (2%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+++R+ KF +VL+G L+ LR+L+WSG+P +RP W+LL GY P N++R+E L RK
Sbjct: 223 EASRLDKFRQVLAGPNTDLEELRKLSWSGIPRQVRPITWKLLSGYLPANAERRESTLQRK 282
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 283 RQEYFGFIEQYYDSRNDEHHQDTY---RQIHIDIPRTNPLIPLFQQASVQEIFERILFIW 339
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VV++ E++E ++++++S L + + N+EAD +WC+SKL
Sbjct: 340 AIRHPASGYVQGINDLVTPFFVVYVFEYIEEEVENFNVSSLQEEVLRNIEADSFWCMSKL 399
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YT+AQPGIQR V L+ELV RIDE + +H++ +E+LQFAFRW N LL+RE+
Sbjct: 400 LDGIQDNYTYAQPGIQRKVKALEELVSRIDETVHRHMQLYEVEYLQFAFRWMNNLLMREL 459
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLLT 402
P RLWDTY + V+++A FLLT
Sbjct: 460 PLRCTIRLWDTYQVRRHS----TVFVYA-FLLT 487
>UniRef100_Q5ZMI3 Hypothetical protein [Gallus gallus]
Length = 518
Score = 282 bits (722), Expect = 1e-74
Identities = 147/322 (45%), Positives = 208/322 (63%), Gaps = 13/322 (4%)
Query: 89 EVQSTSKSVSAANVSKLKSSTSLGENLSEDI--------RKYTIGARATDSARVTKFNKV 140
E ++S S +A+ S ++ S SL + + + T +++R+ KF ++
Sbjct: 146 EGHTSSSSGNASENSSMQRSQSLPQTSTPPLVSGMADFNTSVTPALTEREASRLDKFKQL 205
Query: 141 LSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQY 200
L+G L+ LR+L+WSG+P +RP W+LL GY P N DR+E L RKR EY + QY
Sbjct: 206 LAGPNTDLEELRKLSWSGIPKRVRPIAWKLLSGYLPANVDRRESTLQRKRKEYFAFVEQY 265
Query: 201 YDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQ 260
YD + E D RQI +D PR P+V Q + V + ERIL+ WAIRHPASGYVQ
Sbjct: 266 YDSRNDESHQDTY---RQIHIDIPRMSPEVLRLQPK-VTEIFERILFIWAIRHPASGYVQ 321
Query: 261 GINDLVTPFLVVFISEHLEGG-IDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTF 319
GINDLVTPF VVF+ E++E ++++ +S L + + N+EAD YWC+SKLLDG+QD+YTF
Sbjct: 322 GINDLVTPFFVVFVCEYIEEEEVENFDVSSLPEEVLQNIEADSYWCMSKLLDGIQDNYTF 381
Query: 320 AQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
AQPGIQ+ V L+ELV RIDE + +H++ +++LQFAFRW N LL+RE+P RLWD
Sbjct: 382 AQPGIQKKVKMLEELVSRIDEQVHRHLDQHEVKYLQFAFRWMNNLLMREVPLRCTIRLWD 441
Query: 380 TYLAEGDALPDFLVYIFASFLL 401
TY +E + F +Y+ A+FL+
Sbjct: 442 TYQSEPEGFSHFHLYVCAAFLV 463
>UniRef100_UPI00003A952B UPI00003A952B UniRef100 entry
Length = 438
Score = 280 bits (715), Expect = 8e-74
Identities = 161/375 (42%), Positives = 223/375 (58%), Gaps = 18/375 (4%)
Query: 39 GRSMPGKVLMSRRSEPSDNLNSKVSSTFYKRSFSHNDA---GTSDQISGAVEEEVQSTSK 95
G +PG ++ P+ + K STF + + +DA G D A+ E +T
Sbjct: 15 GTKVPGSLIKPGSKPPTTPVKLKKVSTFQEFESNTSDAWDPGDDDDELLAIAAENLNTEV 74
Query: 96 SVSAA-----NVSKLKSSTS-LGENLSED--IRKYTIGARATDSARVTKFNKVLSGTVVI 147
+ A N SKL+ L E+L E+ + + + A R+ K V G
Sbjct: 75 VMETAHKVIQNHSKLQEQHKFLEEHLQEEKQVESAELTSVACGDNRLVK--SVSEGHTSS 132
Query: 148 LDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSE 207
LR+L+WSG+P +RP W+LL GY P N DR+E L RKR EY + QYYD + E
Sbjct: 133 SSELRKLSWSGIPKRVRPIAWKLLSGYLPANVDRRESTLQRKRKEYFAFVEQYYDSRNDE 192
Query: 208 RSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVT 267
D RQI +D PR P+V Q + V + ERIL+ WAIRHPASGYVQGINDLVT
Sbjct: 193 SHQDTY---RQIHIDIPRMSPEVLRLQPK-VTEIFERILFIWAIRHPASGYVQGINDLVT 248
Query: 268 PFLVVFISEHLEGG-IDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPGIQR 326
PF VVF+ E++E ++++ +S L + + N+EAD YWC+SKLLDG+QD+YTFAQPGIQ+
Sbjct: 249 PFFVVFVCEYIEEEEVENFDVSSLPEEVLQNIEADSYWCMSKLLDGIQDNYTFAQPGIQK 308
Query: 327 LVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGD 386
V L+ELV RIDE + +H++ +++LQFAFRW N LL+RE+P RLWDTY +E +
Sbjct: 309 KVKMLEELVSRIDEQVHRHLDQHEVKYLQFAFRWMNNLLMREVPLRCTIRLWDTYQSEPE 368
Query: 387 ALPDFLVYIFASFLL 401
F +Y+ A+FL+
Sbjct: 369 GFSHFHLYVCAAFLV 383
>UniRef100_UPI00003A952A UPI00003A952A UniRef100 entry
Length = 517
Score = 278 bits (710), Expect = 3e-73
Identities = 147/322 (45%), Positives = 208/322 (63%), Gaps = 14/322 (4%)
Query: 89 EVQSTSKSVSAANVSKLKSSTSLGENLSEDI--------RKYTIGARATDSARVTKFNKV 140
E ++S S +A+ S ++ S SL + + + T +++R+ KF ++
Sbjct: 146 EGHTSSSSGNASENSSMQRSQSLPQTSTPPLVSGMADFNTSVTPALTEREASRLDKFKQL 205
Query: 141 LSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQY 200
L+G L+ LR+L+WSG+P +RP W+LL GY P N DR+E L RKR EY + QY
Sbjct: 206 LAGPNTDLE-LRKLSWSGIPKRVRPIAWKLLSGYLPANVDRRESTLQRKRKEYFAFVEQY 264
Query: 201 YDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQ 260
YD + E D RQI +D PR P+V Q + V + ERIL+ WAIRHPASGYVQ
Sbjct: 265 YDSRNDESHQDTY---RQIHIDIPRMSPEVLRLQPK-VTEIFERILFIWAIRHPASGYVQ 320
Query: 261 GINDLVTPFLVVFISEHLEGG-IDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTF 319
GINDLVTPF VVF+ E++E ++++ +S L + + N+EAD YWC+SKLLDG+QD+YTF
Sbjct: 321 GINDLVTPFFVVFVCEYIEEEEVENFDVSSLPEEVLQNIEADSYWCMSKLLDGIQDNYTF 380
Query: 320 AQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
AQPGIQ+ V L+ELV RIDE + +H++ +++LQFAFRW N LL+RE+P RLWD
Sbjct: 381 AQPGIQKKVKMLEELVSRIDEQVHRHLDQHEVKYLQFAFRWMNNLLMREVPLRCTIRLWD 440
Query: 380 TYLAEGDALPDFLVYIFASFLL 401
TY +E + F +Y+ A+FL+
Sbjct: 441 TYQSEPEGFSHFHLYVCAAFLV 462
>UniRef100_UPI00004311DD UPI00004311DD UniRef100 entry
Length = 432
Score = 277 bits (708), Expect = 5e-73
Identities = 146/331 (44%), Positives = 209/331 (63%), Gaps = 13/331 (3%)
Query: 75 DAGTSDQISGAVEEEVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSARV 134
D+GT+ + S V+E + ++ + ++ ++ NLS + G +++
Sbjct: 57 DSGTNVKSSQRVQEIIPEEKRAEALQRLA-VQPLHLRNVNLSTHSQMNNNG-----ESKI 110
Query: 135 TKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYL 194
KF +L +V+ LD LR+L+WSG+P +R WRLL Y P N +R++ VL RKR +Y
Sbjct: 111 DKFQSLLEASVLNLDELRQLSWSGIPARLRSVTWRLLSEYLPANLERRQHVLERKRLDYW 170
Query: 195 DCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHP 254
+ + QYYD +ER + + RQI +D PR P ++ FQQ VQ ERILY WAIRHP
Sbjct: 171 NLVKQYYD---TERDEGFQDTYRQIHIDIPRMSPLISLFQQTTVQLIFERILYIWAIRHP 227
Query: 255 ASGYVQGINDLVTPFLVVFISEHLEGG----IDDWSMSDLSSDKISNVEADCYWCLSKLL 310
ASGYVQG+NDLVTPF +VF+ E + ++++ ++ L ++ +EAD +WCLSK L
Sbjct: 228 ASGYVQGMNDLVTPFFLVFLQEAVPVSAWQDLENYDVASLQKEQRDIIEADSFWCLSKFL 287
Query: 311 DGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIP 370
DG+QD+Y FAQ GIQ V +LKEL++RID P+ QH+ G+++LQF+FRW N LL REIP
Sbjct: 288 DGIQDNYIFAQLGIQHKVNQLKELIQRIDAPLHQHLHQHGVDYLQFSFRWMNNLLTREIP 347
Query: 371 FDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
RLWDTYLAE D F +Y+ A+FLL
Sbjct: 348 LHCTIRLWDTYLAESDRFASFQLYVCAAFLL 378
>UniRef100_Q61YN6 Hypothetical protein CBG03461 [Caenorhabditis briggsae]
Length = 495
Score = 273 bits (699), Expect = 6e-72
Identities = 135/259 (52%), Positives = 174/259 (67%), Gaps = 5/259 (1%)
Query: 146 VILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPD 205
V LD LRE W G+P +RP+ WRLL GY P N++R+E L KR EY + QY+
Sbjct: 181 VDLDKLREDCWMGIPHKLRPQAWRLLSGYLPTNAERREVTLQCKRDEYWHYVEQYFH--- 237
Query: 206 SERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDL 265
S D + RQI +D PR P + FQQ+ VQ+ ERILY WAIRHPASGYVQGINDL
Sbjct: 238 SRFEDQNADTFRQINIDIPRMCPLIPLFQQKMVQEMFERILYIWAIRHPASGYVQGINDL 297
Query: 266 VTPFLVVFISEHLEGGID--DWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFAQPG 323
VTPF VVF+SE + ++ + +S L ++ +EAD +WC+S LLD +QD+YTFAQPG
Sbjct: 298 VTPFFVVFLSEFIPQDVEVGSFDVSQLPLEQCQLIEADSFWCVSALLDSIQDNYTFAQPG 357
Query: 324 IQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLA 383
IQR V +L+ L+ R+D P+ +H+E G+E+LQFAFRW N LL+REIP RLWDTYL+
Sbjct: 358 IQRKVLQLRHLMSRVDRPLHKHLESNGIEYLQFAFRWMNNLLMREIPLRATIRLWDTYLS 417
Query: 384 EGDALPDFLVYIFASFLLT 402
E D F Y+ A+FL T
Sbjct: 418 EPDGFMQFHNYVCAAFLRT 436
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 747,413,742
Number of Sequences: 2790947
Number of extensions: 30777509
Number of successful extensions: 132065
Number of sequences better than 10.0: 937
Number of HSP's better than 10.0 without gapping: 531
Number of HSP's successfully gapped in prelim test: 408
Number of HSP's that attempted gapping in prelim test: 128564
Number of HSP's gapped (non-prelim): 1985
length of query: 451
length of database: 848,049,833
effective HSP length: 131
effective length of query: 320
effective length of database: 482,435,776
effective search space: 154379448320
effective search space used: 154379448320
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 76 (33.9 bits)
Medicago: description of AC137825.12


