

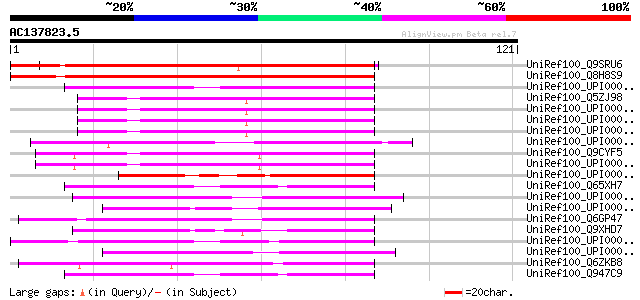
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137823.5 - phase: 0
(121 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SRU6 F14P3.5 protein [Arabidopsis thaliana] 141 4e-33
UniRef100_Q8H8S9 Putative chromosome condensation regulator [Ory... 115 2e-25
UniRef100_UPI00002AFC57 UPI00002AFC57 UniRef100 entry 50 1e-05
UniRef100_Q5ZJ98 Hypothetical protein [Gallus gallus] 49 3e-05
UniRef100_UPI00003AB5C2 UPI00003AB5C2 UniRef100 entry 48 4e-05
UniRef100_UPI00003AB5C1 UPI00003AB5C1 UniRef100 entry 48 4e-05
UniRef100_UPI00003AB5C0 UPI00003AB5C0 UniRef100 entry 48 4e-05
UniRef100_UPI00002D3580 UPI00002D3580 UniRef100 entry 47 1e-04
UniRef100_Q9CYF5 Williams-Beuren syndrome chromosome region 16 p... 46 2e-04
UniRef100_UPI000017E724 UPI000017E724 UniRef100 entry 46 2e-04
UniRef100_UPI000035F228 UPI000035F228 UniRef100 entry 46 2e-04
UniRef100_Q65XH7 Ptative chromosome condensation factor [Oryza s... 46 2e-04
UniRef100_UPI000035F90D UPI000035F90D UniRef100 entry 45 3e-04
UniRef100_UPI00003AA00E UPI00003AA00E UniRef100 entry 45 3e-04
UniRef100_Q6GP47 MGC80684 protein [Xenopus laevis] 45 3e-04
UniRef100_Q9XHD7 UVB-resistance protein UVR8 [Arabidopsis thaliana] 45 3e-04
UniRef100_UPI00003AA019 UPI00003AA019 UniRef100 entry 45 4e-04
UniRef100_UPI000026AC52 UPI000026AC52 UniRef100 entry 45 4e-04
UniRef100_Q6ZKB8 Putative UVB-resistance protein UVR8 [Oryza sat... 45 4e-04
UniRef100_Q947C9 Putative chromosome condensation factor [Tritic... 45 5e-04
>UniRef100_Q9SRU6 F14P3.5 protein [Arabidopsis thaliana]
Length = 456
Score = 141 bits (355), Expect = 4e-33
Identities = 65/87 (74%), Positives = 73/87 (83%), Gaps = 1/87 (1%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
MDI I+G P + SIP KSAIYVWGYNQSGQTGR E++ LRIPKQLPP+LFGCPAG
Sbjct: 1 MDIGEIIGEVAP-SVSIPTKSAIYVWGYNQSGQTGRNEQEKLLRIPKQLPPELFGCPAGA 59
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N+RWLDI+CGREHTAA+ASDG LF WG
Sbjct: 60 NSRWLDISCGREHTAAVASDGSLFAWG 86
Score = 48.5 bits (114), Expect = 3e-05
Identities = 28/82 (34%), Positives = 43/82 (52%), Gaps = 3/82 (3%)
Query: 8 GTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQL-FGCPAGLNTRWLD 66
GT VA S + + WGYN+ GQ GR + L+ P+ + F A + +
Sbjct: 174 GTAHVVALS--EEGLLQAWGYNEQGQLGRGVTCEGLQAPRVINAYAKFLDEAPELVKIMQ 231
Query: 67 IACGREHTAAIASDGLLFTWGL 88
++CG HTAA++ G ++TWGL
Sbjct: 232 LSCGEYHTAALSDAGEVYTWGL 253
Score = 39.7 bits (91), Expect = 0.016
Identities = 22/76 (28%), Positives = 37/76 (47%), Gaps = 11/76 (14%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQLFGCPAGLN-TRWLDIACGR 71
A++ +Y WG GQ G + D+ IP+++ GL+ ++ACG
Sbjct: 240 AALSDAGEVYTWGLGSMGQLGHVSLQSGDKELIPRRV--------VGLDGVSMKEVACGG 291
Query: 72 EHTAAIASDGLLFTWG 87
HT A++ +G L+ WG
Sbjct: 292 VHTCALSLEGALYAWG 307
Score = 35.4 bits (80), Expect = 0.30
Identities = 19/67 (28%), Positives = 27/67 (39%), Gaps = 16/67 (23%)
Query: 21 SAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASD 80
S ++VWG NQ P+LF T ++CG H A++ +
Sbjct: 141 SRLWVWGQNQGSNL----------------PRLFSGAFPATTAIRQVSCGTAHVVALSEE 184
Query: 81 GLLFTWG 87
GLL WG
Sbjct: 185 GLLQAWG 191
>UniRef100_Q8H8S9 Putative chromosome condensation regulator [Oryza sativa]
Length = 505
Score = 115 bits (289), Expect = 2e-25
Identities = 54/87 (62%), Positives = 65/87 (74%), Gaps = 2/87 (2%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
MD+D +LG + A +P KSAIY+WGYN SGQT RK K+ LRIPK LPP+LF G
Sbjct: 1 MDMDDMLGGLR--VAGVPTKSAIYLWGYNHSGQTARKGKECHLRIPKSLPPKLFKWQDGK 58
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
N RW+D+ACGR HTAA+ASDG L+TWG
Sbjct: 59 NLRWIDVACGRGHTAAVASDGSLYTWG 85
Score = 43.5 bits (101), Expect = 0.001
Identities = 24/67 (35%), Positives = 37/67 (54%), Gaps = 1/67 (1%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQL-FGCPAGLNTRWLDIACGREHTAAIASDG 81
+ WGYN+ GQ GR + L+ + L F A + + ++CG HTAAI+ +G
Sbjct: 212 LQAWGYNEYGQLGRGCTSEGLQGARVLNAYARFLDEAPELVKIVRVSCGEYHTAAISENG 271
Query: 82 LLFTWGL 88
++TWGL
Sbjct: 272 EVYTWGL 278
Score = 41.6 bits (96), Expect = 0.004
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 9/75 (12%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGR--KEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGRE 72
A+I +Y WG GQ G + D+ IP+++ A T D++CG
Sbjct: 265 AAISENGEVYTWGLGSMGQLGHCSLQSGDKELIPRRVV-------ALDRTVIRDVSCGGV 317
Query: 73 HTAAIASDGLLFTWG 87
H+ A+ DG L+ WG
Sbjct: 318 HSCAVTEDGALYAWG 332
>UniRef100_UPI00002AFC57 UPI00002AFC57 UniRef100 entry
Length = 259
Score = 50.1 bits (118), Expect = 1e-05
Identities = 26/74 (35%), Positives = 37/74 (49%), Gaps = 6/74 (8%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
AA+I +Y WG N GQ G ++ ++L P G + N R I+CGR H
Sbjct: 62 AAAIASNGEVYTWGLNTFGQLGHGDRMNRLT------PTKVGALSDKNVRIQWISCGRSH 115
Query: 74 TAAIASDGLLFTWG 87
T A++ G +F WG
Sbjct: 116 TVAVSDYGEVFAWG 129
Score = 43.5 bits (101), Expect = 0.001
Identities = 27/68 (39%), Positives = 36/68 (52%), Gaps = 6/68 (8%)
Query: 23 IYVWGYNQSGQTGRKEK-DDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDG 81
+Y WG +SGQ G DD R L PQL + ++D G H AAIAS+G
Sbjct: 15 VYSWGIGESGQLGLGSSVDDPFRA---LRPQLVDALRDVEIVFID--AGEFHAAAIASNG 69
Query: 82 LLFTWGLD 89
++TWGL+
Sbjct: 70 EVYTWGLN 77
>UniRef100_Q5ZJ98 Hypothetical protein [Gallus gallus]
Length = 454
Score = 48.5 bits (114), Expect = 3e-05
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFG-CPAGLNTRWLDIACGREHTA 75
+ + A++VWGY G G+ + +P+ +PP LFG + R + CG H A
Sbjct: 341 LTEEGAVFVWGY---GILGKGPNVTETAVPEMIPPSLFGWSDFSSDVRVARVRCGLSHFA 397
Query: 76 AIASDGLLFTWG 87
A+ + G LF WG
Sbjct: 398 ALTNRGELFVWG 409
Score = 35.8 bits (81), Expect = 0.23
Identities = 20/69 (28%), Positives = 33/69 (46%), Gaps = 6/69 (8%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ G N GQ GRK +D++ L QL ++R + + CG++H+ G
Sbjct: 186 VFTMGNNSYGQCGRKVVEDEIYSESHLIHQL----NDFDSRVVQVVCGQDHSLFRTKKGA 241
Query: 83 LFT--WGLD 89
+F WG D
Sbjct: 242 VFACGWGAD 250
Score = 30.8 bits (68), Expect = 7.4
Identities = 28/88 (31%), Positives = 37/88 (41%), Gaps = 7/88 (7%)
Query: 25 VWGY--NQSGQTG--RKEKDDQLRIPKQLPPQLFGCPAGL--NTRWLDIACGREHTAAIA 78
VWG N+ Q G R +D L P P TR L ++CGR H+ +
Sbjct: 122 VWGLGLNKDSQLGFQRSIRDRTKGYEYVLEPSPIPLPLEKPQKTRVLQVSCGRAHSLVLT 181
Query: 79 SDGLLFTWGLDFSYSLLVLLIVHDFIYS 106
+FT G + SY +V D IYS
Sbjct: 182 DSEGVFTMG-NNSYGQCGRKVVEDEIYS 208
>UniRef100_UPI00003AB5C2 UPI00003AB5C2 UniRef100 entry
Length = 351
Score = 48.1 bits (113), Expect = 4e-05
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFG-CPAGLNTRWLDIACGREHTA 75
+ + A++VWGY G G+ + +P+ +PP LFG + R + CG H A
Sbjct: 240 LTEEGAVFVWGY---GILGKGPNLTETAVPEMIPPSLFGWSDFSSDVRVARVRCGLSHFA 296
Query: 76 AIASDGLLFTWG 87
A+ + G LF WG
Sbjct: 297 ALTNRGELFVWG 308
Score = 32.7 bits (73), Expect = 1.9
Identities = 20/73 (27%), Positives = 31/73 (42%), Gaps = 16/73 (21%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWL----DIACGREHTAAIA 78
++ G N GQ GRK +D++ P Q+ WL + CG++H+
Sbjct: 87 VFTMGNNSYGQCGRKVVEDEIYRHMTFPDQV----------WLSVCVQVVCGQDHSLFRT 136
Query: 79 SDGLLFT--WGLD 89
G +F WG D
Sbjct: 137 KKGAVFACGWGAD 149
>UniRef100_UPI00003AB5C1 UPI00003AB5C1 UniRef100 entry
Length = 374
Score = 48.1 bits (113), Expect = 4e-05
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFG-CPAGLNTRWLDIACGREHTA 75
+ + A++VWGY G G+ + +P+ +PP LFG + R + CG H A
Sbjct: 261 LTEEGAVFVWGY---GILGKGPNLTETAVPEMIPPSLFGWSDFSSDVRVARVRCGLSHFA 317
Query: 76 AIASDGLLFTWG 87
A+ + G LF WG
Sbjct: 318 ALTNRGELFVWG 329
>UniRef100_UPI00003AB5C0 UPI00003AB5C0 UniRef100 entry
Length = 450
Score = 48.1 bits (113), Expect = 4e-05
Identities = 24/72 (33%), Positives = 37/72 (51%), Gaps = 4/72 (5%)
Query: 17 IPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFG-CPAGLNTRWLDIACGREHTA 75
+ + A++VWGY G G+ + +P+ +PP LFG + R + CG H A
Sbjct: 337 LTEEGAVFVWGY---GILGKGPNLTETAVPEMIPPSLFGWSDFSSDVRVARVRCGLSHFA 393
Query: 76 AIASDGLLFTWG 87
A+ + G LF WG
Sbjct: 394 ALTNRGELFVWG 405
Score = 35.8 bits (81), Expect = 0.23
Identities = 20/69 (28%), Positives = 33/69 (46%), Gaps = 6/69 (8%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ G N GQ GRK +D++ L QL ++R + + CG++H+ G
Sbjct: 182 VFTMGNNSYGQCGRKVVEDEIYSESHLIHQL----NDFDSRVVQVVCGQDHSLFRTKKGA 237
Query: 83 LFT--WGLD 89
+F WG D
Sbjct: 238 VFACGWGAD 246
Score = 30.8 bits (68), Expect = 7.4
Identities = 28/88 (31%), Positives = 37/88 (41%), Gaps = 7/88 (7%)
Query: 25 VWGY--NQSGQTG--RKEKDDQLRIPKQLPPQLFGCPAGL--NTRWLDIACGREHTAAIA 78
VWG N+ Q G R +D L P P TR L ++CGR H+ +
Sbjct: 118 VWGMGLNKDSQLGFQRSIRDRTKGYEYVLEPSPIPLPLEKPQKTRVLQVSCGRAHSLVLT 177
Query: 79 SDGLLFTWGLDFSYSLLVLLIVHDFIYS 106
+FT G + SY +V D IYS
Sbjct: 178 DSEGVFTMG-NNSYGQCGRKVVEDEIYS 204
>UniRef100_UPI00002D3580 UPI00002D3580 UniRef100 entry
Length = 233
Score = 46.6 bits (109), Expect = 1e-04
Identities = 32/98 (32%), Positives = 45/98 (45%), Gaps = 17/98 (17%)
Query: 6 ILGTTKPVAASIPRKSA-------IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA 58
I GTT A + SA ++VWG N +G+ G Q P Q+
Sbjct: 98 IPGTTWKQLACLENSSAATKTDGTLWVWGRNGNGELGLNSTATQYSSPVQV--------- 148
Query: 59 GLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLLV 96
G +T W + G +H A+ +DG L+TWGL+ Y LV
Sbjct: 149 GADTDWYKVNGGGDHFMAVKTDGTLWTWGLN-DYGQLV 185
Score = 31.6 bits (70), Expect = 4.3
Identities = 21/72 (29%), Positives = 30/72 (41%), Gaps = 11/72 (15%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG N SG G + + P Q+P G G +H A+ +DG
Sbjct: 22 LFAWGANSSGVLGLNNQT-KYSSPVQIPGTWSTTTVG----------GGDHGGAVNTDGE 70
Query: 83 LFTWGLDFSYSL 94
LF WG + SL
Sbjct: 71 LFMWGSEGGGSL 82
>UniRef100_Q9CYF5 Williams-Beuren syndrome chromosome region 16 protein homolog [Mus
musculus]
Length = 461
Score = 46.2 bits (108), Expect = 2e-04
Identities = 30/88 (34%), Positives = 42/88 (47%), Gaps = 10/88 (11%)
Query: 7 LGTTKPVA------ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA-G 59
+G K VA A + + ++VWGY G G+ K + IP+ +PP LFG
Sbjct: 332 VGKVKQVACGGTGCAVLNAEGHVFVWGY---GILGKGPKLLETAIPEMIPPTLFGLTEFN 388
Query: 60 LNTRWLDIACGREHTAAIASDGLLFTWG 87
+ I CG H AA+ + G LF WG
Sbjct: 389 PEVQVSQIRCGLSHFAALTNKGELFVWG 416
>UniRef100_UPI000017E724 UPI000017E724 UniRef100 entry
Length = 461
Score = 45.8 bits (107), Expect = 2e-04
Identities = 28/88 (31%), Positives = 41/88 (45%), Gaps = 10/88 (11%)
Query: 7 LGTTKPVA------ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPA-G 59
+G K VA A + + ++VWGY G G+ + +P+ +PP LFG
Sbjct: 332 VGKVKQVACGGTGCAILNEEGHVFVWGY---GILGKGPNLSETALPEMIPPTLFGLTEFN 388
Query: 60 LNTRWLDIACGREHTAAIASDGLLFTWG 87
+ I CG H AA+ + G LF WG
Sbjct: 389 PEVKVSQIRCGLSHFAALTNKGELFVWG 416
>UniRef100_UPI000035F228 UPI000035F228 UniRef100 entry
Length = 342
Score = 45.8 bits (107), Expect = 2e-04
Identities = 23/61 (37%), Positives = 38/61 (61%), Gaps = 8/61 (13%)
Query: 27 GYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTW 86
GYN GQ GRK+ +++ K+ P GC GL + +ACG++H+ A+++ G +F+W
Sbjct: 52 GYNSDGQLGRKKNNNK---KKRTP----GCVEGLG-HIVQVACGQDHSLAVSACGRVFSW 103
Query: 87 G 87
G
Sbjct: 104 G 104
Score = 39.7 bits (91), Expect = 0.016
Identities = 26/80 (32%), Positives = 39/80 (48%), Gaps = 8/80 (10%)
Query: 8 GTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDI 67
G T + ++P +Y G NQSGQ G D++ + P L P G+ I
Sbjct: 195 GATHTMVLTLP--GLVYCCGANQSGQLGLNRVDEKGNFNVCMVPALR--PLGVTF----I 246
Query: 68 ACGREHTAAIASDGLLFTWG 87
+CG HTA + +G +FT+G
Sbjct: 247 SCGEAHTAVLTMNGEVFTFG 266
Score = 37.0 bits (84), Expect = 0.10
Identities = 19/65 (29%), Positives = 33/65 (50%), Gaps = 5/65 (7%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG +SGQ G + + R + L Q+ P L + +ACG+ H+ A+ G
Sbjct: 100 VFSWGAAESGQLGTDQMSSRPR--QDLTVQV---PIPLPVPVIQVACGKSHSVALTKGGD 154
Query: 83 LFTWG 87
+ +WG
Sbjct: 155 VLSWG 159
Score = 30.8 bits (68), Expect = 7.4
Identities = 21/73 (28%), Positives = 34/73 (45%), Gaps = 9/73 (12%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A + ++ +G + GQ G D+L P+ + + G PA I+CGR HT
Sbjct: 254 AVLTMNGEVFTFGEGRHGQLGHNFTADELS-PRLV--EHVGRPAS------QISCGRNHT 304
Query: 75 AAIASDGLLFTWG 87
+ S G L+ +G
Sbjct: 305 LVLDSSGQLWAFG 317
>UniRef100_Q65XH7 Ptative chromosome condensation factor [Oryza sativa]
Length = 917
Score = 45.8 bits (107), Expect = 2e-04
Identities = 23/74 (31%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
AA + R ++ WG + G+ G ++D + P+L A N + +ACG H
Sbjct: 89 AALVTRNGDVFTWGEDSGGRLGHGTREDSVH------PRLVESLAACNVDF--VACGEFH 140
Query: 74 TAAIASDGLLFTWG 87
T A+ + G L+TWG
Sbjct: 141 TCAVTTTGELYTWG 154
Score = 33.5 bits (75), Expect = 1.1
Identities = 21/51 (41%), Positives = 28/51 (54%), Gaps = 7/51 (13%)
Query: 45 IPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLL 95
IPK++ L G P ++CG HTA I S G LFT+G D S+ +L
Sbjct: 173 IPKRISGALDGLPVAY------VSCGTWHTALITSMGQLFTFG-DGSFGVL 216
Score = 31.6 bits (70), Expect = 4.3
Identities = 21/71 (29%), Positives = 32/71 (44%), Gaps = 7/71 (9%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWL----DIACGREHTAAIA 78
+YVWG + R D +R ++ F P L +R + + CG H A +
Sbjct: 37 VYVWGEVFCENSVRVGSDTIIRSTEKTD---FLLPKPLESRLVLDVYHVDCGVRHAALVT 93
Query: 79 SDGLLFTWGLD 89
+G +FTWG D
Sbjct: 94 RNGDVFTWGED 104
>UniRef100_UPI000035F90D UPI000035F90D UniRef100 entry
Length = 876
Score = 45.4 bits (106), Expect = 3e-04
Identities = 22/79 (27%), Positives = 38/79 (47%), Gaps = 7/79 (8%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ + ++ WG + GQ G + +RIP+ + + R + CG +H
Sbjct: 97 AVNEQGQVFAWGAGEEGQLGLGTTETAVRIPRLIKRLC-------DHRIAQVMCGNQHCI 149
Query: 76 AIASDGLLFTWGLDFSYSL 94
A++ DG LFTWG + S L
Sbjct: 150 ALSKDGQLFTWGQNTSGQL 168
Score = 42.0 bits (97), Expect = 0.003
Identities = 26/69 (37%), Positives = 38/69 (54%), Gaps = 14/69 (20%)
Query: 22 AIYVWGYNQSGQTGRKEKDDQLRIP---KQLPPQLFGCPAGLNTRWLDIACGREHTAAIA 78
A++ WG N++GQ G + D+ +P K L Q + + I+CG EHTAA+
Sbjct: 209 AVFGWGKNKAGQLGLNDIQDRA-VPCHIKFLRSQ----------KVVHISCGEEHTAALT 257
Query: 79 SDGLLFTWG 87
DG LFT+G
Sbjct: 258 KDGGLFTFG 266
Score = 36.6 bits (83), Expect = 0.13
Identities = 21/72 (29%), Positives = 35/72 (48%), Gaps = 7/72 (9%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ + ++ WG N SGQ G + + P +L PQ AG+ I G +H+
Sbjct: 150 ALSKDGQLFTWGQNTSGQLGLGKGE-----PSKLFPQPLKSLAGIPL--AKITAGGDHSF 202
Query: 76 AIASDGLLFTWG 87
A++ G +F WG
Sbjct: 203 ALSLSGAVFGWG 214
Score = 32.3 bits (72), Expect = 2.5
Identities = 18/67 (26%), Positives = 32/67 (46%), Gaps = 12/67 (17%)
Query: 22 AIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD-IACGREHTAAIASD 80
++Y G N GQ G + + P+ G L+T+ + ++CG+ H+ A+
Sbjct: 53 SVYTCGSNSCGQLGHNKPG--------ITPEFVGA---LDTQKITLVSCGQAHSLAVNEQ 101
Query: 81 GLLFTWG 87
G +F WG
Sbjct: 102 GQVFAWG 108
>UniRef100_UPI00003AA00E UPI00003AA00E UniRef100 entry
Length = 531
Score = 45.4 bits (106), Expect = 3e-04
Identities = 22/69 (31%), Positives = 37/69 (52%), Gaps = 7/69 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N Q G + L +P Q+ L +N + +++ACG H+ + SDG
Sbjct: 101 VYTWGHNAYSQLGNGTTNHGL-VPCQVSTNL------VNKKVIEVACGSHHSMVLTSDGE 153
Query: 83 LFTWGLDFS 91
++TWG + S
Sbjct: 154 VYTWGYNNS 162
Score = 41.2 bits (95), Expect = 0.005
Identities = 21/65 (32%), Positives = 34/65 (52%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WGYN SGQ G +Q +P ++ C N ++IACG+ + A+ +G
Sbjct: 154 VYTWGYNNSGQVGSGSTANQ-----PIPRRVTSCLQ--NKVVVNIACGQMCSMAVVENGE 206
Query: 83 LFTWG 87
++ WG
Sbjct: 207 VYVWG 211
Score = 41.2 bits (95), Expect = 0.005
Identities = 24/80 (30%), Positives = 35/80 (43%), Gaps = 9/80 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ +YVWGYN +GQ G +Q P A R +ACG HT
Sbjct: 200 AVVENGEVYVWGYNGNGQLGLGSSGNQ--------PTPCRIAALQGIRVQRVACGCAHTL 251
Query: 76 AIASDGLLFTWGLDFSYSLL 95
+ +G ++ WG + SY L
Sbjct: 252 VLTDEGQIYAWGAN-SYGQL 270
>UniRef100_Q6GP47 MGC80684 protein [Xenopus laevis]
Length = 1028
Score = 45.4 bits (106), Expect = 3e-04
Identities = 27/85 (31%), Positives = 40/85 (46%), Gaps = 9/85 (10%)
Query: 3 IDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNT 62
+D GT VA + IY WG GQ G + + PK++ L N
Sbjct: 87 VDVSCGTNHSVAMC--DEGNIYSWGDGSEGQLGTGKFSSKNFTPKRITGLL-------NR 137
Query: 63 RWLDIACGREHTAAIASDGLLFTWG 87
+ + I+CG H+ A+A DG +F+WG
Sbjct: 138 KIIQISCGNFHSVALAEDGRVFSWG 162
Score = 38.5 bits (88), Expect = 0.036
Identities = 21/74 (28%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
++ ++ WG N +GQ G K + P L + I+CG EHTA
Sbjct: 204 ALSMSGTVFAWGRNNAGQLGFKNDAKK----GTFKPYAVDSLRDLGVAY--ISCGEEHTA 257
Query: 76 AIASDGLLFTWGLD 89
+ DG+++T+G D
Sbjct: 258 VLNKDGIVYTFGDD 271
Score = 33.1 bits (74), Expect = 1.5
Identities = 19/74 (25%), Positives = 32/74 (42%), Gaps = 7/74 (9%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ ++ WG N+ GQ G +I Q PQL G+ + + G
Sbjct: 149 SVALAEDGRVFSWGQNKCGQLGLGS-----QIINQATPQLVKSLKGIPL--VQVTAGGSQ 201
Query: 74 TAAIASDGLLFTWG 87
T A++ G +F WG
Sbjct: 202 TFALSMSGTVFAWG 215
>UniRef100_Q9XHD7 UVB-resistance protein UVR8 [Arabidopsis thaliana]
Length = 440
Score = 45.4 bits (106), Expect = 3e-04
Identities = 26/73 (35%), Positives = 41/73 (55%), Gaps = 10/73 (13%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLF-GCPAGLNTRWLDIACGREHT 74
++ + + WG NQ+GQ G + +D L +P+++ Q F G P + +A G EHT
Sbjct: 134 AVTMEGEVQSWGRNQNGQLGLGDTEDSL-VPQKI--QAFEGIPIKM------VAAGAEHT 184
Query: 75 AAIASDGLLFTWG 87
AA+ DG L+ WG
Sbjct: 185 AAVTEDGDLYGWG 197
Score = 40.4 bits (93), Expect = 0.009
Identities = 25/72 (34%), Positives = 38/72 (52%), Gaps = 8/72 (11%)
Query: 16 SIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTA 75
S+ A+Y +G+++ GQ G + +D L IP +L A N+ I+ G HT
Sbjct: 238 SVSYSGALYTYGWSKYGQLGHGDLEDHL-IPHKLE-------ALSNSFISQISGGWRHTM 289
Query: 76 AIASDGLLFTWG 87
A+ SDG L+ WG
Sbjct: 290 ALTSDGKLYGWG 301
Score = 35.4 bits (80), Expect = 0.30
Identities = 18/73 (24%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 15 ASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
A++ +Y WG+ + G G ++ D+L +P+++ + + +ACG HT
Sbjct: 185 AAVTEDGDLYGWGWGRYGNLGLGDRTDRL-VPERVT-------STGGEKMSMVACGWRHT 236
Query: 75 AAIASDGLLFTWG 87
+++ G L+T+G
Sbjct: 237 ISVSYSGALYTYG 249
Score = 34.3 bits (77), Expect = 0.67
Identities = 18/65 (27%), Positives = 29/65 (43%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N+ GQ G DQ + P + + + ++CG HT A+
Sbjct: 297 LYGWGWNKFGQVGVGNNLDQCSPVQVRFPD--------DQKVVQVSCGWRHTLAVTERNN 348
Query: 83 LFTWG 87
+F WG
Sbjct: 349 VFAWG 353
Score = 32.0 bits (71), Expect = 3.3
Identities = 17/63 (26%), Positives = 30/63 (46%), Gaps = 9/63 (14%)
Query: 26 WGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL-LF 84
WG + GQ G + +D + P QL + + + CG +HT A + G+ ++
Sbjct: 39 WGRGEDGQLGHGDAED-----RPSPTQLSALDGH---QIVSVTCGADHTVAYSQSGMEVY 90
Query: 85 TWG 87
+WG
Sbjct: 91 SWG 93
>UniRef100_UPI00003AA019 UPI00003AA019 UniRef100 entry
Length = 536
Score = 45.1 bits (105), Expect = 4e-04
Identities = 29/87 (33%), Positives = 39/87 (44%), Gaps = 10/87 (11%)
Query: 1 MDIDAILGTTKPVAASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGL 60
M + G T +A + +Y WGYN +GQ G +QL P G+
Sbjct: 187 MVVGIACGQTSSMA--VVNNGEVYGWGYNGNGQLGLGNNGNQLT------PCRVAALHGV 238
Query: 61 NTRWLDIACGREHTAAIASDGLLFTWG 87
L IACG HT A+ +GLL+ WG
Sbjct: 239 CI--LQIACGYAHTLALTDEGLLYAWG 263
Score = 38.9 bits (89), Expect = 0.027
Identities = 20/65 (30%), Positives = 33/65 (50%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WG+N Q G + + P Q+ C L + +++ACG H+ A++ DG
Sbjct: 101 VYAWGHNGYSQLGNGATNQGVT-----PVQV--CTNLLLKKVIEVACGSHHSMALSFDGD 153
Query: 83 LFTWG 87
L+ WG
Sbjct: 154 LYAWG 158
Score = 36.6 bits (83), Expect = 0.13
Identities = 19/65 (29%), Positives = 32/65 (49%), Gaps = 7/65 (10%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
+Y WGYN GQ G +Q P ++ C + + IACG+ + A+ ++G
Sbjct: 154 LYAWGYNNCGQVGSGSTANQ-----PTPRRVSNCLQ--DKMVVGIACGQTSSMAVVNNGE 206
Query: 83 LFTWG 87
++ WG
Sbjct: 207 VYGWG 211
>UniRef100_UPI000026AC52 UPI000026AC52 UniRef100 entry
Length = 216
Score = 45.1 bits (105), Expect = 4e-04
Identities = 23/70 (32%), Positives = 38/70 (53%), Gaps = 6/70 (8%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG SG+ G D + + +L +L G + +ACG +H+AA+ SDG
Sbjct: 85 LFAWGSADSGKLGLGGGTDDVVLKPKLVSKLEGVDI------VSVACGDKHSAAVDSDGN 138
Query: 83 LFTWGLDFSY 92
L+TWG + S+
Sbjct: 139 LYTWGYNGSF 148
Score = 32.7 bits (73), Expect = 1.9
Identities = 23/81 (28%), Positives = 35/81 (42%), Gaps = 7/81 (8%)
Query: 14 AASIPRKSA---IYVWGYNQSGQTG----RKEKDDQLRIPKQLPPQLFGCPAGLNTRWLD 66
A+S +K A +++WG GQ G K + P L P + + ++
Sbjct: 9 ASSTVQKGATRSLFIWGNGNDGQLGLGAINKVGTAFPSYDELTPMPLSTIPLEEDEKVVE 68
Query: 67 IACGREHTAAIASDGLLFTWG 87
A G H+ A+ S G LF WG
Sbjct: 69 FAGGMNHSLAVTSAGRLFAWG 89
>UniRef100_Q6ZKB8 Putative UVB-resistance protein UVR8 [Oryza sativa]
Length = 395
Score = 45.1 bits (105), Expect = 4e-04
Identities = 29/94 (30%), Positives = 44/94 (45%), Gaps = 11/94 (11%)
Query: 3 IDAILGTTKPVAA-------SIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQL 53
+DA+ G + AA ++ K Y WG N+ GQ G + K+D R ++ P
Sbjct: 91 VDALAGVSIVQAAIGGWHCLAVDDKGRAYAWGGNEYGQCGEEAERKEDGTRALRRDIPIP 150
Query: 54 FGCPAGLNTRWLDIACGREHTAAIASDGLLFTWG 87
GC L R +A G H+ + DG ++TWG
Sbjct: 151 QGCAPKLKVR--QVAAGGTHSVVLTQDGHVWTWG 182
Score = 35.0 bits (79), Expect = 0.39
Identities = 24/78 (30%), Positives = 35/78 (44%), Gaps = 14/78 (17%)
Query: 12 PVAASIPRKSAIYVWGYNQSGQTGRK--EKDDQLRIPKQLPPQLFGCPAGLNTRWLDIAC 69
P A+S+P +A+ WG + GQ G E++D R L +
Sbjct: 4 PPASSLP--TAVLAWGSGEDGQLGMGGYEEEDWARGVAALDALAVSA----------VVA 51
Query: 70 GREHTAAIASDGLLFTWG 87
G ++ AI +DG LFTWG
Sbjct: 52 GSRNSLAICADGRLFTWG 69
Score = 35.0 bits (79), Expect = 0.39
Identities = 16/74 (21%), Positives = 38/74 (50%), Gaps = 7/74 (9%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
+ ++ ++ +Y WG + G+ G + +P+++ AG + + ++CG H
Sbjct: 267 STALTKEGEVYAWGRGEHGRLGFGDDKSSHMVPQKVE-----LLAGEDI--IQVSCGGTH 319
Query: 74 TAAIASDGLLFTWG 87
+ A+ DG +F++G
Sbjct: 320 SVALTRDGRMFSYG 333
Score = 34.3 bits (77), Expect = 0.67
Identities = 18/65 (27%), Positives = 32/65 (48%), Gaps = 8/65 (12%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREHTAAIASDGL 82
++ WG N+ GQ G + P+ P ++ G + +DIA G H+ A+ +G
Sbjct: 224 LWAWGNNEYGQLGTGDTQ-----PRSQPIRVEGLS---DLSLVDIAAGGWHSTALTKEGE 275
Query: 83 LFTWG 87
++ WG
Sbjct: 276 VYAWG 280
>UniRef100_Q947C9 Putative chromosome condensation factor [Triticum monococcum]
Length = 907
Score = 44.7 bits (104), Expect = 5e-04
Identities = 22/74 (29%), Positives = 37/74 (49%), Gaps = 8/74 (10%)
Query: 14 AASIPRKSAIYVWGYNQSGQTGRKEKDDQLRIPKQLPPQLFGCPAGLNTRWLDIACGREH 73
A+ + R ++ WG + G+ G ++D + P+L A N + +ACG H
Sbjct: 89 ASLVTRNGEVFTWGEDSGGRLGHGTREDSVH------PRLVESLAACNIDF--VACGEFH 140
Query: 74 TAAIASDGLLFTWG 87
T A+ + G L+TWG
Sbjct: 141 TCAVTTTGELYTWG 154
Score = 32.7 bits (73), Expect = 1.9
Identities = 22/75 (29%), Positives = 32/75 (42%), Gaps = 15/75 (20%)
Query: 23 IYVWGYNQSGQTGRKEKDDQLR--------IPKQLPPQLFGCPAGLNTRWLDIACGREHT 74
+YVWG + R D +R +PK L L L+ +D CG H
Sbjct: 37 VYVWGEVVCDNSARTSSDTVIRSTGKTDFLLPKPLESNLV-----LDVYHVD--CGVRHA 89
Query: 75 AAIASDGLLFTWGLD 89
+ + +G +FTWG D
Sbjct: 90 SLVTRNGEVFTWGED 104
Score = 31.2 bits (69), Expect = 5.7
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 3/47 (6%)
Query: 49 LPPQLFGCPAGLNTRWLDIACGREHTAAIASDGLLFTWGLDFSYSLL 95
+P ++ G GL + ++CG HTA I S G LFT+G D S+ +L
Sbjct: 173 IPRRISGPLEGLQIAY--VSCGTWHTALITSMGQLFTFG-DGSFGVL 216
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.326 0.143 0.458
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 210,207,637
Number of Sequences: 2790947
Number of extensions: 8207253
Number of successful extensions: 24776
Number of sequences better than 10.0: 520
Number of HSP's better than 10.0 without gapping: 261
Number of HSP's successfully gapped in prelim test: 259
Number of HSP's that attempted gapping in prelim test: 22346
Number of HSP's gapped (non-prelim): 2258
length of query: 121
length of database: 848,049,833
effective HSP length: 97
effective length of query: 24
effective length of database: 577,327,974
effective search space: 13855871376
effective search space used: 13855871376
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137823.5


