

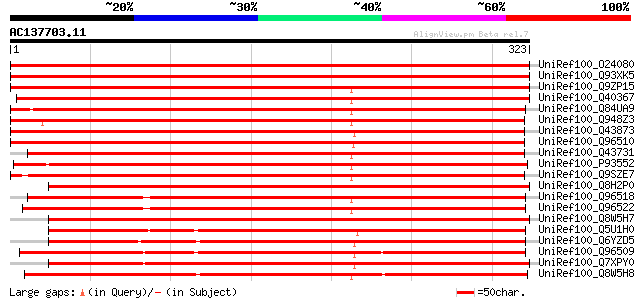
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137703.11 + phase: 0
(323 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O24080 Peroxidase2 precursor [Medicago sativa] 649 0.0
UniRef100_Q93XK5 Peroxidase2 precursor [Medicago sativa] 646 0.0
UniRef100_Q9ZP15 Peroxidase precursor [Trifolium repens] 602 e-171
UniRef100_Q40367 Peroxidase precursor [Medicago sativa] 479 e-134
UniRef100_Q84UA9 Peroxidase 1 [Artemisia annua] 470 e-131
UniRef100_Q948Z3 Putative peroxidase [Solanum tuberosum] 464 e-129
UniRef100_Q43873 Peroxidase 73 precursor [Arabidopsis thaliana] 448 e-125
UniRef100_Q96510 Peroxidase 35 precursor [Arabidopsis thaliana] 446 e-124
UniRef100_Q43731 Peroxidase 50 precursor [Arabidopsis thaliana] 446 e-124
UniRef100_P93552 Peroxidase precursor [Spinacia oleracea] 444 e-123
UniRef100_Q9SZE7 Peroxidase 51 precursor [Arabidopsis thaliana] 442 e-123
UniRef100_Q8H2P0 Putative peroxidase [Oryza sativa] 421 e-116
UniRef100_Q96518 Peroxidase 16 precursor [Arabidopsis thaliana] 417 e-115
UniRef100_Q96522 Peroxidase 45 precursor [Arabidopsis thaliana] 417 e-115
UniRef100_Q8W5H7 Putative peroxidase [Oryza sativa] 406 e-112
UniRef100_Q5U1H0 Class III peroxidase 123 precursor [Oryza sativa] 357 2e-97
UniRef100_Q6YZD5 Putative peroxidase [Oryza sativa] 340 2e-92
UniRef100_Q96509 Peroxidase 55 precursor [Arabidopsis thaliana] 337 3e-91
UniRef100_Q7XPY0 OSJNBa0071I13.13 protein [Oryza sativa] 318 2e-85
UniRef100_Q8W5H8 Putative peroxidase [Oryza sativa] 309 6e-83
>UniRef100_O24080 Peroxidase2 precursor [Medicago sativa]
Length = 323
Score = 649 bits (1673), Expect = 0.0
Identities = 320/323 (99%), Positives = 322/323 (99%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA
Sbjct: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR
Sbjct: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF
Sbjct: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA 240
ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA
Sbjct: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA 240
Query: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM 300
INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM
Sbjct: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM 300
Query: 301 TKLGRVGVKNARNGKIRTDCSVL 323
TKLGR+GVK ARNGKIRTDC+VL
Sbjct: 301 TKLGRIGVKTARNGKIRTDCTVL 323
>UniRef100_Q93XK5 Peroxidase2 precursor [Medicago sativa]
Length = 323
Score = 646 bits (1667), Expect = 0.0
Identities = 319/323 (98%), Positives = 321/323 (98%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA
Sbjct: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR
Sbjct: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF
Sbjct: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA 240
ANNGLTQTDMIALSGAHT GFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA
Sbjct: 181 ANNGLTQTDMIALSGAHTSGFSHCDRFSNRIQTPVDPTLNKQYAAQLQQMCPRNVDPRIA 240
Query: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM 300
INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM
Sbjct: 241 INMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAM 300
Query: 301 TKLGRVGVKNARNGKIRTDCSVL 323
TKLGR+GVK ARNGKIRTDC+VL
Sbjct: 301 TKLGRIGVKTARNGKIRTDCTVL 323
>UniRef100_Q9ZP15 Peroxidase precursor [Trifolium repens]
Length = 329
Score = 602 bits (1551), Expect = e-171
Identities = 301/329 (91%), Positives = 313/329 (94%), Gaps = 6/329 (1%)
Query: 1 MGRYN-VILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVP 59
MGR+N VILVWSL L LC IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVP
Sbjct: 1 MGRFNNVILVWSLLLMLCFIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVP 60
Query: 60 ATLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQC 119
ATLRLFFHDCFVQGCDASVLVASSG N+AEKD+P+NLSLAGDGFDTVIKAKAALDAVPQC
Sbjct: 61 ATLRLFFHDCFVQGCDASVLVASSGGNQAEKDNPDNLSLAGDGFDTVIKAKAALDAVPQC 120
Query: 120 RNKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTL 179
RNKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQP FNLNQLN+L
Sbjct: 121 RNKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPGFNLNQLNSL 180
Query: 180 FANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRN 234
FA+NGLTQTDMIALSGAHTLGFSHC+RFSNRI Q+PVDPTLNKQYAAQLQQMCPRN
Sbjct: 181 FASNGLTQTDMIALSGAHTLGFSHCNRFSNRIFNFNNQSPVDPTLNKQYAAQLQQMCPRN 240
Query: 235 VDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNA 294
VDPRIAINMDPTTPR FDN YY+NLQQGKGLFTSDQILFTDTRSR TVNSFA++GNVFNA
Sbjct: 241 VDPRIAINMDPTTPRQFDNAYYQNLQQGKGLFTSDQILFTDTRSRATVNSFASSGNVFNA 300
Query: 295 NFITAMTKLGRVGVKNARNGKIRTDCSVL 323
NFI AMTKLGR+GVK ARNGKIRTDCSVL
Sbjct: 301 NFINAMTKLGRIGVKTARNGKIRTDCSVL 329
>UniRef100_Q40367 Peroxidase precursor [Medicago sativa]
Length = 325
Score = 479 bits (1232), Expect = e-134
Identities = 232/324 (71%), Positives = 278/324 (85%), Gaps = 5/324 (1%)
Query: 5 NVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRL 64
N+ILV L LTL L T AQLS +HY N CPNV++IVR AV+KKF QTF TVPATLRL
Sbjct: 2 NLILVPLLFLTLFLHSCRTHAQLSRHHYKNSCPNVENIVRQAVKKKFHQTFTTVPATLRL 61
Query: 65 FFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVS 124
FFHDCFVQGCD S+LVAS+ +N+AE+DHP+NLSLAGDGFDTVI+AKAA+DAVP C+NKVS
Sbjct: 62 FFHDCFVQGCDGSILVASTPHNRAERDHPDNLSLAGDGFDTVIQAKAAVDAVPLCQNKVS 121
Query: 125 CADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNG 184
CADILA+ATRDVI LAGGP Y VELGRFDGL S+ SDVNG+LP+P FNLNQLNTLF ++G
Sbjct: 122 CADILAMATRDVIALAGGPYYEVELGRFDGLRSKDSDVNGKLPEPGFNLNQLNTLFKHHG 181
Query: 185 LTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRI 239
LTQT+MIALSGAHT+GFSHC++F+NR+ + VDPTL+ YAA+L+ MCPR+VDPR+
Sbjct: 182 LTQTEMIALSGAHTVGFSHCNKFTNRVYNFKTTSRVDPTLDLHYAAKLKSMCPRDVDPRV 241
Query: 240 AINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITA 299
A++MDP TP FDNVY+KNLQ+GKGLFTSDQ+LFTD+RS+ VN+FA++ +F ANF+ A
Sbjct: 242 AVDMDPITPHAFDNVYFKNLQKGKGLFTSDQVLFTDSRSKAAVNAFASSNKIFRANFVAA 301
Query: 300 MTKLGRVGVKNARNGKIRTDCSVL 323
MTKLGRVGVKN+ NG IRTDCSV+
Sbjct: 302 MTKLGRVGVKNSHNGNIRTDCSVI 325
>UniRef100_Q84UA9 Peroxidase 1 [Artemisia annua]
Length = 328
Score = 470 bits (1209), Expect = e-131
Identities = 234/326 (71%), Positives = 265/326 (80%), Gaps = 6/326 (1%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
MGR V V +L +L + P FAQL N+YANICPNV+SIV+ AV K +QTFVT+P
Sbjct: 1 MGRIIVFQVLALC-SLLVFPNIAFAQLKQNYYANICPNVESIVQKAVAAKVKQTFVTIPG 59
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFVQGCDASV++ SSG+N AEKDHP+NLSLAGDGFDTVIKAKAA+DA P CR
Sbjct: 60 TLRLFFHDCFVQGCDASVMIQSSGSNTAEKDHPDNLSLAGDGFDTVIKAKAAVDANPSCR 119
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADIL +ATRDV+ +AGGPSY+VELGR DGL S ++ V G LP+P+ NL+QLN LF
Sbjct: 120 NKVSCADILTMATRDVVKIAGGPSYSVELGRLDGLSSTAASVGGNLPKPNQNLDQLNALF 179
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNV 235
A NGLTQ DMIALSGAHTLGFSHC++FSNRI Q PVDPTLN YA QLQQ CP+NV
Sbjct: 180 AANGLTQADMIALSGAHTLGFSHCNQFSNRIYNFSKQNPVDPTLNPSYATQLQQQCPKNV 239
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPRIAINMDP TPRTFDNVYYKNLQ G+GLFTSDQ+LFTDTRS+ TV S+A + FN
Sbjct: 240 DPRIAINMDPNTPRTFDNVYYKNLQNGQGLFTSDQVLFTDTRSKQTVISWANSPTAFNNA 299
Query: 296 FITAMTKLGRVGVKNARNGKIRTDCS 321
FITAMTKLGRVGVK G IR DC+
Sbjct: 300 FITAMTKLGRVGVKTGTKGNIRKDCA 325
>UniRef100_Q948Z3 Putative peroxidase [Solanum tuberosum]
Length = 331
Score = 464 bits (1195), Expect = e-129
Identities = 226/327 (69%), Positives = 267/327 (81%), Gaps = 7/327 (2%)
Query: 1 MGRYNVILVWSLALTLCLI--PYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTV 58
MG V +V L+++ I P AQL N YA CPNV+SIVR+ V +KF+QTFVT+
Sbjct: 1 MGHLQVFIVSFLSISCVSIFMPNLVAAQLKTNFYAQTCPNVESIVRNVVNQKFKQTFVTI 60
Query: 59 PATLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQ 118
PA LRLFFHDCFV+GCDASV++AS+ N AEKDHP+NLSLAGDGFDTVIKAKAA+DA+P
Sbjct: 61 PAVLRLFFHDCFVEGCDASVIIASTSGNTAEKDHPDNLSLAGDGFDTVIKAKAAVDAIPS 120
Query: 119 CRNKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNT 178
C+NKVSCADILALATRDVI L+GGP Y VELGR DGL S+SS+V G LP+P+FNL+QLNT
Sbjct: 121 CKNKVSCADILALATRDVIQLSGGPGYAVELGRLDGLTSKSSNVGGNLPKPTFNLDQLNT 180
Query: 179 LFANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPR 233
+FA++GL Q DMIALS AHTLGFSHCD+FSNRI + PVDP++NK YAAQLQQMCP+
Sbjct: 181 MFASHGLNQADMIALSAAHTLGFSHCDQFSNRIFNFSPKNPVDPSVNKTYAAQLQQMCPK 240
Query: 234 NVDPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFN 293
NVDPRIAINMDP TPR FDNVY++NLQ+G GLFTSDQ+LFTD RS+ TV+ +A+N VF
Sbjct: 241 NVDPRIAINMDPITPRAFDNVYFQNLQKGMGLFTSDQVLFTDQRSKGTVDLWASNSKVFQ 300
Query: 294 ANFITAMTKLGRVGVKNARNGKIRTDC 320
F+ AMTKLGRVGVK +NG IR DC
Sbjct: 301 TAFVNAMTKLGRVGVKTGKNGNIRIDC 327
>UniRef100_Q43873 Peroxidase 73 precursor [Arabidopsis thaliana]
Length = 329
Score = 448 bits (1153), Expect = e-125
Identities = 219/325 (67%), Positives = 256/325 (78%), Gaps = 5/325 (1%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M R+++++V +L+L + + P TT AQL N Y N CPNV+ IV+ VQ+K +QTFVT+PA
Sbjct: 1 MARFSLVVVVTLSLAISMFPDTTTAQLKTNFYGNSCPNVEQIVKKVVQEKIKQTFVTIPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFV GCDASV++ S+ NKAEKDHP+N+SLAGDGFD VIKAK ALDA+P C+
Sbjct: 61 TLRLFFHDCFVNGCDASVMIQSTPTNKAEKDHPDNISLAGDGFDVVIKAKKALDAIPSCK 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADILALATRDV+ A GPSY VELGRFDGLVS ++ VNG LP P+ + +LN LF
Sbjct: 121 NKVSCADILALATRDVVVAAKGPSYAVELGRFDGLVSTAASVNGNLPGPNNKVTELNKLF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNV 235
A N LTQ DMIALS AHTLGF+HC + NRI VDPTLNK YA +LQ CP+ V
Sbjct: 181 AKNKLTQEDMIALSAAHTLGFAHCGKVFNRIYNFNLTHAVDPTLNKAYAKELQLACPKTV 240
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPRIAINMDPTTPR FDN+Y+KNLQQGKGLFTSDQ+LFTD RS+ TVN +A N FN
Sbjct: 241 DPRIAINMDPTTPRQFDNIYFKNLQQGKGLFTSDQVLFTDGRSKPTVNDWAKNSVAFNKA 300
Query: 296 FITAMTKLGRVGVKNARNGKIRTDC 320
F+TAMTKLGRVGVK RNG IR DC
Sbjct: 301 FVTAMTKLGRVGVKTRRNGNIRRDC 325
>UniRef100_Q96510 Peroxidase 35 precursor [Arabidopsis thaliana]
Length = 329
Score = 446 bits (1148), Expect = e-124
Identities = 219/325 (67%), Positives = 256/325 (78%), Gaps = 5/325 (1%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M R++++L+ L L + + P TT AQLS Y+ CPNV+ IVR+AVQKK ++TFV VPA
Sbjct: 1 MARFDIVLLIGLCLIISVFPDTTTAQLSRGFYSKTCPNVEQIVRNAVQKKIKKTFVAVPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRLFFHDCFV GCDASV++ S+ NKAEKDHP+N+SLAGDGFD VI+AK ALD+ P CR
Sbjct: 61 TLRLFFHDCFVNGCDASVMIQSTPKNKAEKDHPDNISLAGDGFDVVIQAKKALDSNPSCR 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADIL LATRDV+ AGGPSY VELGRFDGLVS +S V G LP PS N+++LN LF
Sbjct: 121 NKVSCADILTLATRDVVVAAGGPSYEVELGRFDGLVSTASSVEGNLPGPSDNVDKLNALF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRIQ-----TPVDPTLNKQYAAQLQQMCPRNV 235
N LTQ DMIALS AHTLGF+HC + RI VDPTLNK YA +LQ+ CP+NV
Sbjct: 181 TKNKLTQEDMIALSAAHTLGFAHCGKVFKRIHKFNGINSVDPTLNKAYAIELQKACPKNV 240
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPRIAINMDP TP+TFDN Y+KNLQQGKGLFTSDQ+LFTD RSR TVN++A+N FN
Sbjct: 241 DPRIAINMDPVTPKTFDNTYFKNLQQGKGLFTSDQVLFTDGRSRPTVNAWASNSTAFNRA 300
Query: 296 FITAMTKLGRVGVKNARNGKIRTDC 320
F+ AMTKLGRVGVKN+ NG IR DC
Sbjct: 301 FVIAMTKLGRVGVKNSSNGNIRRDC 325
>UniRef100_Q43731 Peroxidase 50 precursor [Arabidopsis thaliana]
Length = 329
Score = 446 bits (1146), Expect = e-124
Identities = 218/314 (69%), Positives = 251/314 (79%), Gaps = 5/314 (1%)
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
L L+LCL + AQL N YA CPNV+ IVR+AVQKK QQTF T+PATLRL+FHDCFV
Sbjct: 12 LLLSLCLTLDLSSAQLRRNFYAGSCPNVEQIVRNAVQKKVQQTFTTIPATLRLYFHDCFV 71
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
GCDASV++AS+ NNKAEKDH ENLSLAGDGFDTVIKAK ALDAVP CRNKVSCADIL +
Sbjct: 72 NGCDASVMIASTNNNKAEKDHEENLSLAGDGFDTVIKAKEALDAVPNCRNKVSCADILTM 131
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMI 191
ATRDV+NLAGGP Y VELGR DGL S ++ V G+LP P+ ++N+L +LFA NGL+ DMI
Sbjct: 132 ATRDVVNLAGGPQYDVELGRLDGLSSTAASVGGKLPHPTDDVNKLTSLFAKNGLSLNDMI 191
Query: 192 ALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPT 246
ALSGAHTLGF+HC + NRI T VDPT+NK Y +L+ CPRN+DPR+AINMDPT
Sbjct: 192 ALSGAHTLGFAHCTKVFNRIYTFNKTTKVDPTVNKDYVTELKASCPRNIDPRVAINMDPT 251
Query: 247 TPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRV 306
TPR FDNVYYKNLQQGKGLFTSDQ+LFTD RS+ TV+ +A NG +FN FI +M KLGRV
Sbjct: 252 TPRQFDNVYYKNLQQGKGLFTSDQVLFTDRRSKPTVDLWANNGQLFNQAFINSMIKLGRV 311
Query: 307 GVKNARNGKIRTDC 320
GVK NG IR DC
Sbjct: 312 GVKTGSNGNIRRDC 325
>UniRef100_P93552 Peroxidase precursor [Spinacia oleracea]
Length = 329
Score = 444 bits (1142), Expect = e-123
Identities = 215/326 (65%), Positives = 260/326 (78%), Gaps = 7/326 (2%)
Query: 3 RYNVILVWSLALTLCL-IPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPAT 61
++N + + +L + LCL PYT LS +YA CPNV+ IVR AVQKK QQTFVT+PAT
Sbjct: 3 QFNNVSISALLIILCLSFPYTA-TSLSTTYYAKTCPNVEKIVRQAVQKKIQQTFVTIPAT 61
Query: 62 LRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRN 121
LRLFFHDCFV GCDAS+++ S+G N AEKDHP+NLSLAGDGFDTVIKAKAA+DAVP C N
Sbjct: 62 LRLFFHDCFVSGCDASIIIQSTGTNTAEKDHPDNLSLAGDGFDTVIKAKAAVDAVPGCTN 121
Query: 122 KVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFA 181
VSCADILALATRDV+NL+GGP + VELGRFDGLVS++S VNGRLPQP+ LN+LN+LFA
Sbjct: 122 NVSCADILALATRDVVNLSGGPFWEVELGRFDGLVSKASSVNGRLPQPTDELNRLNSLFA 181
Query: 182 NNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVD 236
+NGLTQ +M+ALSGAHT+GFSHC +FS RI + P+DPTLN Q+A QLQ MCP+NVD
Sbjct: 182 SNGLTQAEMVALSGAHTVGFSHCSKFSKRIYGFTPKNPIDPTLNAQFATQLQTMCPKNVD 241
Query: 237 PRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANF 296
PRIA+NMD +PR FDN YY+NL GKGLFTSDQ+L+TD R++ V +A + + F F
Sbjct: 242 PRIAVNMDVQSPRIFDNAYYRNLINGKGLFTSDQVLYTDPRTKGLVTGWAQSSSSFKQAF 301
Query: 297 ITAMTKLGRVGVKNARNGKIRTDCSV 322
+M KLGRVGVKN++NG IR C V
Sbjct: 302 AQSMIKLGRVGVKNSKNGNIRVQCDV 327
>UniRef100_Q9SZE7 Peroxidase 51 precursor [Arabidopsis thaliana]
Length = 329
Score = 442 bits (1136), Expect = e-123
Identities = 217/325 (66%), Positives = 257/325 (78%), Gaps = 8/325 (2%)
Query: 1 MGRYNVILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPA 60
M + N++L L L+L L + AQL + YA CPNV+ IVR+AVQKK QQTF T+PA
Sbjct: 4 MNKTNLLL---LILSLFLAINLSSAQLRGDFYAGTCPNVEQIVRNAVQKKIQQTFTTIPA 60
Query: 61 TLRLFFHDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCR 120
TLRL+FHDCFV GCDASV++AS+ NKAEKDH +NLSLAGDGFDTVIKAK A+DAVP CR
Sbjct: 61 TLRLYFHDCFVNGCDASVMIASTNTNKAEKDHEDNLSLAGDGFDTVIKAKEAVDAVPNCR 120
Query: 121 NKVSCADILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLF 180
NKVSCADIL +ATRDV+NLAGGP Y VELGR DGL S +S V G+LP+P+F+LNQLN LF
Sbjct: 121 NKVSCADILTMATRDVVNLAGGPQYAVELGRRDGLSSSASSVTGKLPKPTFDLNQLNALF 180
Query: 181 ANNGLTQTDMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNV 235
A NGL+ DMIALSGAHTLGF+HC + NR+ VDPT+NK Y +L+ CP+N+
Sbjct: 181 AENGLSPNDMIALSGAHTLGFAHCTKVFNRLYNFNKTNNVDPTINKDYVTELKASCPQNI 240
Query: 236 DPRIAINMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNAN 295
DPR+AINMDP TPR FDNVYYKNLQQGKGLFTSDQ+LFTD+RS+ TV+ +A NG +FN
Sbjct: 241 DPRVAINMDPNTPRQFDNVYYKNLQQGKGLFTSDQVLFTDSRSKPTVDLWANNGQLFNQA 300
Query: 296 FITAMTKLGRVGVKNARNGKIRTDC 320
FI++M KLGRVGVK NG IR DC
Sbjct: 301 FISSMIKLGRVGVKTGSNGNIRRDC 325
>UniRef100_Q8H2P0 Putative peroxidase [Oryza sativa]
Length = 330
Score = 421 bits (1083), Expect = e-116
Identities = 207/300 (69%), Positives = 243/300 (81%), Gaps = 1/300 (0%)
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
AQL N+YA +CPNV+SIVR AV +K Q+TF TV AT+RLFFHDCFV GCDASV+VAS+G
Sbjct: 30 AQLRRNYYAGVCPNVESIVRGAVARKVQETFATVGATVRLFFHDCFVDGCDASVVVASAG 89
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
NN AEKDHP NLSLAGDGFDTVIKAKAA+DAVP CR++VSCADILA+ATRD I LAGGPS
Sbjct: 90 NNTAEKDHPNNLSLAGDGFDTVIKAKAAVDAVPGCRDRVSCADILAMATRDAIALAGGPS 149
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
Y VELGR DGL S +S VNGRLP P+FNL+QL LFA NGL+Q DMIALS HT+GF+HC
Sbjct: 150 YAVELGRLDGLRSTASSVNGRLPPPTFNLDQLTALFAANGLSQADMIALSAGHTVGFAHC 209
Query: 205 DRFSNRIQ-TPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGK 263
+ F RI+ + VDPT++ +YAAQLQ+ CP NVDPRIA+ MDP TPR FDN Y+KNLQ G
Sbjct: 210 NTFLGRIRGSSVDPTMSPRYAAQLQRSCPPNVDPRIAVTMDPVTPRAFDNQYFKNLQNGM 269
Query: 264 GLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDCSVL 323
GL SDQ+L++D RSR V+S+A + FN F+TAMTKLGRVGVK G IR +C+VL
Sbjct: 270 GLLGSDQVLYSDPRSRPIVDSWAQSSAAFNQAFVTAMTKLGRVGVKTGSQGNIRRNCAVL 329
>UniRef100_Q96518 Peroxidase 16 precursor [Arabidopsis thaliana]
Length = 323
Score = 417 bits (1073), Expect = e-115
Identities = 210/315 (66%), Positives = 249/315 (78%), Gaps = 9/315 (2%)
Query: 12 LALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFV 71
+AL L + FAQL N Y CPNV++IVR+AV++KFQQTFVT PATLRLFFHDCFV
Sbjct: 10 VALLLIFFSSSVFAQLQTNFYRKSCPNVETIVRNAVRQKFQQTFVTAPATLRLFFHDCFV 69
Query: 72 QGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILAL 131
+GCDAS+L+AS +EKDHP++ SLAGDGFDTV KAK ALD P CRNKVSCADILAL
Sbjct: 70 RGCDASILLASP----SEKDHPDDKSLAGDGFDTVAKAKQALDRDPNCRNKVSCADILAL 125
Query: 132 ATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMI 191
ATRDV+ L GGP+Y VELGR DG +S + V LPQPSF L+QLNT+FA +GL+QTDMI
Sbjct: 126 ATRDVVVLTGGPNYPVELGRRDGRLSTVASVQHSLPQPSFKLDQLNTMFARHGLSQTDMI 185
Query: 192 ALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPT 246
ALSGAHT+GF+HC +FS RI + P+DPTLN +YA QL+QMCP VD RIAINMDPT
Sbjct: 186 ALSGAHTIGFAHCGKFSKRIYNFSPKRPIDPTLNIRYALQLRQMCPIRVDLRIAINMDPT 245
Query: 247 TPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRV 306
+P TFDN Y+KNLQ+G GLFTSDQ+LF+D RSR+TVNSFA++ F FI+A+TKLGRV
Sbjct: 246 SPNTFDNAYFKNLQKGMGLFTSDQVLFSDERSRSTVNSFASSEATFRQAFISAITKLGRV 305
Query: 307 GVKNARNGKIRTDCS 321
GVK G+IR DCS
Sbjct: 306 GVKTGNAGEIRRDCS 320
>UniRef100_Q96522 Peroxidase 45 precursor [Arabidopsis thaliana]
Length = 325
Score = 417 bits (1071), Expect = e-115
Identities = 206/318 (64%), Positives = 249/318 (77%), Gaps = 9/318 (2%)
Query: 9 VWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHD 68
++S L L+ AQL Y N CPNV++IVR+AV++KFQQTFVT PATLRLFFHD
Sbjct: 9 IFSNFFLLLLLSSCVSAQLRTGFYQNSCPNVETIVRNAVRQKFQQTFVTAPATLRLFFHD 68
Query: 69 CFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADI 128
CFV+GCDAS+++AS +E+DHP+++SLAGDGFDTV+KAK A+D+ P CRNKVSCADI
Sbjct: 69 CFVRGCDASIMIASP----SERDHPDDMSLAGDGFDTVVKAKQAVDSNPNCRNKVSCADI 124
Query: 129 LALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQT 188
LALATR+V+ L GGPSY VELGR DG +S + V +LPQP FNLNQLN +F+ +GL+QT
Sbjct: 125 LALATREVVVLTGGPSYPVELGRRDGRISTKASVQSQLPQPEFNLNQLNGMFSRHGLSQT 184
Query: 189 DMIALSGAHTLGFSHCDRFSNRI-----QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINM 243
DMIALSGAHT+GF+HC + S RI T +DP++N+ Y QL+QMCP VD RIAINM
Sbjct: 185 DMIALSGAHTIGFAHCGKMSKRIYNFSPTTRIDPSINRGYVVQLKQMCPIGVDVRIAINM 244
Query: 244 DPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKL 303
DPT+PRTFDN Y+KNLQQGKGLFTSDQILFTD RSR+TVNSFA + F FITA+TKL
Sbjct: 245 DPTSPRTFDNAYFKNLQQGKGLFTSDQILFTDQRSRSTVNSFANSEGAFRQAFITAITKL 304
Query: 304 GRVGVKNARNGKIRTDCS 321
GRVGV G+IR DCS
Sbjct: 305 GRVGVLTGNAGEIRRDCS 322
>UniRef100_Q8W5H7 Putative peroxidase [Oryza sativa]
Length = 324
Score = 406 bits (1043), Expect = e-112
Identities = 200/301 (66%), Positives = 244/301 (80%), Gaps = 2/301 (0%)
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
AQL ++YA++CP+V++IVR AV KK Q+T V V AT+RLFFHDCFV+GCDASV+V SSG
Sbjct: 23 AQLRRDYYASVCPDVETIVRDAVTKKVQETSVAVGATVRLFFHDCFVEGCDASVIVVSSG 82
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
NN AEKDHP NLSLAGDGFDTVIKA+AA+DAVPQC N+VSCADIL +ATRDVI LAGGPS
Sbjct: 83 NNTAEKDHPNNLSLAGDGFDTVIKARAAVDAVPQCTNQVSCADILVMATRDVIALAGGPS 142
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
Y VELGR DGL S +S V+G+LP PSFNL+QL +LFA N L+QTDMIALS AHT+GF+HC
Sbjct: 143 YAVELGRLDGLSSTASSVDGKLPPPSFNLDQLTSLFAANNLSQTDMIALSAAHTVGFAHC 202
Query: 205 DRFSNRIQ-TPVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQQGK 263
F++RIQ + VDPT++ YA+QLQ CP VDP IA+ +DP TPR FDN Y+ NLQ+G
Sbjct: 203 GTFASRIQPSAVDPTMDAGYASQLQAACPAGVDPNIALELDPVTPRAFDNQYFVNLQKGM 262
Query: 264 GLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVK-NARNGKIRTDCSV 322
GLFTSDQ+L++D RSR TV+++A N + F F+ AMT LGRVGVK + G IR DC++
Sbjct: 263 GLFTSDQVLYSDDRSRPTVDAWAANSSDFELAFVAAMTNLGRVGVKTDPSQGNIRRDCAM 322
Query: 323 L 323
L
Sbjct: 323 L 323
>UniRef100_Q5U1H0 Class III peroxidase 123 precursor [Oryza sativa]
Length = 331
Score = 357 bits (917), Expect = 2e-97
Identities = 179/301 (59%), Positives = 221/301 (72%), Gaps = 7/301 (2%)
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
A+LSP HY + CP V+S+VRS V +K ++TFVTVPATLRLFFHDCFV+GCDASV++AS G
Sbjct: 31 ARLSPEHYRSTCPGVESVVRSVVARKVKETFVTVPATLRLFFHDCFVEGCDASVMIASRG 90
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
N+ AEKD P+NLSLAGDGFDTV++AKAA++ +C VSCADILA+A RDV+ ++ GP
Sbjct: 91 ND-AEKDSPDNLSLAGDGFDTVVRAKAAVEK--KCPGVVSCADILAIAARDVVAMSSGPR 147
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
+TVELGR DGLVS+S V G+LP P + L +FA N LT DM+ALSGAHT+GF+HC
Sbjct: 148 WTVELGRLDGLVSKSGGVAGKLPGPDMRVKDLAAIFAKNNLTVLDMVALSGAHTVGFAHC 207
Query: 205 DRFSNRIQTPV----DPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNLQ 260
RF+ R+ V DP+ + YA QL CPR+V P IA+NMDP TP FDN YY NL
Sbjct: 208 TRFAGRLYGRVGGGVDPSYDPAYARQLMAACPRDVAPTIAVNMDPITPAAFDNAYYANLA 267
Query: 261 QGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTDC 320
G GLFTSDQ L+TD SR V FA N +F F AM KLGRVGVK+ ++G+IR DC
Sbjct: 268 GGLGLFTSDQELYTDAASRPAVTGFAKNQTLFFEAFKEAMVKLGRVGVKSGKHGEIRRDC 327
Query: 321 S 321
+
Sbjct: 328 T 328
>UniRef100_Q6YZD5 Putative peroxidase [Oryza sativa]
Length = 333
Score = 340 bits (873), Expect = 2e-92
Identities = 167/302 (55%), Positives = 218/302 (71%), Gaps = 8/302 (2%)
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
A LS +Y++ CP ++SIVR V +K +T VT+PA LRLFFHDC V GCDAS L+ SS
Sbjct: 31 ADLSAGYYSSSCPKLESIVRYEVSRKINETVVTIPAVLRLFFHDCLVTGCDASALI-SSP 89
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
N+ AEKD P+N+SLAGDGFDTV + K A++ C VSCADILALA RDV++LA GP
Sbjct: 90 NDDAEKDAPDNMSLAGDGFDTVNRVKTAVEKA--CPGVVSCADILALAARDVVSLASGPW 147
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
++VELGR DGLVS++SDV+G+LP P + +L +F +GL+ DM+ALSGAHT+GF+HC
Sbjct: 148 WSVELGRLDGLVSKASDVDGKLPGPDMRVTKLAAVFDKHGLSMRDMVALSGAHTVGFAHC 207
Query: 205 DRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNL 259
RF+ R+ DP++NK YAAQL + CPR+V IA+NMDP +P FDNVYY NL
Sbjct: 208 TRFTGRLYNYSAGEQTDPSMNKDYAAQLMEACPRDVGKTIAVNMDPVSPIVFDNVYYSNL 267
Query: 260 QQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTD 319
G GLFTSDQ+L+TD SR TV FA N F F+++M +LGR+GVK ++G++R D
Sbjct: 268 VNGLGLFTSDQVLYTDGASRRTVEEFAVNQTAFFDAFVSSMVRLGRLGVKAGKDGEVRRD 327
Query: 320 CS 321
C+
Sbjct: 328 CT 329
>UniRef100_Q96509 Peroxidase 55 precursor [Arabidopsis thaliana]
Length = 330
Score = 337 bits (864), Expect = 3e-91
Identities = 179/320 (55%), Positives = 226/320 (69%), Gaps = 9/320 (2%)
Query: 7 ILVWSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFF 66
+++W L + L + + AQLS N+YA+ CP+V+ IV+ AV KF+QT T PATLR+FF
Sbjct: 12 MMMWFLGMLLFSMVAESNAQLSENYYASTCPSVELIVKQAVTTKFKQTVTTAPATLRMFF 71
Query: 67 HDCFVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCA 126
HDCFV+GCDASV +AS N AEKD +N SLAGDGFDTVIKAK A+++ QC VSCA
Sbjct: 72 HDCFVEGCDASVFIASE-NEDAEKDADDNKSLAGDGFDTVIKAKTAVES--QCPGVVSCA 128
Query: 127 DILALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLT 186
DILALA RDV+ L GGP + VELGR DGLVS++S V G+LP+P ++ L +FA+NGL+
Sbjct: 129 DILALAARDVVVLVGGPEFKVELGRRDGLVSKASRVTGKLPEPGLDVRGLVQIFASNGLS 188
Query: 187 QTDMIALSGAHTLGFSHCDRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIAI 241
TDMIALSGAHT+G SHC+RF+NR+ PVDPT++ YA QL Q C + +P +
Sbjct: 189 LTDMIALSGAHTIGSSHCNRFANRLHNFSTFMPVDPTMDPVYAQQLIQAC-SDPNPDAVV 247
Query: 242 NMDPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMT 301
++D T+ TFDN YY+NL KGLFTSDQ LF D S+ TV FA N F + F +AM
Sbjct: 248 DIDLTSRDTFDNSYYQNLVARKGLFTSDQALFNDLSSQATVVRFANNAEEFYSAFSSAMR 307
Query: 302 KLGRVGVKNARNGKIRTDCS 321
LGRVGVK G+IR DCS
Sbjct: 308 NLGRVGVKVGNQGEIRRDCS 327
>UniRef100_Q7XPY0 OSJNBa0071I13.13 protein [Oryza sativa]
Length = 337
Score = 318 bits (814), Expect = 2e-85
Identities = 159/304 (52%), Positives = 205/304 (67%), Gaps = 6/304 (1%)
Query: 25 AQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDCFVQGCDASVLVASSG 84
AQL N+Y + CPN +S VRS + + QQ+F P TLRLFFHDCFV+GCDASV++ +
Sbjct: 34 AQLRQNYYGSTCPNAESTVRSVISQHLQQSFAVGPGTLRLFFHDCFVRGCDASVMLMAP- 92
Query: 85 NNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADILALATRDVINLAGGPS 144
N E + +L+ D + + KAKAA++A+P C KVSCADILA+A RDV++L GGPS
Sbjct: 93 NGDDESHSGADATLSPDAVEAINKAKAAVEALPGCAGKVSCADILAMAARDVVSLTGGPS 152
Query: 145 YTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTDMIALSGAHTLGFSHC 204
Y+VELGR DG + V LP P FNL+QLN+LFA+NGLTQTDMIALSGAHT+G +HC
Sbjct: 153 YSVELGRLDGKTFNRAIVKHVLPGPGFNLDQLNSLFASNGLTQTDMIALSGAHTIGVTHC 212
Query: 205 DRFSNRIQT-----PVDPTLNKQYAAQLQQMCPRNVDPRIAINMDPTTPRTFDNVYYKNL 259
D+F RI T +P +N + ++++CP N P +D +TPR FDN Y+ NL
Sbjct: 213 DKFVRRIYTFKQRLGYNPPMNLDFLRSMRRVCPINYSPTAFAMLDVSTPRAFDNAYFNNL 272
Query: 260 QQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKLGRVGVKNARNGKIRTD 319
+ KGL SDQILFTD RSR TVN FA N F F+ AM KLGR+GVK +G+IR
Sbjct: 273 RYNKGLLASDQILFTDRRSRPTVNLFAANSTAFFDAFVAAMAKLGRIGVKTGSDGEIRRV 332
Query: 320 CSVL 323
C+ +
Sbjct: 333 CTAV 336
>UniRef100_Q8W5H8 Putative peroxidase [Oryza sativa]
Length = 326
Score = 309 bits (792), Expect = 6e-83
Identities = 157/319 (49%), Positives = 214/319 (66%), Gaps = 9/319 (2%)
Query: 10 WSLALTLCLIPYTTFAQLSPNHYANICPNVQSIVRSAVQKKFQQTFVTVPATLRLFFHDC 69
W L + + + L ++Y + CPNV+SIV V+ K Q T T+ +T+RLFFHDC
Sbjct: 9 WMALLVVAAVAQLGASDLRTDYYNSTCPNVESIVLGVVKDKMQATIRTIGSTVRLFFHDC 68
Query: 70 FVQGCDASVLVASSGNNKAEKDHPENLSLAGDGFDTVIKAKAALDAVPQCRNKVSCADIL 129
FV GCD SVL+ S+ N AE+D P+NLSLA +GF+TV AKAA++A C ++VSC D+L
Sbjct: 69 FVDGCDGSVLITSTAGNTAERDAPDNLSLAFEGFETVRSAKAAVEAA--CPDQVSCTDVL 126
Query: 130 ALATRDVINLAGGPSYTVELGRFDGLVSRSSDVNGRLPQPSFNLNQLNTLFANNGLTQTD 189
A+ATRD I L+GGP + VELGR DG+ S +S+V G+LPQP+ L++L +F +NGL +D
Sbjct: 127 AIATRDAIALSGGPFFPVELGRLDGMRSSASNVAGKLPQPNNTLSELVAIFKSNGLNMSD 186
Query: 190 MIALSGAHTLGFSHCDRFSNRI------QTPVDPTLNKQYAAQLQQMCPRNVDPRIAINM 243
M+ALS AH++G +HC +FS+R+ P DPTLN++YAA L+ CP + P + + M
Sbjct: 187 MVALSAAHSVGLAHCSKFSDRLYRYNPPSQPTDPTLNEKYAAFLKGKCP-DGGPDMMVLM 245
Query: 244 DPTTPRTFDNVYYKNLQQGKGLFTSDQILFTDTRSRNTVNSFATNGNVFNANFITAMTKL 303
D TP FDN YY+NLQ G GL SD++L+TD R+R TV+S A + F F A+ KL
Sbjct: 246 DQATPALFDNQYYRNLQDGGGLLASDELLYTDNRTRPTVDSLAASTPDFYKAFADAIVKL 305
Query: 304 GRVGVKNARNGKIRTDCSV 322
GRVGVK+ G IR C V
Sbjct: 306 GRVGVKSGGKGNIRKQCDV 324
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 517,749,037
Number of Sequences: 2790947
Number of extensions: 20687036
Number of successful extensions: 51380
Number of sequences better than 10.0: 662
Number of HSP's better than 10.0 without gapping: 582
Number of HSP's successfully gapped in prelim test: 80
Number of HSP's that attempted gapping in prelim test: 49170
Number of HSP's gapped (non-prelim): 763
length of query: 323
length of database: 848,049,833
effective HSP length: 127
effective length of query: 196
effective length of database: 493,599,564
effective search space: 96745514544
effective search space used: 96745514544
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC137703.11


